Introduction
Head and neck squamous cell carcinoma represents the
sixth most common malignancy worldwide (1). Laryngeal carcinoma is the second most
common malignancy of the head and neck cancers (2). In the United States, it is estimated
that there would be 13,560 new cases and 3,640 mortalities due to
laryngeal carcinoma in 2016 (3).
The most common type of laryngeal carcinomais comprised of
laryngeal squamous cell carcinoma (LSCC), which accounts for ~95%
of laryngeal carcinoma cases (4).
Until now, tobacco smoking, alcohol drinking, air pollution and
unhealthy diet are the major risk factors for LSCC (5,6).
Currently, the main therapeutic treatments for LSCC are surgery or
total laryngectomy, followed by radiotherapy and chemotherapy
(7). Despite great progress in
diagnostic and therapeutic techniques over the last few decades,
the prognosis for patients with LSCC remains poor with a 5-year
survival rate of 64% (8). Most
LSCC patients diagnosed with advanced-stage die of recurrence
and/ormetastasis (9). Therefore,
fully understanding the molecular mechanism underlying LSCC would
provide effective therapeutic targets to improve outcomes for
patients with this disease.
MicroRNAs (miRNAs/miRs), ~22–25 nucleotides
inlength, are the most characterized of the non coding RNAs and
endogenously expressed in animal and plant cells (10,11).
They regulate the expression of protein-coding genes at the
translational level and post-translational level through
interaction with the 3′-untranslated region of their target genes
in sequence-specific base pairing manner, modulating mRNA stability
and/or translation inhibition (12,13).
A number of studies have demonstrated that miRNAs serve critical
roles in many physiological and pathological processes, including
cell proliferation, differentiation metabolism, apoptosis, cell
cycle, invasion, migration and death (14–16).
The dysregulation of miRNAs are significantly correlated with many
diverse diseases, such as neuronal disorders (17), inflammation (18) and cancer (19). Accumulated studies reported thata
large number of miRNAs are dysregulated in LSCC. For example,
miR-153 was downregulated in LSCC and functioned as a tumor
suppressor through inhibiting cell proliferation and invasion via
targeting KLF5 (20). miR-365a-3p
was upregulated in LSCC and promoted cell growth and metastasis
through regulating the PI3K/AKT pathway (21). Therefore, miRNAs may be molecular
therapeutic targets for cancer diagnosis and treatments.
In the present study, the authors measured miR-195
expression in LSCC tissues and cell lines. In addition, they
explored the functional roles of miR-195 in LSCC and its underlying
molecular mechanism. The purpose of the present study was to
validate the anticancer effects of miR-195 in LSCC.
Materials and methods
Tissue samples
A total of 51 pairs of LSCC tissues and adjacent
normal epithelial tissues were obtained from patients who received
primary surgical resection of LSCC between September 2012 and July
2015 in the Department of Otolaryngology, Head and Neck Surgery,
Tianjin Union Medical Center (Tianjin, China). None of the LSCC
patients were treated with radiotherapy or chemotherapy prior to
surgery. Tissue samples were snap-frozen in liquid nitrogen
immediately following resection and stored at −80°C until use. The
present study was approved by the Ethics Committee of Tianjin Union
Medical Center (Tianjin, China), and all patients gave their
informed written consent.
Cell lines, culture condition and
transfection
Three human LSCC cell lines (Hep-2, AMC-HN-8 and
Tu-177), a normal human keratinocyte cell line (HaCaT) and 293T
cell line were purchased from American Type Culture Collection
(Manassas, VA, USA). Cells were cultured in RPMI-1640 or Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and were grown in a
humidified atmosphere at 37°C with 5% CO2.
miR-195 mimics and miRNA mimics negative control
(miR-NC) were obtained from Shanghai GenePharma Co., Ltd.
(Shanghai, China). Small interfering (si)RNA targeting
Rho-associated kinase (ROCK)1 (si-ROCK1) and its control siRNA
(si-NC) were chemical synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). For cell transfection, cells (8×105
cells/well) were seeded in 6-well plates. Following overnight
incubationat 37°C with 5% CO2, cells were transfected
with miRNA mimics or siRNA by using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) following to the
manufacturer' sinstructions.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer' sprotocol. The concentration and purity of total RNA
was measured using a NanoDrop® ND-1000 spectrophotometer
(NanoDrop; Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
For miR-195 expression, the One Step PrimeScript miRNA cDNA
Synthesis kit (Takara Bio, Inc., Otsu, Japan) was used to perform
reverse transcription, followed by qPCR with SYBR®
Premix Ex Taq™ II (Takara Bio, Inc.). The temperature protocol for
reverse transcription was as follows: 37°C for 15 min and 85°C for
5 sec. qPCR was performed with the following thermo cycling
conditions: 5 min at 95°C, followed by 40 cycles of 95°C for 30 sec
and 65°C for 45 sec.
For ROCK1 mRNA expression, cDNA was synthesized from
RNA by using cDNA Synthesis kit (Takara Bio, Inc.). qPCR was
carried out using SYBR® Premix Ex Taq™ II (Takara Bio,
Inc.). The temperature protocol for reverse transcription was as
follows: 37°C for 60 min and 85°C for 5 sec. The thermocycling
conditions for qPCR was as follows: 5 min at 95°C, followed by 40
cycles of 95°C for 30 sec and 65°C for 45 sec. U6 and GAPDH were
used as control for miR-195 and ROCK1 mRNA expression,
respectively. The primers were designed as follows: miR-195,
5′-ACACTCCAGCTGGGTAGCAGCACAGAAAT-3′ (forward) and
5′-TGGTGTCGTGGAGTCG-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); ROCK1,
5′-AGGAAGGCGGACATATTGATCCCT-3′ (forward) and
5′-AGACGATAGTTGGGTCCCGGC-3′ (reverse); and GAPDH,
5′-CCCCTTCATTGACCTCAACT-3′ (forward) and 5′-ATGAGTCCTTCCACGATACC-3′
(reverse). The data were calculated using the 2−ΔΔCq
method (22).
MTT assay
At 24 h post-transfection, cells were collected and
seeded into 96-well plates at a density of 2,000 cells/well. Cells
were then cultured for 24, 48, 72 and 96 h. At each time point, MTT
(5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) assay was
carried out. A total volume of 20 µl MTT solution was added to each
well and incubated at 37°C with 5% CO2 for another 4 h.
The culture medium was then removed and 150 µl DMSO was added to
each well. Following incubation at 37°C for 10 min with a constant
shaking, the absorbance at 490 nm was determined by using a
microtiter plate reader (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Cell migration and invasion assay
Cell migration and invasion assays were performed
using Trans well chambers (Corning Life Sciences, Corning, NY, USA)
and Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) coated Trans
well chambers, respectively. At 48 h post-transfection, cells were
collected and suspended in FBS-free culture medium. A total of
1×105 cells were seeded into the upper chamber, whereas
500 µl culture medium containing 20% FBS was added in the lower
chamber. After incubation at 37°C with 5% CO2 for 48 h,
those cells left on the upperchamber were removed with cotton
swabs. The migrated or invaded cells were fixed with 95% ethanol
for 20 min and stained with 0.1% crystal violet for 10 min. After
washing with PBS (Gibco; Thermo Fisher Scientific, Inc.), the
migrated and invaded cells were photographed and quantified using
an inverted microscope.
Luciferase reporter assay
The putative target genes of miR-195 were predicted
using the TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org/microrna/). Based
onbioinformatics analysis, ROCK1 was identified as a potential
target of miR-195. Thewild type (Wt) or mutant (Mut)
3′-untranslated region (UTR) of ROCK1 harboring the miR-195 binding
site was cloned into pGL3 control vector. For luciferase reporter
assay, pGL3-ROCK1-3′UTR Wt or pGL3-ROCK1-3′UTR Mut together with
miR-195 mimics or miR-NC were injected into 293T cells by using
Lipofectamine 2000, according to the manufacturer's instructions.
Luciferaseactivities were determined 48 h post-transfection using
the Dual-Luciferase Reporter Assay system (Promega Corporation,
Madison, WI, USA). The results were expressed as relative
luciferase activities (firefly luciferase/Renilla
luciferase).
Western blot analysis
At 72 h after transfection, total protein was
extracted from transfected cells using radioimmunoprecipitation
assay buffer (Beyotime Institute of Biotechnology, Haimen, China).
The protein concentration was determined by using the bicinchoninic
acid assay (Thermo Fisher Scientific, Inc.). Equal amounts of
protein (20 µg) were separated by electrophoresis on a 10% SDS-PAGE
and then transferred onto polyvinylidene fluoride membrane (EMD
Millipore Corporation, Billerica, MA, USA). The membrane was
blocked with 5% non-fat milk in 0.1% TBS and 0.05% Tween-20 (TBST;
Beyotime Institute of Biotechnology) for 2 h and incubated with
primary antibodies at 4°C overnight, including mouse anti-human
monoclonal ROCK1 antibody (1:1,000 dilution; sc-365628; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and mouse anti-human
monoclonal GADPH antibody (1:1,000 dilution, sc-365062; Santa Cruz
Biotechnology, Inc.). After washing three times with TBST, the
membrane was incubated with corresponding horseradish
peroxidase-conjugated secondary antibody (1:5,000 dilution;
sc-2005; Santa Cruz Biotechnology, Inc.) at room temperature for 1
h. The protein bands were visualized with enhanced
chemiluminescence detection system (Amersham; GE Healthcare Life
Sciences, Chalfont, UK). ROCK1 protein expression was normalized to
total GAPDH.
Statistical analysis
The SPSS software (version, 17.0; SPSS Inc.,
Chicago, IL, USA) was used for statistical analysis. All data were
expressed aspresented as mean ± standard deviation, and were
compared by a Student's t test to determine the statistical
significance. P<0.05 was considered to indicate a statistically
significant difference.
Results
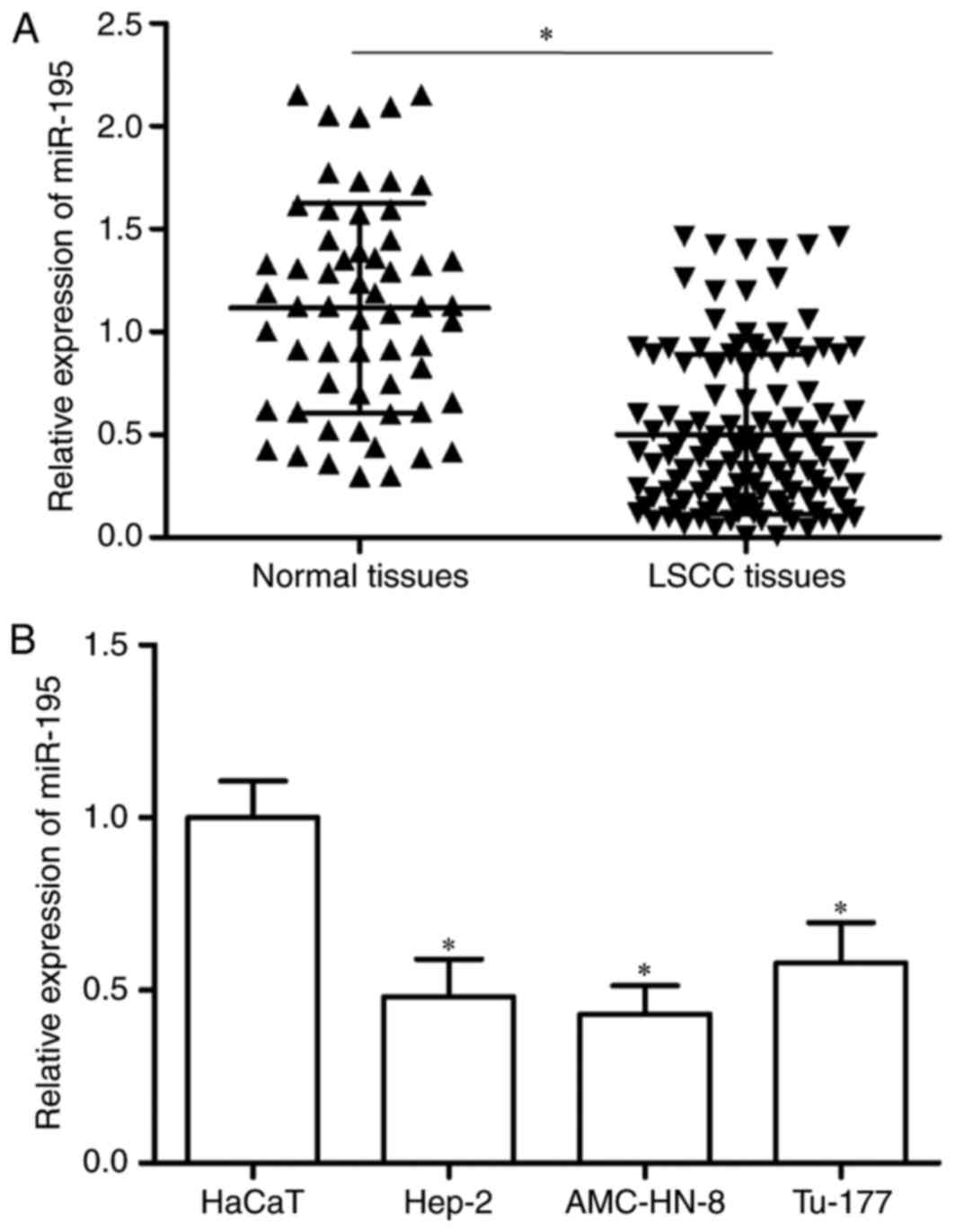
miR-195 was downregulated in LSCC and
was correlated with cancer progression
The expression levels of miR-195 in LSCC tissues and
adjacent normal epithelial tissues were determined using RT-qPCR.
The results indicated that miR-195 was lower in LSCC tissues than
adjacent normal epithelial tissues (Fig. 1A, P<0.05). Furthermore, the
authors analyzed miR-195 expression in LSCC cell lines. The data
indicated that miR-195 was significantly downregulated in LSCC cell
lines compared with normal human keratinocyte cell line (Fig. 1B, P<0.05).
To further investigate whether there was an
association between miR-195 expression and LSCC prognosis,
statistical analysis was performed to analyze the correlation
between miR-195 expression and the clinicopathological factors of
LSCC. As presented in Table I,
there were significantly association of miR-195 expression with
lymph node metastasis (P = 0.011) and TNM stage (P = 0.015).
However, there were no statistically correlation between miR-195
expression and other clinicopathological features, including sex
distribution, age, location, alcohol history, pathological
differentiation and T classification (all P>0.05).
 | Table I.Relationship between miR-195
expression level and clinicopathological factors in laryngeal
squamous cell carcinoma. |
Table I.
Relationship between miR-195
expression level and clinicopathological factors in laryngeal
squamous cell carcinoma.
|
|
| miR-195
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Case number | Low | High | P-value |
|---|
| Sex
distribution |
|
|
|
|
|
Male | 25 | 13 | 12 | 0.447 |
|
Female | 26 | 15 | 11 |
|
| Age (year) |
|
|
|
|
|
<65 | 25 | 16 | 9 | 0.264 |
|
≥65 | 26 | 12 | 14 |
|
| Location |
|
|
| 0.253 |
|
Supraglottic | 30 | 14 | 16 |
|
|
Glottic | 21 | 14 | 7 |
|
| Alcohol
history |
|
|
| 0.404 |
|
Negative | 25 | 12 | 13 |
|
|
Positive | 26 | 16 | 10 |
|
| Pathological
differentiation |
|
|
| 0.779 |
|
Moderately and highly
differentiated | 32 | 17 | 15 |
|
| Poorly
differentiated | 19 | 11 | 8 |
|
| T
classification |
|
|
| 0.264 |
|
T1-2 | 26 | 12 | 14 |
|
|
T3-4 | 25 | 16 | 9 |
|
| Lymph node
metastasis |
|
|
| 0.011 |
|
Positive | 24 | 18 | 6 |
|
|
Negative | 27 | 10 | 17 |
|
| TNM stage |
|
|
| 0.015 |
|
I–II | 28 | 11 | 17 |
|
|
III–IV | 23 | 17 | 6 |
|
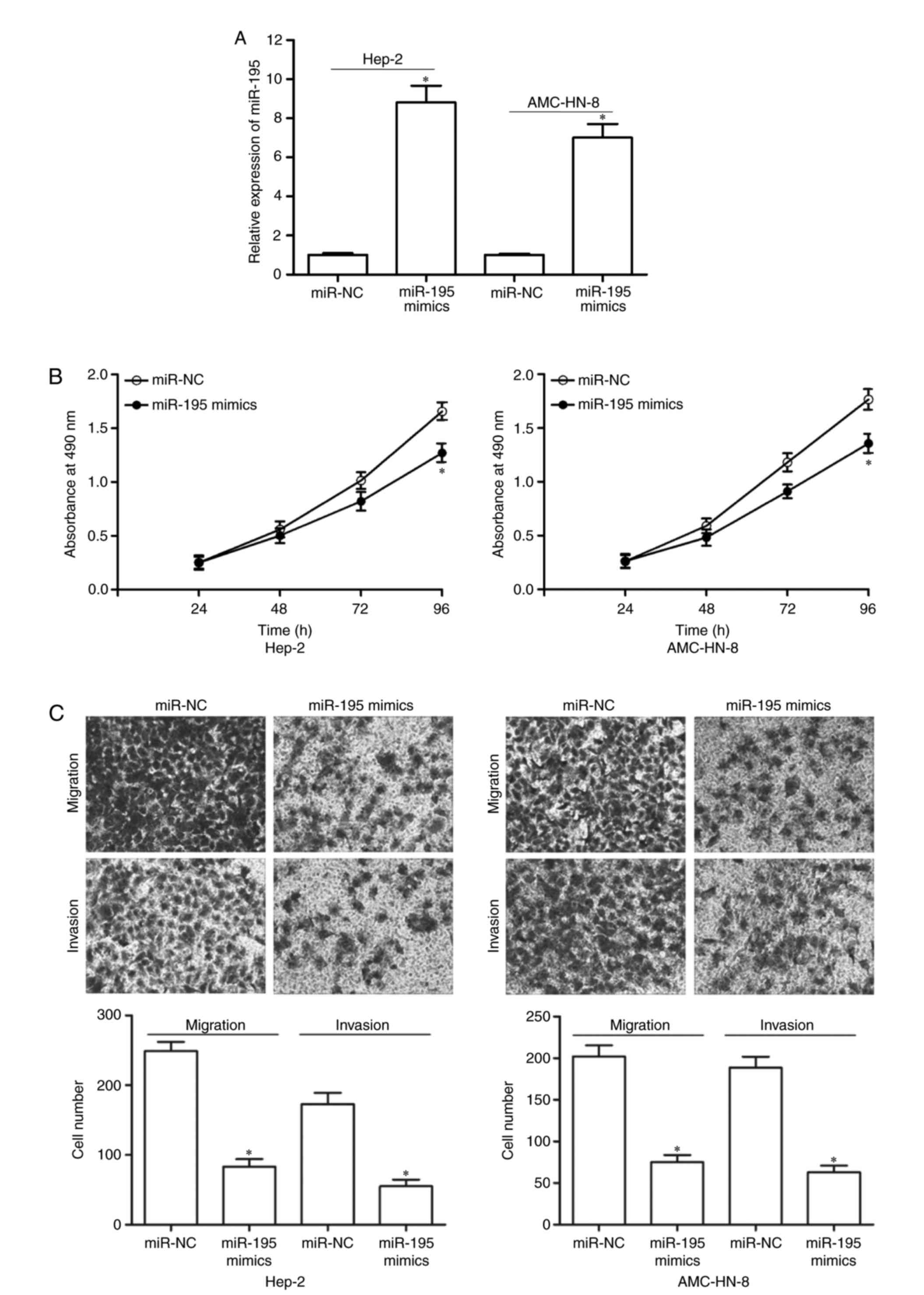
miR-195 suppressed proliferation,
migration and invasion of LSCC cells
To investigate the biological roles of miR-195 on
LSCC cancer cells, Hep-2 and AMC-HN-8 cells were transfected with
miR-195 mimics or miR-NC. RT-qPCR showed that miR-195 was markedly
upregulated in Hep-2 and AMC-HN-8 cells transfected with miR-195
mimics (Fig. 2A, P<0.05). The
effect of miR-195 on proliferation of LSCC cells was assessed using
MTT assay. As demonstrated in Fig.
2B, upregulation of miR-195 suppressed Hep-2 and AMC-HN-8 cell
proliferation (P<0.05). Then, the author sex amined the effects
of miR-195 on the migration and invasion capacities of LSCC cells
by using cell migration and invasion assays. The results revealed
that restoration of miR-195 obviously decreased the migration and
invasion abilities of Hep-2 and AMC-HN-8 cells compared with miR-NC
groups (Fig. 2C, P<0.05). These
data suggested that overexpression of miR-195 suppressed growth and
metastasis of LSCC cells.
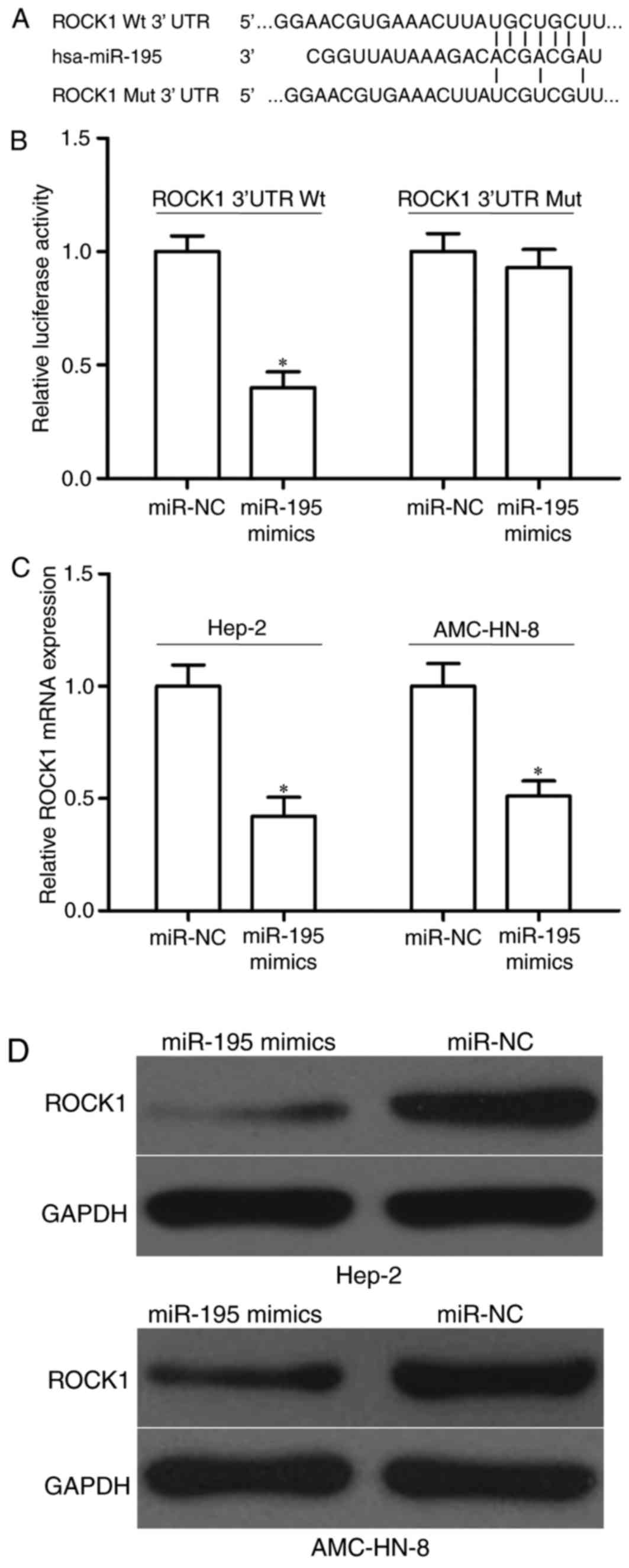
miR-195 directly targeted ROCK1
The potential molecular mechanism on how miR-195
suppressed cell growth and metastasis of LSCC was analyzed by
exploring its direct target genes. Based onbioinformatics analysis
with public databases, ROCK1 was identified as a potential target
of miR-195 (Fig. 3A).
To verify whether ROCK1 was a direct target gene of
miR-195, aluciferase reporter assay was performed. 293T cells were
transfected with miR-195 mimics or miR-NC as well as
pGL3-ROCK1-3′UTR Wt or pGL3-ROCK1-3′UTR Mut. The results indicated
that miR-195 overexpression reduced luciferase activities of vector
containing wild type ROCK1 3′UTR (Fig.
3B, P<0.05), while miR-195 had no regulation effect on
mutant type of ROCK1 3′UTR, suggesting that this binding site in
ROCK1 3′UTR was essential for the regulation by miR-195.
Moreover, the authors assessed the effects of
miR-195 overexpression on the expression of ROCK1. RT-qPCR and
western blotting indicated that ectopic of miR-195 expression
suppressed ROCK1 mRNA (Fig. 3C,
P<0.05) and protein (Fig. 3D,
P<0.05) expression level in Hep-2 and AMC-HN-8 cells. Taken
together, miR-195 can directly decrease ROCK1 expression through
targeting the binding site in the 3′UTR of ROCK1.
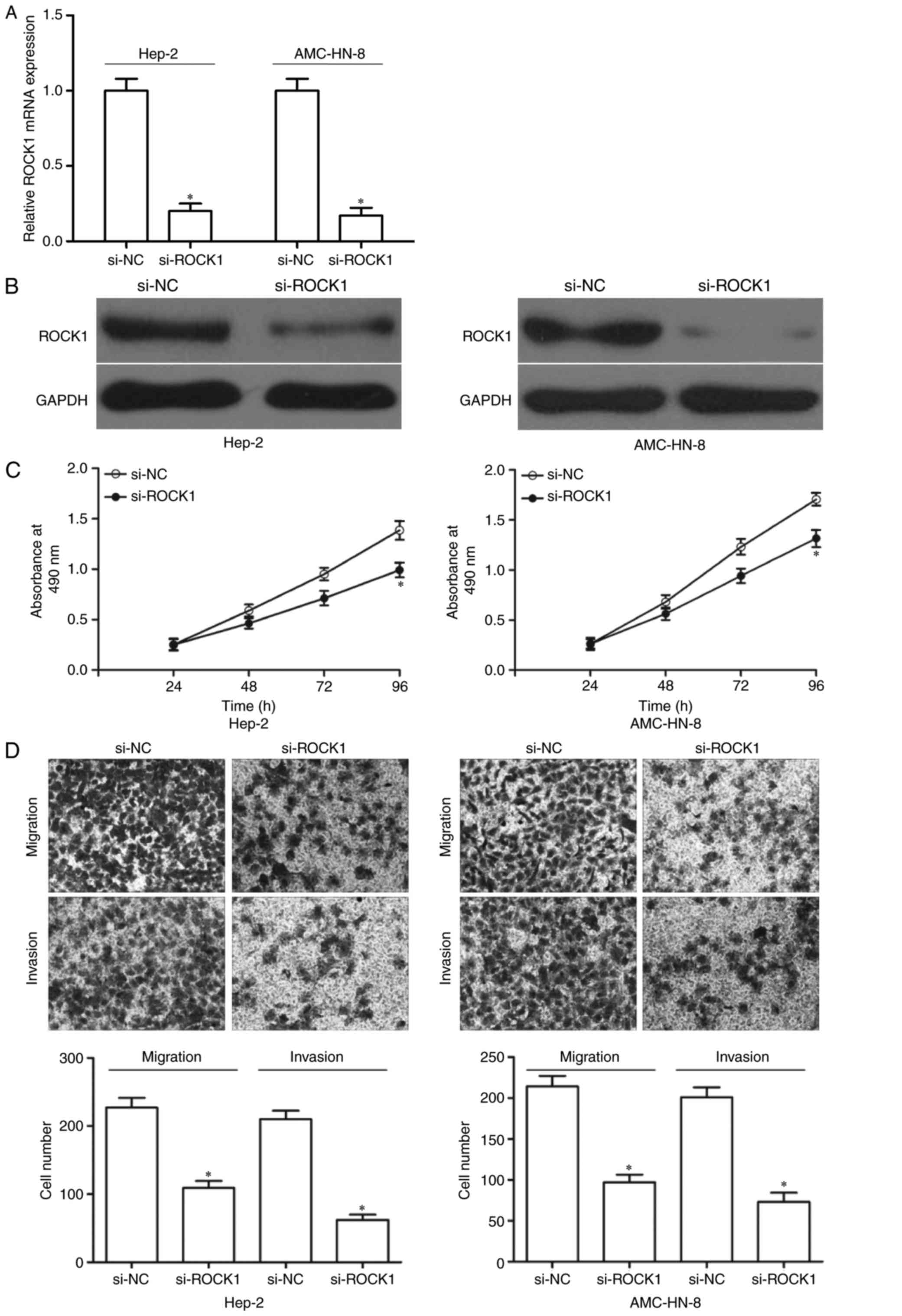
Inhibition of ROCK1 exerted similar
roles to that of miR-195 overexpression in LSCC
To study the effects of ROCK1 on LSCC, Hep-2 and
AMC-HN-8 cells were transfected with si-ROCK1 or si-NC. After
transfection, RT-qPCR and western blotting were used to evaluate
its transfection efficiency. The results demonstrated that si-ROCK1
significantly decreased ROCK1 expression in Hep-2 and AMC-HN-8
cells at both mRNA (Fig. 4A,
P<0.05) and protein (Fig. 4B,
P<0.05) levels. Moreover, MTT assay, cell migration and invasion
assays were used to investigate the effects of ROCK1
underexpression on LSCC cell proliferation, migration and invasion,
respectively. The data revealed that inhibition of ROCK1 has
similar effects to that of miR-195 overexpression, since it
obviously suppressed growth (Fig.
4C, P<0.05) and metastasis (Fig. 4D, P<0.05) of Hep-2 and AMC-HN-8
cells. The results suggested that miR-195 overexpression suppressed
proliferation, migration and invasion of LSCC cells through
downregulation of ROCK1.
Discussion
miR-195, one of the miR-16/15/195/424/497 family
members, has been reported to be downregulated in various kinds of
human cancer. For example, in hepatocellular carcinoma, miR-195
expression was markedly impaired in tumor tissues (23–25).
Wang et al (26) found that
miR-195 was downregulated in colorectal cancer. Its low expression
was significantly associated with lymph node metastasis and
advanced tumor stage. Kaplan-Meier survival analysis indicated that
colorectal cancer patients with reduced miR-195 had a poor overall
survival. Song et al (27)
showed that expression level of miR-195 was reduced in breast
cancer and obviously correlated with histological grade, tumor
size, lymph nodal involvement and vessel invasion. In addition,
Kaplan-Meier survival analysis indicated that breast cancer
patients with high miR-195 level showed a positive association
towards a longer survival. Downregulation of miR-195 was also
observed in bladder cancer (28),
glioblastoma (29), gastric cancer
(30), glioma (31), tongue squamous cell carcinoma
(32) and non-small cell lung
cancer (33). Consistent with
these results, it was identified that miR-195 was downregulated in
LSCC and correlated with lymph node metastasis and TNM stage. These
results suggested that miR-195 serves important roles in these
cancer types, and may therefore serve as a potential diagnostic and
prognosis marker for these cancers.
To date, numerous studies have provided sufficient
evidences to demonstrate that miR-195 functions as a tumor
suppressor in human cancer. For example, in hepatocellular
carcinoma, restoration of miR-195 dramatically suppressed cell
migration, invasion, proliferation, angiogenesis, enhanced
apoptosis and decreased tumor growth in vivo (23–25,34,35).
Zhou et al (36) reported
that ectopic of miR-195 decreased metastasis of cervical cancer. In
colorectal cancer, introduction of miR-195 suppressed cell
viability, colony formation, invasion induced apoptosis and
increased the chemosensitivity of cells to the chemotherapeutic
drug doxorubicin (37–39). Zhang et al (40) determined that miR-195
overexpression inhibited cell proliferation, cell cycle
progression, migration, invasion, EMT and tumorigenesis in prostate
cancer (41,42). In breast cancer, upregulation of
miR-195 repressed breast cancer cells proliferation, cell colony
formation, migration, invasion, enhanced apoptosis,
radiosensitivity and chemosensitivity of cells to adriamycin
(43–47). Liu et al (48) revealed that miR-195 inhibited
growth and metastasis of non-small cell lung cancer. In present
study, it was found that enforced miR-195 expression inhibited
proliferation, migration and invasion of LSCC cells. These findings
suggested that miR-195 could be a potential candidate therapeutic
target for cancer treatments.
The present study further elucidated the molecular
mechanism on how miR-195 regulated cell biological functions during
the development of LSCC. Bioinformatics analysis predicted that
ROCK1 is the potential target gene of miR-195. Luciferase reporter
assays then confirmed that miR-195 decreased luciferase activities
of vector containing wild type ROCK1 3′UTR, while miR-195 had no
regulation effect on mutant type of ROCK1 3′UTR, suggesting that
this binding site in ROCK1 3′UTR was essential for the regulation
by miR-195. RT-qPCR and western blotting were performed to evaluate
the regulation effect of miR-195 on ROCK1 expression. Results
confirmed that miR-195 reduced ROCK1 expression at both mRNA and
protein level. Finally, downregulation of ROCK1 had similar effects
to that of miR-195 overexpression, since it obviously suppressed
growth and metastasis. These results validated that ROCK1 was a
direct functional downstream target of miR-195 in LSCC.
ROCK, an essential downstream effect or of the Rho
small GTPase, acts as a molecular switch that binds GTP (active)
and GDP (inactive) to regulate cell survival, proliferation and
cytoskeleton organization, inducing alterations in cell
shape/morphology, invasion and movement (49–51).
ROCK1, located at chromosome 18 (18q11.1) (52), is frequently highly expressed in
human cancers (53). A study by
Zhang et al (54) found
that ROCK1 expression was increased in LSCC tissues. Its expression
was correlated with tumor size and lymph node metastasis.
Functional study revealed that downregulation of ROCK1 inhibited
cell proliferation, migration and invasion in LSCC. Combined with
these findings, the authors speculated that the miR-195/ROCK1 axis
could be developed as a therapeutic target for suppression of human
LSCC rapidly growth and metastasis.
In summary, a downregulation of miR-195 was observed
in LSCC tissues and cell lines. In addition, reduced miR-195
expression was significantly correlated with lymph node metastasis
and TNM stage. Moreover, it was demonstrated that miR-195 may act
as a tumor suppressor in LSCC tumorigenesis and tumor development
through directly targeting ROCK1, suggesting that miR-195 could
potentially serve as a therapeutic target for the treatment of
LSCC.
References
|
1
|
Marcu LG and Yeoh E: A review of risk
factors and genetic alterations in head and neck carcinogenesis and
implications for current and future approaches to treatment. J
Cancer Res Clin Oncol. 135:1303–1314. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bodnar M, Szylberg Ł, Kaźmierczak W and
Marszalek A: Immunohistochemical expression of p27(kip1) in
metastatic laryngeal squamous cell carcinoma. Adv Med Sci.
59:206–212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li M, Tian L, Ren H, Chen X, Wang Y, Ge J,
Wu S, Sun Y, Liu M and Xiao H: MicroRNA-101 is a potential
prognostic indicator of laryngeal squamous cell carcinoma and
modulates CDK8. J Transl Med. 13:2712015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Alcohol drinking. Epidemiological studies
of cancer in humans. IARC Monogr Eval Carcinog Risks Hum.
44:153–250. 1988.PubMed/NCBI
|
|
6
|
Edefonti V, Bravi F, Garavello W, La
Vecchia C, Parpinel M, Franceschi S, Dal Maso L, Bosetti C,
Boffetta P, Ferraroni M and Decarli A: Nutrient-based dietary
patterns and laryngeal cancer: Evidence from an exploratory factor
analysis. Cancer Epidemiol Biomarkers Prev. 19:18–27. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yungang W, Xiaoyu L, Pang T, Wenming L and
Pan X: miR-370 targeted FoxM1 functions as a tumor suppressor in
laryngeal squamous cell carcinoma (LSCC). Biomed Pharmacother.
68:149–154. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Boyle P and Ferlay J: Cancer incidence and
mortality in Europe, 2004. Ann Oncol. 16:481–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ramroth H, Schoeps A, Rudolph E, Dyckhoff
G, Plinkert P, Lippert B, Feist K, Delank KW, Scheuermann K, Baier
G, et al: Factors predicting survival after diagnosis of laryngeal
cancer. Oral Oncol. 47:1154–1158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iorio MV and Croce CM: MicroRNA
dysregulation in cancer: Diagnostics, monitoring and therapeutics.
A comprehensive review. EMBO Mol Med. 9:8522017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gibcus JH, Tan LP, Harms G, Schakel RN, de
Jong D, Blokzijl T, Möller P, Poppema S, Kroesen BJ and van den
Berg A: Hodgkin lymphoma cell lines are characterized by a specific
miRNA expression profile. Neoplasia. 11:167–176. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leaman D, Chen PY, Fak J, Yalcin A, Pearce
M, Unnerstall U, Marks DS, Sander C, Tuschl T and Gaul U:
Antisense-mediated depletion reveals essential and specific
functions of microRNAs in Drosophila development. Cell.
121:1097–1108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brennecke J, Hipfner DR, Stark A, Russell
RB and Cohen SM: Bantam encodes a developmentally regulated
microRNA that controls cell proliferation and regulates the
proapoptotic gene hid in Drosophila. Cell. 113:25–36. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fiore R, Siegel G and Schratt G: MicroRNA
function in neuronal development, plasticity and disease. Biochim
Biophys Acta. 1779:471–478. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
O'Connell RM, Taganov KD, Boldin MP, Cheng
G and Baltimore D: MicroRNA-155 is induced during the macrophage
inflammatory response. Proc Natl Acad Sci USA. 104:1604–1609. 2007;
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Calin GA and Croce CM: MicroRNA-cancer
connection: The beginning of a new tale. Cancer Res. 66:7390–7394.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu JY, Lu JB and Xu Y: MicroRNA-153
inhibits the proliferation and invasion of human laryngeal squamous
cell carcinoma by targeting KLF5. Exp Ther Med. 11:2503–2508. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Geng J, Liu Y, Jin Y, Tai J, Zhang J, Xiao
X, Chu P, Yu Y, Wang SC, Lu J, et al: MicroRNA-365a-3p promotes
tumor growth and metastasis in laryngeal squamous cell carcinoma.
Oncol Rep. 35:2017–2026. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu H, Hu YW, Zhao JY, Hu XM, Li SF, Wang
YC, Gao JJ, Sha YH, Kang CM, Lin L, et al: MicroRNA-195-5p acts as
an anti-oncogene by targeting PHF19 in hepatocellular carcinoma.
Oncol Rep. 34:175–182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zheng C, Li J, Wang Q, Liu W, Zhou J, Liu
R, Zeng Q, Peng X, Huang C, Cao P and Cao K: MicroRNA-195 functions
as a tumor suppressor by inhibiting CBX4 in hepatocellular
carcinoma. Oncol Rep. 33:1115–1122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Wang J, Ma H, Zhang J and Zhou X:
Downregulation of miR-195 correlates with lymph node metastasis and
poor prognosis in colorectal cancer. Med Oncol. 29:919–927. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song CG, Wu XY, Wang C, Fu FM and Shao ZM:
Correlation of miR-195 with invasiveness and prognosis of breast
cancer. Zhonghua Wai Ke Za Zhi. 50:353–356. 2012.(In Chinese).
PubMed/NCBI
|
|
28
|
Fei X, Qi M, Wu B, Song Y, Wang Y and Li
T: MicroRNA-195-5p suppresses glucose uptake and proliferation of
human bladder cancer T24 cells by regulating GLUT3 expression. FEBS
Lett. 586:392–397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang QQ, Xu H, Huang MB, Ma LM, Huang QJ,
Yao Q, Zhou H and Qu LH: MicroRNA-195 plays a tumor-suppressor role
in human glioblastoma cells by targeting signaling pathways
involved in cellular proliferation and invasion. Neuro Oncol.
14:278–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deng H, Guo Y, Song H, Xiao B, Sun W, Liu
Z, Yu X, Xia T, Cui L and Guo J: MicroRNA-195 and microRNA-378
mediate tumor growth suppression by epigenetical regulation in
gastric cancer. Gene. 518:351–359. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hui W, Yuntao L, Lun L, WenSheng L,
ChaoFeng L, HaiYong H and Yueyang B: MicroRNA-195 inhibits the
proliferation of human glioma cells by directly targeting cyclin D1
and cyclin E1. PLoS One. 8:e549322013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jia LF, Wei SB, Gong K, Gan YH and Yu GY:
Prognostic implications of micoRNA miR-195 expression in human
tongue squamous cell carcinoma. PLoS One. 8:e566342013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yongchun Z, Linwei T, Xicai W, Lianhua Y,
Guangqiang Z, Ming Y, Guanjian L, Yujie L and Yunchao H:
MicroRNA-195 inhibits non-small cell lung cancer cell
proliferation, migration and invasion by targeting MYB. Cancer
Lett. 347:65–74. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang Y, Li M, Chang S, Wang L, Song T, Gao
L, Hu L, Li Z, Liu L, Yao J and Huang C: MicroRNA-195 acts as a
tumor suppressor by directly targeting Wnt3a in HepG2
hepatocellular carcinoma cells. Mol Med Rep. 10:2643–2648. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang X, Yu J, Yin J, Xiang Q, Tang H and
Lei X: MiR-195 regulates cell apoptosis of human hepatocellular
carcinoma cells by targeting LATS2. Pharmazie. 67:645–651.
2012.PubMed/NCBI
|
|
36
|
Zhou Q, Han LR, Zhou YX and Li Y: MiR-195
suppresses cervical cancer migration and invasion through targeting
Smad3. Int J Gynecol Cancer. 26:817–824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu L, Chen L, Xu Y, Li R and Du X:
microRNA-195 promotes apoptosis and suppresses tumorigenicity of
human colorectal cancer cells. Biochem Biophys Res Commun.
400:236–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang L, Qian L, Li X and Yan J:
MicroRNA-195 inhibits colorectal cancer cell proliferation,
colony-formation and invasion through targeting CARMA3. Mol Med
Rep. 10:473–478. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Qu J, Zhao L, Zhang P, Wang J, Xu N, Mi W,
Jiang X, Zhang C and Qu J: MicroRNA-195 chemosensitizes colon
cancer cells to the chemotherapeutic drug doxorubicin by targeting
the first binding site of BCL2L2 mRNA. J Cell Physiol. 230:535–545.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang X, Tao T, Liu C, Guan H, Huang Y, Xu
B and Chen M: Downregulation of miR-195 promotes prostate cancer
progression by targeting HMGA1. Oncol Rep. 36:376–382. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu C, Guan H, Wang Y, Chen M, Xu B, Zhang
L, Lu K, Tao T, Zhang X and Huang Y: miR-195 inhibits EMT by
targeting FGF2 in prostate cancer cells. PLoS One. 10:e01440732015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo J, Wang M and Liu X: MicroRNA-195
suppresses tumor cell proliferation and metastasis by directly
targeting BCOX1 in prostate carcinoma. J Exp Clin Cancer Res.
34:912015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang
Y, Zou C, Zhang X, Liu S, Wang X, et al: Analysis of MiR-195 and
MiR-497 expression, regulation and role in breast cancer. Clin
Cancer Res. 17:1722–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang G, Wu D, Zhu J, Jiang O, Shi Q, Tian
J and Weng Y: Upregulation of miR-195 increases the sensitivity of
breast cancer cells to Adriamycin treatment through inhibition of
Raf-1. Oncol Rep. 30:877–889. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhu J, Ye Q, Chang L, Xiong W, He Q and Li
W: Upregulation of miR-195 enhances the radiosensitivity of breast
cancer cells through the inhibition of BCL-2. Int J Clin Exp Med.
8:9142–9148. 2015.PubMed/NCBI
|
|
46
|
Luo Q, Wei C, Li X, Li J, Chen L, Huang Y,
Song H, Li D and Fang L: MicroRNA-195-5p is a potential diagnostic
and therapeutic target for breast cancer. Oncol Rep. 31:1096–1102.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Singh R, Yadav V, Kumar S and Saini N:
MicroRNA-195 inhibits proliferation, invasion and metastasis in
breast cancer cells by targeting FASN HMGCR, ACACA and CYP27B1. Sci
Rep. 5:174542015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu B, Qu J, Xu F, Guo Y, Wang Y, Yu H and
Qian B: MiR-195 suppresses non-small cell lung cancer by targeting
CHEK1. Oncotarget. 6:9445–9456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang C, Zhang S, Zhang Z, He J, Xu Y and
Liu S: ROCK has a crucial role in regulating prostate tumor growth
through interaction with c-Myc. Oncogene. 33:5582–5591. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rossman KL, Der CJ and Sondek J: GEF means
go: Turning on RHO GTPases with guanine nucleotide-exchange
factors. Nat Rev Mol Cell Biol. 6:167–180. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Patel RA, Forinash KD, Pireddu R, Sun Y,
Sun N, Martin MP, Schönbrunn E, Lawrence NJ and Sebti SM: RKI-1447
is a potent inhibitor of the Rho-associated ROCK kinases with
anti-invasive and antitumor activities in breast cancer. Cancer
Res. 72:5025–5034. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lock FE, Ryan KR, Poulter NS, Parsons M
and Hotchin NA: Differential regulation of adhesion complex
turnover by ROCK1 and ROCK2. PLoS One. 7:e314232012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou X, Wei M and Wang W: MicroRNA-340
suppresses osteosarcoma tumor growth and metastasis by directly
targeting ROCK1. Biochem Biophys Res Commun. 437:653–658. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhang J, He X, Ma Y, Liu Y, Shi H, Guo W
and Liu L: Overexpression of ROCK1 and ROCK2 inhibits human
laryngeal squamous cell carcinoma. Int J Clin Exp Pathol.
8:244–251. 2015.PubMed/NCBI
|