Introduction
Long non-coding RNAs (lncRNAs) are characterized as
a subgroup of RNAs>200 nucleotides in length without, or with
limited, protein-coding potential (1–3).
Previous studies have demonstrated that certain lncRNAs have
regulatory roles in diverse biological processes at the epigenetic,
transcriptional and post-transcriptional levels due to their
various structural and biochemical characteristics (4–9). The
aberrant expression of lncRNAs has been shown in various human
diseases, including cancer (Table
I).
 | Table I.Effects of taurine-upregulated gene
1in various types of cancer. |
Table I.
Effects of taurine-upregulated gene
1in various types of cancer.
| Author, year | Cancer | Expression | Clinical
significance | Function | Associated
factors | (Refs.) |
|---|
| Zhang et al,
2014 | Non-small cell lung
cancer | Downregulated | Prognosis | Proliferation; cell
cycle | P53; PRC2;
HOXB7 | (15) |
| Li et al,
2016 | Glioma | Downregulated | Pathological stage;
tumor size | Not reported | Not reported | (16) |
| Zhang et al,
2013 | Osteosarcoma | Upregulated | Not reported | Not reported | Not reported | (17) |
| Han et al,
2013; Tan et al, 2015 | Bladder cancer | Upregulated | TNM stage; overall
survival | Invasion;
radioresistance | miR145; ZEB2 | (18,19) |
| Sun et al,
2016 | Colorectal
cancer | Upregulated | Prognosis | Proliferation;
invasion; metastasis | HDAC1 | (20) |
| Xu et al,
2015 | Esophageal squamous
cell carcinoma | Upregulated | Family history;
tumor location | Proliferation;
migration | Not reported | (21) |
| Zhang et al,
2016 | Gastric | Upregulated | Invasion depth; TNM
stage; overall survival | Proliferation;
apoptosis; cell cycle | EZH2; p57 | (22) |
| Huang et al,
2015 | Hepatocellular
carcinoma | Upregulated | Tumor size; BCLC
stage | Proliferation;
apoptosis; cell cycle | SP1; KLF2 | (23) |
| Isin et al,
2014 | Malignant
melanoma | Upregulated | Stage | Not reported | Not reported | (35) |
Taurine-upregulated gene 1 (TUG1), a 7.1-kb lncRNA,
was originally detected in a genomic screen for genes upregulated
in response to taurine treatment in developing mouse retinal cells
(10). It is located on chromosome
22q12.2 in the human genome, and has been reported to be expressed
in a tissue-specific pattern and to exert oncogenic or tumor
suppressive functions in different types of cancer in humans
(11–14). The downregulation of TUG1 has been
detected in glioma and non-small-cell lung cancer (NSCLC), with
TUG1 shown to induce apoptosis as a tumor suppressor (15,16).
By contrast, the overexpression of TUG1 has been reported in
osteosarcoma (17), bladder cancer
(18,19), colorectal cancer (CRC) (20), esophageal squamous cell carcinoma
(ESCC) (21), gastric cancer
(22) and hepatocellular cancer
(HCC) (23). TUG1 was shown to
function as an oncogene by promoting cell proliferation and was
correlated with a poor prognosis (17–23).
Polycomb repressive complex 2 (PRC2) is a
methyltransferase, which is composed of enhancer of zeste homolog 2
(EZH2), suppressor of zeste 12 (SUZ12) and embryonic ectoderm
development, and is capable of catalyzing the di- and
trimethylation of lysine residue 27 of histone 3 (H3K27me3), which
regulates gene expression. Various lncRNAs, including TUG1,
modulate specific genetic loci by recruiting and binding to PRC2
protein complexes, and PRC2-mediated epigenetic regulation is vital
in tumorigenesis and development (24–27).
The knockdown of TUG1 results in wide changes in gene expression,
particularly the upregulation of cell-cycle genes, indicating that
TUG1 is important in cell proliferation and apoptosis through
effects on the cell cycle (22).
However, the comprehensive mechanisms remain to be fully
elucidated.
TUG1 in human cancer
Colorectal cancer (CRC)
It was previously reported that the expression of
TUG1 was significantly enhanced in CRC tumor tissues, compared with
that in paratumor tissues. Further analysis showed that the
expression of TUG1 was negatively correlated with overall survival
rates in patients (20). The
stable knockdown of histone deacetylase 1 (HDAC1) induced the
expression of TUG1, indicating that the expression of TUG1 is
regulated by histone modification (20,28).
In vitro experiments have confirmed that the knockdown of
TUG1 suppresses the colony formation, migration and invasion of CRC
cells in vitro. In addition, an in vivo liver
metastasis model revealed that the overexpression of TUG1 increased
the number of metastatic tumor nodules in the liver, indicating
that TUG1 promoted CRC metastasis (20). The molecular mechanism by which
TUG1 promotes the invasion and metastasis of CRC has also been
investigated. It was demonstrated that the overexpression of TUG1
reduced the expression of E-cadherin, and upregulated the
expression levels of N-cadherin, vimentin and fibronectin, whereas
knockdown of the expression of TUG1 showed the opposite effects.
This suggested that TUG1 may affect CRC metastasis and invasion
through mediating epithelial-mesenchymal transition
(EMT)-associated gene expression (20,29).
However, the mechanisms by which HDAC1 affects TUG1 and regulates
EMT require further investigation.
Bladder cancer
The expression of TUG1 was also found to be
upregulated in bladder cancer tissues and cell lines. A higher
expression of TUG1 was found to be associated with poorer
tumor-necrosis-metastasis (TNM) staging and shorter overall
survival rates (18,19). Subsequent investigations revealed
that TUG1 promoted bladder cancer cell invasion and
radioresistance. The expression level of epithelial markers
increased whereas those of mesenchymal markers decreased following
the overexpression of TUG1, indicating that TUG1 was involved in
bladder cancer through EMT (19,29).
TUG1 acted as a microRNA (miRNA) sponge, as miRNA (miR)-145 was
able to bind to TUG1 and exhibit reciprocal regulatory effects. In
addition, Zinc finger E-box binding homeobox 2 (ZEB2), a
transcription factor regulating the EMT marker E-cadherin (30), has been identified as a direct
target of miR-145 (31). The
evidence above indicates a possible mechanism by which TUG1 is
involved in EMT and the radioresistance in bladder cancer through
the miR-145/ZEB2 axis.
Hepatocellular cancer (HCC)
A previous study detected an upregulation in the
expression of TUG1 in HCC tissues, which was confirmed to be
associated with tumor size and Barcelona Clinic Liver Cancer stage
(23). The transcription factor,
stimulatory protein 1 (SP1) was later confirmed to directly bind to
TUG1 promoter regions and positively regulate the expression of
TUG1. In previous in vitro and xenograft model experiments,
the functions of TUG1 in inhibiting cell proliferation and inducing
cell apoptosis in HCC were demonstrated. Kruppel-like factor 2
(KLF2) was identified as a novel downstream gene of TUG1, which was
found to be involved in HCC cell G0/G1 arrest, suppression of cell
proliferation and the induction of apoptosis. TUG1 inhibited the
transcription of KLF2 through binding to EZH2/SUZ12, the core
subunits of PRC2, to the KLF2 gene promoter locus in HCC cells,
thus acting as an oncogenic factor (32).
Gastric cancer (GC)
A previous study observed that TUG1 was
overexpressed in GC cells, and was positively correlated with
invasion depth and TNM stage, but negatively correlated with
overall survival rates (22).
In vitro and in vivo experiments confirmed that TUG1
suppressed GC cell proliferation through its effects on cell cycle
progression in a pattern of G0/G1 arrest. Subsequent investigation
of the mechanism demonstrated that TUG1 specifically targeted EZH2
and epigenetically regulated the expression of cyclin-dependent
kinase inhibitor (CKI) family members, including p15, p16, p21, p27
and p57 (22,33,34).
Analysis of the mechanism showed that TUG1 epigenetically repressed
CKIs by binding to PRC2, and thereby regulated the cell cycle to
promote GC cell proliferation.
ESCC
It was previously found that TUG1 was overexpressed
in ESCC. Patients with a family history of esophageal cancer and
upper segment ESCC were shown to express higher levels of TUG1.
TUG1 also promoted the proliferation and migration of ESCC cell
lines in vitro (21).
Osteosarcoma
TUG1 is upregulated in osteosarcoma tissues and
cells (17). Experiments have
demonstrated that TUG1 acts as an oncogenic gene via increasing
osteosarcoma cell proliferation and affecting apoptosis. However,
the detailed mechanisms require further investigation.
Multiple myeloma (MM)
It has been reported that, in the plasma of patients
with MM, the expression of TUG1 was upregulated and showed marked
association with clinical stages (35).
Glioma
Unlike the overexpression of TUG1 found in the types
of cancer described above, TUG1 was downregulated in human glioma
tissues and cells, and negatively associated with advanced
pathological progression, serving as an indicator of poor prognosis
(16). TUG1 exerts its antitumor
function in glioma through promoting cell apoptosis via intrinsic
pathways mediated by caspase-3 together with caspase-9, and
simultaneously suppressing anti-apoptotic pathways mediated by
B-cell lymphoma 2 (36–39).
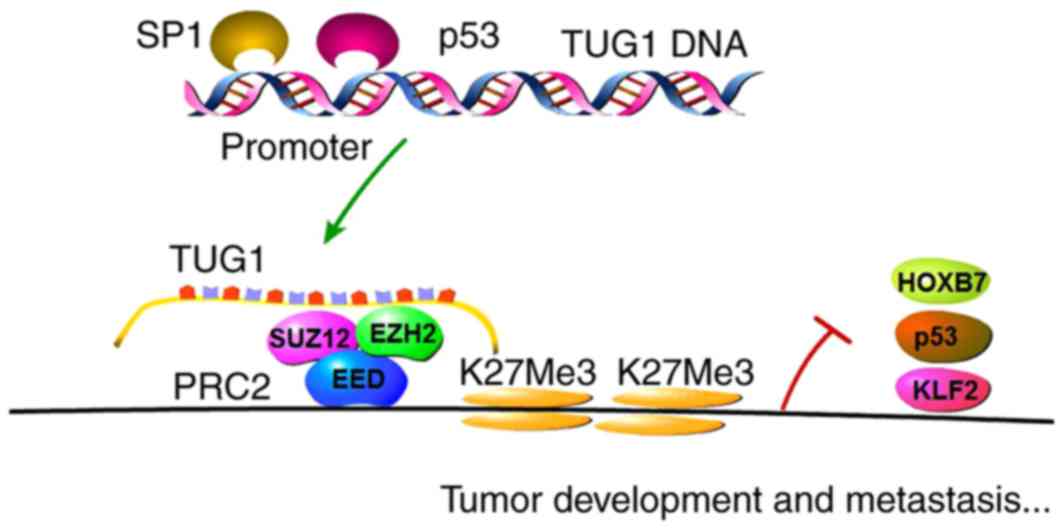
Non-small-cell lung cancer
(NSCLC)
The expression of TUG1 was also shown to be
downregulated in human NSCLC tissues and correlated with poor
prognosis of patients (15). TUG1
has been shown to modulate NSCLC cell proliferation in vitro
and in vivo through alterations in cell cycle progression.
Further analysis demonstrated that p53 was able to directly bind to
the promoter region of TUG1 and regulate the expression of TUG1. In
addition, investigations have suggested that homeobox B7 (HOXB7), a
known oncogene, is a downstream gene of TUG1, and it was suggested
that TUG1 targets PRC2 to regulate HOXB7 at the transcriptional
level (40,41). The role of HOXB7 has also been
investigated, which showed that HOXB7 promoted NSCLC cell
proliferation via activating the AKT and mitogen-activated protein
kinase pathways (15,38,39,42,43).
Therefore, the p53/TUG1/PRC2/HOXB7 axis was found to be vital in
the tumorigenesis and progression of NSCLC, which may be a target
for future therapy.
Conclusions and perspectives
In previous years, widespread investigations have
been performed on the biological roles and clinical significance of
lncRNAs. The deregulation of lncRNAs is capable of affecting tumor
development, functioning as tumor inducers or suppressors. One of
the most comprehensively investigated lncRNAs is the hox transcript
antisense intergenic RNA (HOTAIR). HOTAIR has been identified as an
oncogenic gene, recruiting PRC2 and interacting with
lysine-specific demethylase1 (LSD1), regulating the expression of
its downstream targets (8,11,42).
This review discusses TUG1 and its association with
cancer in humans. TUG1 is expressed in a tissue-specific pattern,
showing oncogenic or tumor inhibiting capacities in different types
of cancer in humans (Table I). The
downregulation of TUG1 is observed in glioma and NSCLC, showing
that TUG1 serves as a tumor suppressor. By contrast, the
overexpression of TUG1 has been reported in ESCC, CRC, HCC, bladder
cancer and osteosarcoma, indicating that TUG1 functions as an
oncogene. The mechanisms underlying the biological functions of
TUG1 are shown in Figs. 1 and
2. These findings indicate that
the expression of TUG1 can be enhanced or weakened by upstream or
downstream interference to slow down or alter tumor progression.
The level of TUG1 in tumor tissues correlates with tumor stage and
prognosis, which may be utilized as a diagnostic and prognostic
biomarker clinically. In a previous study, TUG1 was moderately
elevated in secreted exosomes (44), and this may enable the isolation of
exosomes from blood plasma or serum to assist in monitoring the
state of disease dynamically (45). However, further investigations are
required in order to fully elucidate the mechanisms underlying TUG1
and cancer development. TUG1 may offer potential as a novel
diagnostic biomarker and therapeutic approach for clinical
utilization.
Acknowledgements
This review was supported by grants from National
Natural Science Foundation of China (grant no. 81272532).
References
|
1
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
An integrated encyclopedia of DNA elements
in the human genome. Nature. 489:57–74. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amaral PP and Mattick JS: Noncoding RNA in
development. Mamm Genome. 19:454–492. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blackshaw S, Harpavat S, Trimarchi J, Cai
L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al:
Genomic analysis of mouse retinal development. PLoS Biol.
2:e2472004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dinger ME, Amaral PP, Mercer TR, Pang KC,
Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C,
et al: Long noncoding RNAs in mouse embryonic stem cell
pluripotency and differentiation. Genome Res. 18:1433–1445. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E and
Chang HY: Functional demarcation of active and silent chromatin
domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ginger MR, Shore AN, Contreras A, Rijnkels
M, Miller J, Gonzalez-Rimbau MF and Rosen JM: A noncoding RNA is a
potential marker of cell fate during mammary gland development.
Proc Natl Acad Sci USA. 103:5781–5786. 2006; View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Young TL, Matsuda T and Cepko CL: The
noncoding RNA taurine upregulated gene 1 is required for
differentiation of the murine retina. Curr Biol. 15:501–512. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Zhang M, An G and Ma Q: LncRNA TUG1
acts as a tumor suppressor in human glioma by promoting cell
apoptosis. Exp Biol Med (Maywood). 241:644–649. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Q, Geng PL, Yin P, Wang XL, Jia JP
and Yao J: Down-regulation of long non-coding RNA TUG1 inhibits
osteosarcoma cell proliferation and promotes apoptosis. Asian Pac J
Cancer Prev. 14:2311–2315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han Y, Liu Y, Gui Y and Cai Z: Long
intergenic non-coding RNA TUG1 is overexpressed in urothelial
carcinoma of the bladder. J Surg Oncol. 107:555–559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan J, Qiu K, Li M and Liang Y:
Double-negative feedback loop between long non-coding RNA TUG1 and
miR-145 promotes epithelial to mesenchymal transition and
radioresistance in human bladder cancer cells. FEBS Lett.
589:3175–3181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun J, Ding C, Yang Z, Liu T, Zhang X,
Zhao C and Wang J: The long non-coding RNA TUG1 indicates a poor
prognosis for colorectal cancer and promotes metastasis by
affecting epithelial-mesenchymal transition. J Transl Med.
14:422016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu Y, Wang J, Qiu M and Xu L, Li M, Jiang
F, Yin R and Xu L: Upregulation of the long noncoding RNA TUG1
promotes proliferation and migration of esophageal squamous cell
carcinoma. Tumour Biol. 36:1643–1651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang E, He X, Yin D, Han L, Qiu M, Xu T,
Xia R, Xu L, Yin R and De W: Increased expression of long noncoding
RNA TUG1 predicts a poor prognosis of gastric cancer and regulates
cell proliferation by epigenetically silencing of p57. Cell Death
Dis. 7:e21092016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma
P and Shu YQ: Long non-coding RNA TUG1 is up-regulated in
hepatocellular carcinoma and promotes cell growth and apoptosis by
epigenetically silencing of KLF2. Mol Cancer. 14:1652015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Paul TA, Bies J, Small D and Wolff L:
Signatures of polycomb repression and reduced H3K4 trimethylation
are associated with p15INK4b DNA methylation in AML. Blood.
115:3098–3108. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aoki R, Chiba T, Miyagi S, Negishi M,
Konuma T, Taniguchi H, Ogawa M, Yokosuka O and Iwama A: The
polycomb group gene product Ezh2 regulates proliferation and
differentiation of murine hepatic stem/progenitor cells. J Hepatol.
52:854–863. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen H, Gu X, Su IH, Bottino R, Contreras
JL, Tarakhovsky A and Kim SK: Polycomb protein Ezh2 regulates
pancreatic beta-cell Ink4a/Arf expression and regeneration in
diabetes mellitus. Genes Dev. 23:975–985. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan T, Jiang S, Chung N, Alikhan A, Ni C,
Lee CC and Hornyak TJ: EZH2-dependent suppression of a cellular
senescence phenotype in melanoma cells by inhibition of p21/CDKN1A
expression. Mol Cancer Res. 9:418–429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zilio N, Codlin S, Vashisht AA, Bitton DA,
Head SR, Wohlschlegel JA, Bähler J and Boddy MN: A novel histone
deacetylase complex in the control of transcription and genome
stability. Mol Cell Biol. 34:3500–3514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vandewalle C, Comijn J, De Craene B,
Vermassen P, Bruyneel E, Andersen H, Tulchinsky E, Van Roy F and
Berx G: SIP1/ZEB2 induces EMT by repressing genes of different
epithelial cell-cell junctions. Nucleic Acids Res. 33:6566–6578.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fernandez-Zapico ME, Lomberk GA, Tsuji S,
DeMars CJ, Bardsley MR, Lin YH, Almada LL, Han JJ, Mukhopadhyay D,
Ordog T, et al: A functional family-wide screening of SP/KLF
proteins identifies a subset of suppressors of KRAS-mediated cell
growth. Biochem J. 435:529–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kotake Y, Nakagawa T, Kitagawa K, Suzuki
S, Liu N, Kitagawa M and Xiong Y: Long non-coding RNA ANRIL is
required for the PRC2 recruitment to and silencing of p15(INK4B)
tumor suppressor gene. Oncogene. 30:1956–1962. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Isin M, Ozgur E, Cetin G, Erten N, Aktan
M, Gezer U and Dalay N: Investigation of circulating lncRNAs in
B-cell neoplasms. Clin Chim Acta. 431:255–259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ghavami S, Hashemi M, Ande SR, Yeganeh B,
Xiao W, Eshraghi M, Bus CJ, Kadkhoda K, Wiechec E, Halayko AJ and
Los M: Apoptosis and cancer: Mutations within caspase genes. J Med
Genet. 46:497–510. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brentnall M, Rodriguez-Menocal L, De
Guevara RL, Cepero E and Boise LH: Caspase-9, caspase-3 and
caspase-7 have distinct roles during intrinsic apoptosis. BMC Cell
Biol. 14:322013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liao WT, Jiang D, Yuan J, Cui YM, Shi XW,
Chen CM, Bian XW, Deng YJ and Ding YQ: HOXB7 as a prognostic factor
and mediator of colorectal cancer progression. Clin Cancer Res.
17:3569–3578. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jin K, Kong X, Shah T, Penet MF, Wildes F,
Sgroi DC, Ma XJ, Huang Y, Kallioniemi A, Landberg G, et al: The
HOXB7 protein renders breast cancer cells resistant to tamoxifen
through activation of the EGFR pathway. Proc Natl AcadSci USA.
109:2736–2741. 2012; View Article : Google Scholar
|
|
40
|
Storti P, Donofrio G, Colla S, Airoldi I,
Bolzoni M, Agnelli L, Abeltino M, Todoerti K, Lazzaretti M, Mancini
C, et al: HOXB7 expression by myeloma cells regulates their
pro-angiogenic properties in multiple myeloma patients. Leukemia.
25:527–537. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yuan W, Zhang X, Xu Y, Li S, Hu Y and Wu
S: Role of HOXB7 in regulation of progression and metastasis of
human lung adenocarcinoma. Mol Carcinog. 53:49–57. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
di Pietro M, Lao-Sirieix P, Boyle S,
Cassidy A, Castillo D, Saadi A, Eskeland R and Fitzgerald RC:
Evidence for a functional role of epigenetically regulated
midcluster HOXB genes in the development of Barrett esophagus. Proc
Natl Acad Sci USA. 109:9077–9082. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu X, Chen H, Parker B, Rubin E, Zhu T,
Lee JS, Argani P and Sukumar S: HOXB7, a homeodomain protein, is
overexpressed in breast cancer and confers epithelial-mesenchymal
transition. Cancer Res. 66:9527–9534. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gezer U, Özgür E, Cetinkaya M, Isin M and
Dalay N: Long non-coding RNAs with low expression levels in cells
are enriched in secreted exosomes. Cell Biol Int. 38:1076–1079.
2014.PubMed/NCBI
|
|
45
|
Bang C and Thum T: Exosomes: New players
in cell-cell communication. Int J Biochem Cell Biol. 44:2060–2064.
2012. View Article : Google Scholar : PubMed/NCBI
|