Introduction
Glioblastoma multiforme (GBM) is one of the most
aggressive and lethal type of malignant tumor of the central
nervous system (CNS) (1–3), which is characterized by its fast
growth and unlimited self-renewal potential. Despite development of
therapeutic strategies for GBM, the overall survival of patients
improved marginally, with a 5-year survival rate of just 9%
(2). Therefore, more precise
prognostic predictors and more effective therapeutic approaches are
urgently required for patients.
MicroRNAs (miRNAs) are single-stranded non-coding
RNAs with 19–23 oligonucliotides, which bind to the 3′-untranslated
region of target genes, induce degradation and partake in virtually
all biopathological steps. Increasing evidence indicates that
miRNAs regulate the diverse biological steps of carcinogenesis and
progression in cancer (4).
Additionally, dysregulation of miRNA exhibits oncogeneic and tumor
suppressor properties (4,5). Human microRNA-132
(has-miR-132), located in ch.17, is aberrantly expressed in
gastric cancer, chronic lymphocytic leukemia, hepatocellular
carcinoma and colorectal cancer (CRC) (6–9).
Furthermore, as a central nervous system-specific miRNA, miR-132
displays vital roles in neurogenesis, neuron stem cell
differentiation and development (10–13).
Its dysregulation results in various types of brain-associated
disease, including Huntington's disease, Parkinson's disease and
schizophrenia (14,15). Previously, miR-132 was detected to
be highly expressed in glioma, serving as a biomarker of a poor
prognosis in patients (16).
However, the functions of miR-132 in GBM stemness are complex and
require further exploration.
In the present study, the potential bias from sample
size was minimized by enrolling the GBM specimens from The Cancer
Genome Atlas (TCGA) Research Network and investigated the clinical
significance of miR-132. A high level of miR-132 was identified to
be significantly correlated with neural subtype of GBM and a poor
outcome for patients. Furthermore, a Gene-Cloud of Biotechnology
Information (GCBI) bioinformatics analysis was performed to
investigate the GEO datasets and the results revealed that miR-132
fuels proliferation and self-renewal potential potentially by
targeting polypyrimidine tract-binding protein 2 (PTBP2) in GBM
cells.
Materials and methods
Cell culture and sphere culture
The U87 GBM cell line was purchased from the Chinese
Academy of Sciences Cell Bank (Shanghai, China) and cultured in
Gibco Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum
(FBS). The conditional culture medium, Gibco DMEM/F12 (Thermo
Fisher Scientific, Inc.) was supplemented with Invitrogen B27 (1X;
Thermo Fisher Scientific, Inc.), 20 ng/ml basic fibroblast growth
factor and 20 ng/ml epidermal growth factor (both from PeproTech,
Inc., Rocky Hill, NJ, USA). First generation U87-neurospheres were
observed in all wells of a 6-well plate 72 h later. All cultures
were maintained at 37°C in an atmosphere of 5% CO2.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The miRNA was extracted with RNAiso for small RNA
(Takara Bio, Inc., Otsu, Japan) and miR-132 was examined using a
TaqMan microRNA Assay (Thermo Fisher Scientific, Inc.) with U6
serving as the internal control. Total mRNA extraction was
performed using TRIzol (Takara Bio, Inc.) from U87 cells and fresh
glioma specimens. The fresh specimens were obtained from resected
samples from glioma patients in the Chongqing Cancer Hospital
(Chongqing, China). Expression levels of PTBP2 and GAPDH were
measured using an RT-PCR kit (cat no. RR055A; Takara Bio, Inc.)
according to the manufacturer's instructions and a CFX 96 system
(Bio-Rad Laboratories, Inc., CA, USA). Each sample was examined in
triplicate and analyzed according to the 2−ΔΔCq method
(17) and GAPDH served as the
internal control in the assay. The PCR reaction was run as follows:
95°C for 30 sec, 39 cycles of 95°C for 5 sec and 60°C for 30 sec.
The primer sequences for qPCR were as follows: Forward,
5′-TGGATCCCCCCCAGTCCCCGTCCCTCAG-3′ and reverse,
5′-TGAATTCGGATACCTTGGCCGGGAGGAC-3′ for miR-132; forward,
5′-GCGCGTCGTGAAGCGTTC−3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′ for
U6; forward, 5′-GCAACCGAGGAAGCAGCTATT-3′ and reverse,
5′-GCCTGAGCACGTTGGTTTAATG-3′ for PTBP2; forward,
5′-TGTGGGCATCAATGGATTTGG-3′ and reverse,
5′-ACACCATGTATTCCGGGTCAAT-3′ for GAPDH.
miRNA reagent transfection
The miR-132 mimic and control reagents were obtained
from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). U87 cells with
75% confluence were transfected with Invitrogen Lipofectamine 2000
(Thermo Fisher Scientific, Inc.). The culture medium with DMEM
containing 10% FBS was replaced within 6 h. The miR-132 mimic was
as follows: UAACAGUCUACAGCCAUGGUCGACCAUGGCUGUAGACUGUUAUU; and the
miR-132 control: UUCUCCGAACGUGUCACGUTTACGUGACACGUUCGGAGAATT.
Cell proliferation assay
The proliferation ability of glioma cells was
measured using a Cell Counting Kit-8 (CCK8; Beyotime Institute of
Biotechnology, Haimen, China). U87 cells (5×104) with
miRNA reagent transfection or untreated cells were seeded into
96-well plates and cultured for 0, 12, 24, 48 and 72 h. At each
time interval, CCK8 (20 µl) was added to each well. Following 2 h
incubation at 37°C, the absorbance was examined with a Multiskan
(Thermo Fisher Scientific, Inc.) at a wavelength of 450 nm.
Sphere formation assay and colony
formation assay
Both sphere formation assay and colony formation
assay were performed to evaluate the self-renewal ability of U87
cells. According to our previous study (18), different numbers (20, 50 or 100) of
cells were seeded into a 96-well plate. Serum-free medium (25 µl)
was added to each well every 2 days. Plates were incubated for 2
weeks at 37°C in an atmosphere of 5% CO2 until
neurospheres formed and the number of spheroid cells was counted
for statistical analysis.
Colony formation ability was investigated by seeding
U87 cells into a 6-well plate 6 h after transfection. Plates were
incubated for 14–21 days until colonies were large enough to be
visualized. Subsequently, colonies were fixed and stained in
crystal violet (1%) for 10 min. The number of colonies was counted
manually under an inverted microscope (Leica Microsystems GmbH,
Wetzlar, Germany).
Bioinformatics analysis
TCGA (http://cancergenome.nih.gov) Research Network, a huge
tumor profiling data set for a very large collection of tumor
types, was mined in order to evaluate the predictive value of
miR-132 in GBM specimens (n=526). A comprehensive bioinformatics
analysis approach (GCBI; https://www.gcbi.com.cn/gclib/html/index), which was
deeply integrated with the Affymetrix Gene Chip in GEO database,
was used to enrich the dataset for genes, including the heat map
analysis, volcano map analysis, gene ontology (GO) analysis and
pathway analysis (19,20). In order to compare the different
expression genes between groups of neurosphere cell lines with
miR-132 transfection (GCS24468, GCS24458 and GCS24463) or control
transfection (GCS24456, GCS24465 and GCS24464) (19), heat mapping was performed. To
further analyze the functions of different expression genes on the
basis of biological processes and molecular function, GO analysis
was performed. In addition, pathway analysis was used to establish
the significant pathway of differential genes according to the
Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg/), BioCarta's pathways (http://www.biocarta.com/genes/index.asp)
and Reatome pathway databases (http://www.reactome.org). In order to establish the
downstream targets of miR-132, five renowned miRNA prediction
databases (TargetScan, www.targetscan.org/vert_71; miRanda, www.microrna.org/microrna/home.do;
miRDB, www.mirdb.org; miRWalk, zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk/index.html;
RNA22, cm.jefferson.edu/rna22) were
bioinformatically scanned as previously described (18,21).
Patient specimens
Newly diagnosed GBM patients who had received no
previous treatment from 01/2015 to 01/2016 were enrolled in the
present study. A total of 13 fresh GBM specimens were obtained from
these patients straight after surgery at the Department of
Neurosurgery, The Chongqing Cancer Hospital (Chongqing, China).
Specimens were pre-mixed with liquid nitrogen at −80°C and ground.
Following TRIzol (Takara Bio, Inc.) was added, the samples were
prepared for the extraction of miRNA/mRNA following the related
protocol. Written informed consent was obtained from each patient
according to the national regulations of clinical samples and the
study was approved by the Ethics Board of The Chongqing Cancer
Hospital.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 20.0; IBM Corp., Armonk, NY, USA). The expression
levels of miR-132 in different subtypes of GBM were compared using
Student's t-test, and the χ2 test was used to analyze
the correlation between miR-132 and the GBM subtypes. X-tile
software (version 3.6.1; Yale University, New Haven, CT, USA) was
used to determine the cutoff value of miR-181c according to
previously reported instructions (18). Kaplan-Meier survival curve and the
log-rank test were performed to compare the overall survival (OS)
in patient groups. COX's proportional hazard regression model was
established for multivariate analysis of the prognostic value of
each factors. P<0.05 was considered to indicate a statistically
significant difference.
Results
miR-132 is correlated with molecular
subtypes and predicts patient outcomes
It has been reported that miR-132 was overexpressed
in a cohort of 43 patients with glioma and correlated with
unfavorable clinical outcomes (16). In order to minimize the system bias
caused by sample scale, a GBM cohort (n=526) was included, which
was obtained from TCGA database to analyze the expression level of
miR-132. GBM is divided into four molecular subtypes according to
gene expression patterns; classical, mesenchymal, neural and
proneural (22). The results
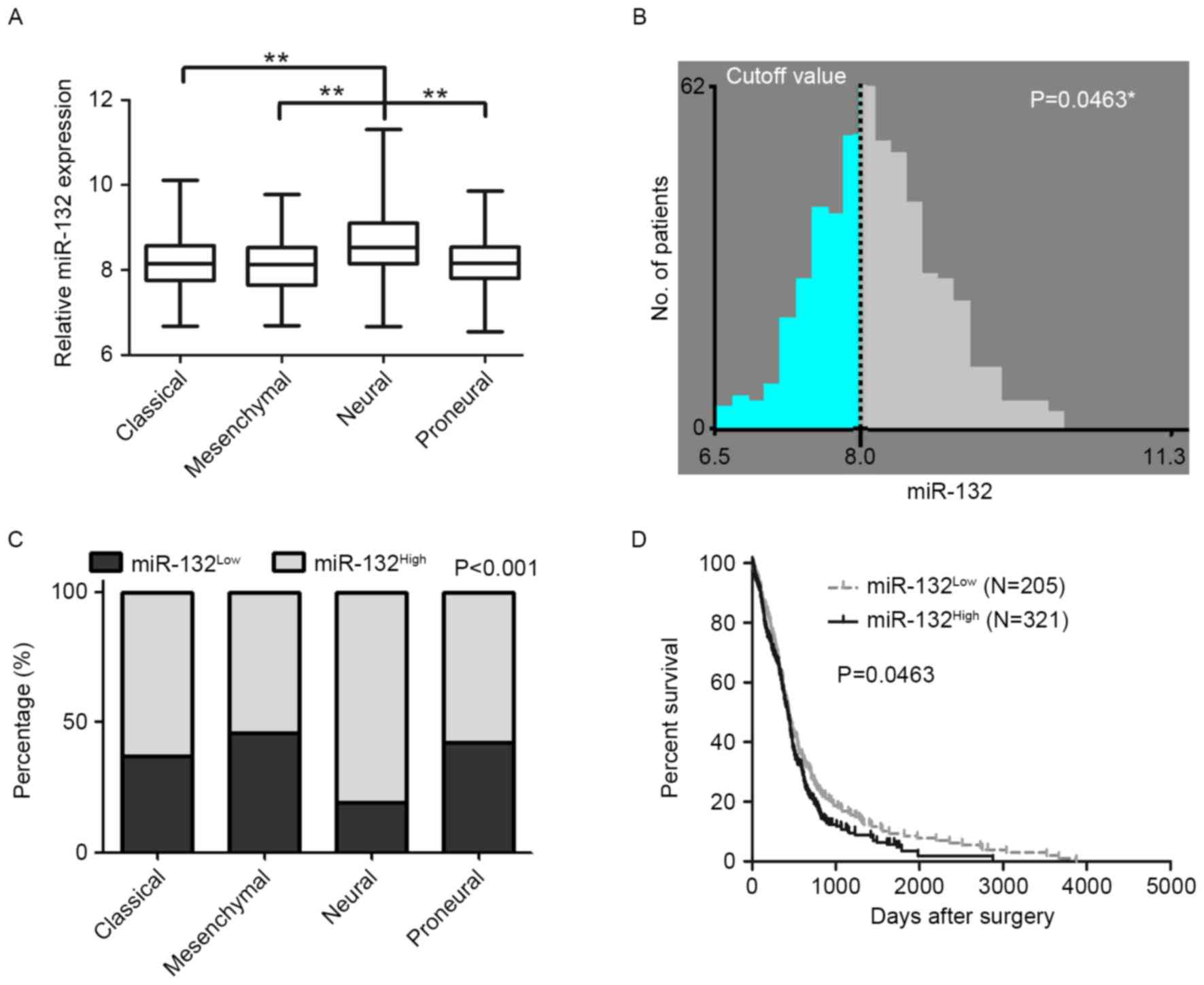
indicated that miR-132 was markedly higher in the neural subtype
(8.625±0.077; n=84) than that in the three other subtypes
[8.625±0.055 for classical subtype (n=138), 8.132±0.051 for
mesenchymal subtype (n=154) and 8.183±0.056 for proneural subtype
(n=125; P<0.01) as presented in Fig. 1A]. To examine the association
between miR-132 expression and GBM patient outcome, a meaningful
approach to classify the cutoff value (8.0) for the miR-132
expression level was set using X-tile software (P=0.0463; Fig. 1B). Results revealed that >80% of
patients with the neural subtype of GBM demonstrated a higher
expression level of miR-132, as compared with patients with the
classical subtype (63.04%), the mesenchymal subtype (53.90%) and
the proneural subtype (57.60%) (P<0.01; Fig. 1C). According to the cutoff value,
patients were then split into two groups (miR-132High
and miR-132Low). High miR-132 expression (>8.0)
indicated a worse prognosis, with a median OS of 425.0 days (n=205)
vs. 441.0 days (n=321) in the low-expression group (<8.0;
P<0.05; Fig. 1D). In addition,
the Cox hazard regression model was applied to evaluate the
predictive value of miR-132 for GBM patients. Results demonstrated
that miR-132 was an independent prognostic factor for predicting
the patient outcome (P<0.05; hazard ratio, 1.204; 95% confidence
interval, 1.040–1.395; Table I).
Collectively, these results indicate that miR-132 was correlated
with the GBM subtypes and serve as a robust prognostic indicator
for GBM patients.
 | Table I.Univariate and multivariate analysis
of the predictive value of miR-132 and clinical features for The
Cancer Genome Atlas glioblastoma multiforme patients. |
Table I.
Univariate and multivariate analysis
of the predictive value of miR-132 and clinical features for The
Cancer Genome Atlas glioblastoma multiforme patients.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
| Factor | P-value | HR (95% CI) | P-value | HR (95% CI) |
|---|
| Gender | 0.237 | 0.885
(0.724–1.083) | 0.280 | 0.894
(0.731–1.095) |
| Subtypes | 0.113 | 0.934
(0.858–1.016) | 0.088 | 0.929
(0.853–1.011) |
| miR-132 | 0.013 | 1.205
(1.041–1.395) | 0.013 | 1.204
(1.040–1.395) |
miR-132 fuels proliferation, as well
as self-renewal of U87 cells
miR-132 was reported to inhibit U87 cell invasion
and metastasis (23), indicating
its anticancer potential. Furthermore, the data demonstrated that
miR-132 acted as an oncogenic miRNA during GBM progression. miR-132
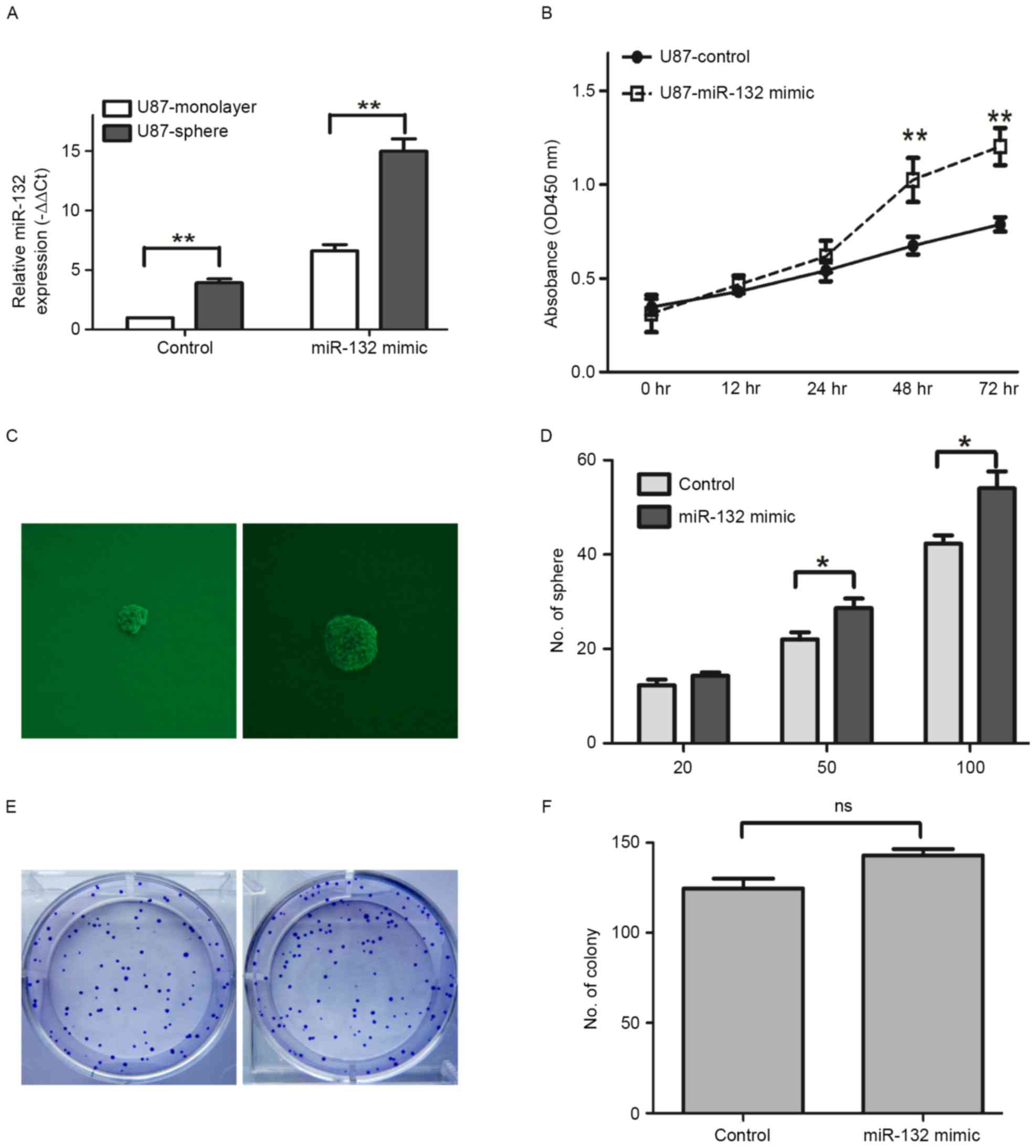
expression levels were identified to be higher in U87 sphere, as
compared with U87-monolayer cells (P<0.01; Fig. 2A). The miR-132 mimic was
transfected into the U87 cells to elevate the expression level of
endogenous miR-132 (Fig. 2A).
Following pre-treatment of the U87 cells with miR-132 mimic, the
proliferation ability was significantly enhanced at 48 and 72 h, as
compared with U87 cells in the control group (P<0.01; Fig. 2B). To further investigate the
effect of miR-132 on self-renewal in U87 cells, two representative
assays (sphere formation and colony formation assays) were
performed. The sphere formation assay indicated that miR-132
overexpression significantly enhanced the sphere formation
potential of the cells when compared with the control group, which
was treated with PBS (P<0.01; Fig.
2C and D). For the colony formation assay, no significant
increase/decrease was observed within the groups of U87 with or
without miR-132 treatment (P>0.05; Fig. 2E and F). These results demonstrated
that a high expression level of miR-132 promotes proliferation, as
well as self-renewal potential of U87 cells.
Microarray based bioinformatics
analysis revealed the role of miR-132 in GBM cells
To further elucidate the mechanisms underlying the
method by which miR-132 promotes GBM proliferation and maintains
the sphere formation properties of U87 cells, a genetic
bioinformatics database, the GCBI was searched, which provides a
web-lab with bioinformatics approaches to manage numerous
microarray results (19). The
Affymetrix Gene Chip was obtained (control group vs. miR-132
transfected group) from the GEO database and run in the GCBI
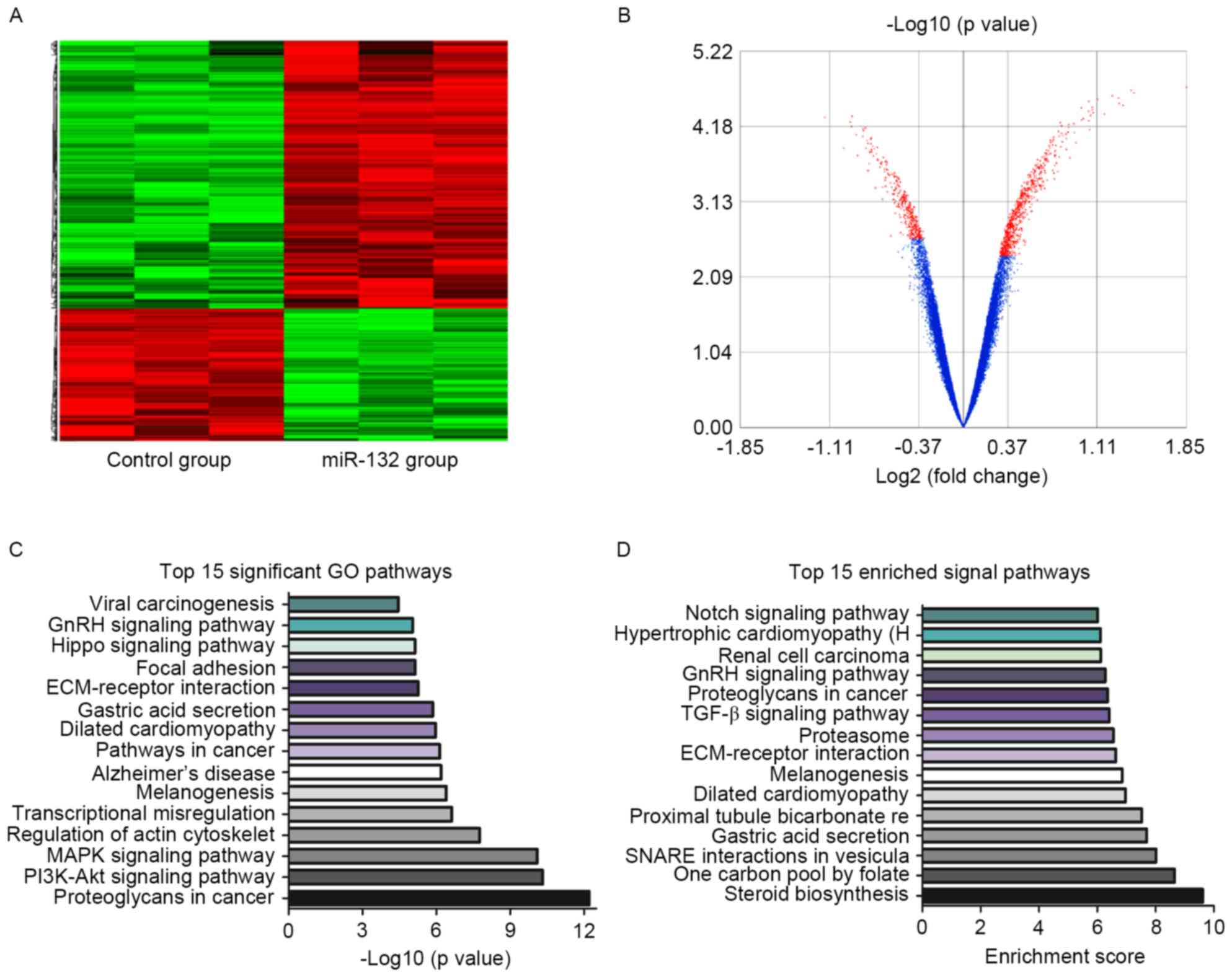
web-lab. Following the unsupervised hierarchical clustering
analysis, differently expressed genes were selected according to
their P-value threshold and represented in a heat map, as well as
in a volcano map (Fig. 3A and B).
The results revealed that 3,121 genes were overexpressed and 2,742
genes were downregulated following miR-132 overexpression in GBM
cells (data not shown). To elucidate the miR-132-associated
biological processes and underlying mechanisms, GO analysis was
applied and the results demonstrated that significant GO pathways
were screened according to their P-values. The representative top
15 significant GO pathways are presented in Fig. 3C. An additional method to identify
the associated signaling pathways is evaluation by their enrichment
score (24). The representative
top 15 enriched signaling pathways are presented in Fig. 3D. To investigate the pathway
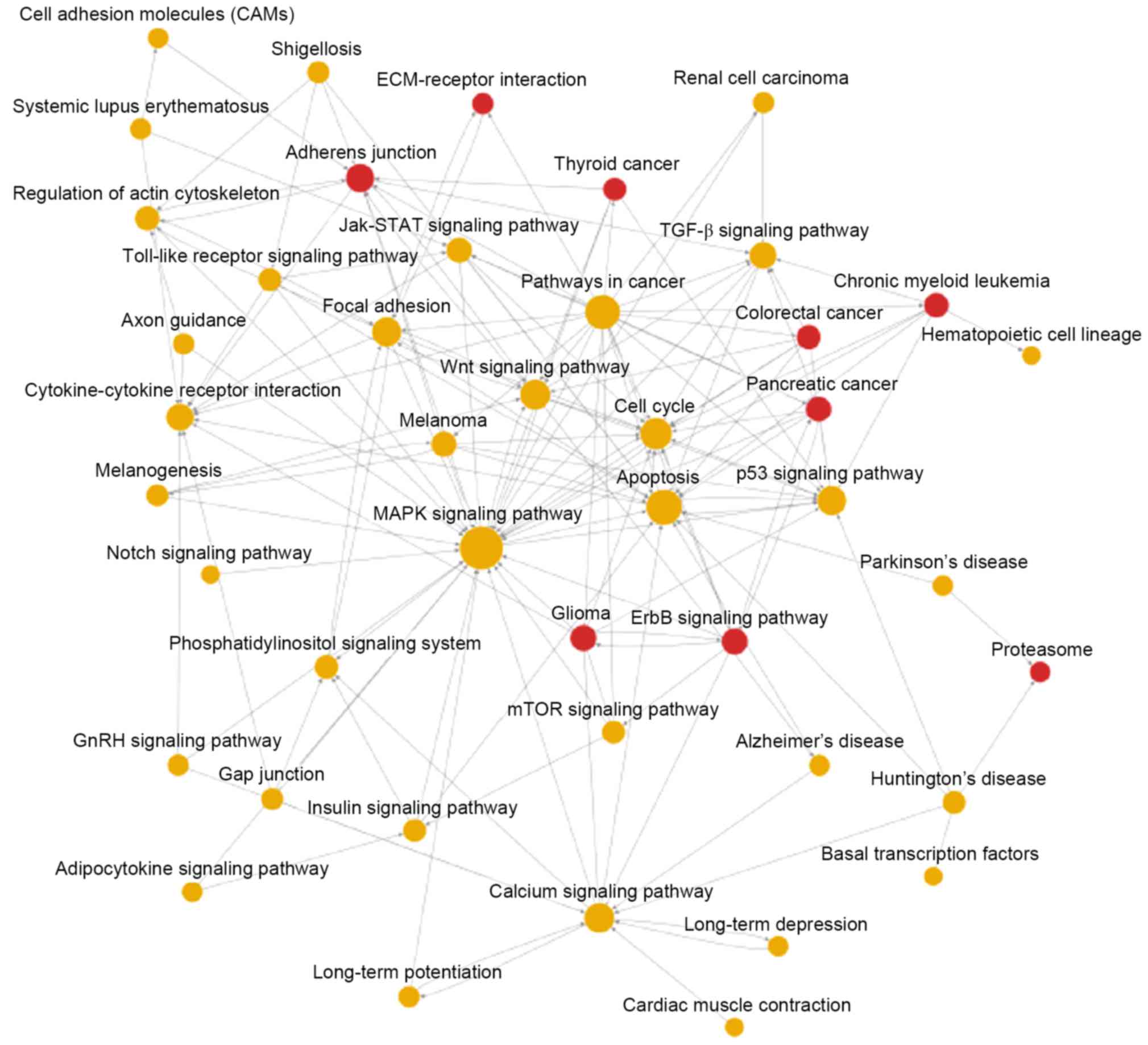
interactions in miR-132 overexpressed GBM cells, the pathway
interaction-based network was also applied. The results indicated
that following miR-132 transfection in GBM cells, only gliomas, the
ErbB signaling pathway, the adherens junction, proteasomes, CRC,
pancreatic cancer, chronic myeloid leukemia, thyroid cancer and the
extracellular matrix-receptor (ECM-receptor) interaction were
definitely upregulated (as demonstrated by red spheres; Fig. 4). Furthermore, other signaling
pathways/functional pathways are represented by yellow spheres,
which indicates that these pathways may be upregulated by certain
pathways but downregulated by some other signaling pathways
(Fig. 4). Taken together, the
results demonstrated that miR-132 transfection in GBM cells
significantly altered a great number of genes and induced
activation of various downstream signals, leading to sustained
proliferation and sphere formation. However, the detailed
mechanisms require further investigation.
PTBP2 was the downstream target of
miR-132 in GBM cells
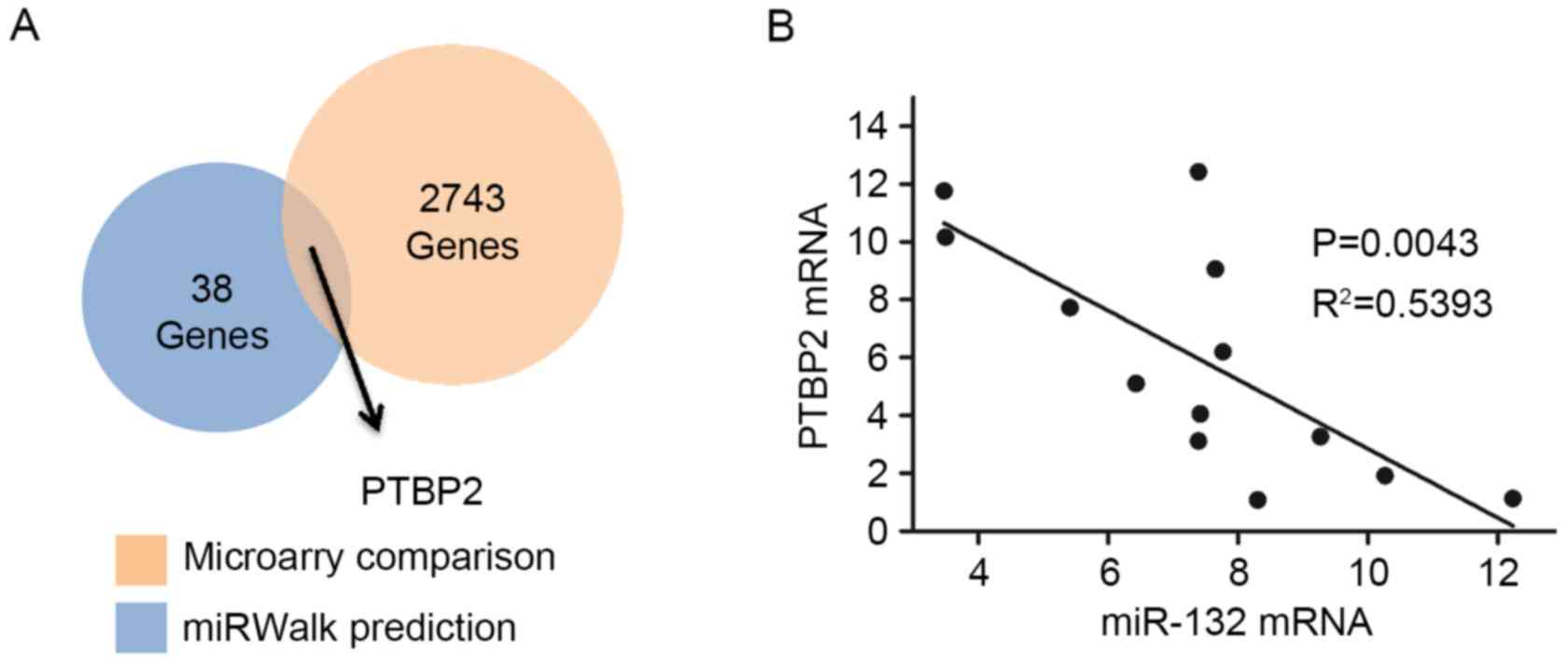
The GCBI successfully revealed 2,743 genes, which
where downregulated in miR-132 transfected GBM cells. miRWalk was
used to predict the miR-132 target genes (21). A total of 38 genes were commonly
predicted in at least five of the six miRNA prediction databases
(miRanda, miRDB, miRWalk, PICTAR5, RNA22 and Targetscan). Notably,
PTBP2 was the only common gene of the 38 predicted genes and the
2,743 downregulated genes (Fig.
5A). To further elucidate the miR-132/PTBP2 regulation
mechanism, 13 fresh GBM specimens were obtained from GBM patients
and the expression levels of miR-132 and PTBP2 were examined.
Subsequently, a linear regression model demonstrated that miR-132
and PTBP2 were negatively correlated in the GBM specimens
(R2=0.5393, P<0.01; Fig.
5B). Thus, these results indicate that PTBP2 was a putative
downstream target of miR-132 in GBM cells.
Discussion
Despite the availability of surgical treatment and
chemoradiotherapy, patients with GBM continue to experience
unfavorable outcomes (2,3). A notably malignant behavior of GBM is
its unlimited proliferation potential, which leads to recurrence
following surgery. Increasing evidence demonstrates that miR-132 is
essential during tumorigenesis and progression. While, the role of
miR-132 remains unclear, it may serves as either an oncogene or
tumor suppressor depending on the tumor type (8,9,16,25).
In ovarian cancer cells, miR-132 suppresses cell
proliferation, invasion and migration by targeting E2F
transcription factor 5 (25).
Downregulated miR-132 was detected in CRC specimens and associated
with a poor prognosis in patients (8). In addition, miR-132 inhibits
proliferation of hepatic carcinoma cells by targeting
yes-associated protein (6).
Furthermore, miR-132 expression levels were significantly increased
in gastric cancer (GC) specimens and resulted in enhanced GC cell
growth, which was mediated by the suppression of Forkhead box
protein O1 (7). In human glioma
tissues, high expression levels of miR-132 were widely detected and
positively correlated with the WHO glioma grade (16), which indicated an oncogenic role in
glioma cells. However, miR-132 was reported to inhibit the invasion
and metastasis of U87, which is indicates a tumor suppressive
function (23). Therefore, a
comprehensive evaluation of miR-132 is required to evaluate its
expression in GBM specimens. A total of 526 GBM specimens were
obtained from the TCGA database to minimize the system bias
resulting from sample size, and revealed miR-132 as a promising and
potential independent prognostic indicator for GBM patients. The
clinical functions of miR-132 in GBM cells were quickly verified
in vitro with an interesting result, which indicated that
miR-132 may serve as an oncogenic miRNA during GBM progression.
The identified molecular subtypes may underlie
differences in patient sensitivity to therapy and prognosis
(22,26). Notably, miR-132 was significantly
elevated in the neural subtype of GBM specimens. According to
previous reports, neural subtype is not sensitive to concurrent
chemoradiotherapy or temozolomide (26). In addition, according to the
classification approach used by Phillips et al (26), the neural subtype belongs to a
proliferative subclass, which has a much shorter median survival
time when compared with proneural and mesenchymal subclassess
(26). The present study
demonstrated that miR-132 promoted the proliferation and
self-renewal potential of U87 cells, which is consistent with a
previous study (26).
To further elucidate the underlying mechanisms of
how miR-132 regulates proliferation and self-renewal of U87 cells,
the GEO microarray database was searched and the miRWalk web tool
was used to predict miR-132 target genes. PTBP2 was identifies as
the only common gene within the downregulated gene pool in the GEO
microarray and miRWalk prediction gene pool.
PTBP2 belongs to the polypyrimidine tract binding
(PTB) proteins, is primarily detected in brain tissues and
regulates tissue-specific post-transcriptional functions during
neuron development and pathological processes (27). In osteosarcoma, the combination of
PTBP2/PTB-associated splicing factor inhibited cell proliferation,
migration, invasion and the epithelial-mesenchymal transition
processes (28). However, its
function in the CNS is quite specific and different. A previous
study revealed that the PTBP2 level induced various splicing
programs including the differentiation of neuron stem cell, early
differentiating neuron splicing, and synaptic maturation (29). PTBP2 was demonstrated to promote
proliferation and migration in the human glioma cell lines, U251
and LN229 (30). However, in
another glioma cell, T98 G, the expression level of PTBP2 was lower
than that of healthy brain tissues (31). In the present study, PTBP2 was
downregulated in glioma stem cells and served as a tumor
suppressor, which was revealed as a promising downstream target of
miR-132 during GBM progression.
In conclusion, the results indicate that the
miR-132/PTBP2 signaling pathway may sustain U87 cell proliferation
and self-renewal, and elucidate the potential role exerted by
miRNAs in GBM. In addition to highlighting the ability of miRNAs,
the present study demonstrated the complexity of the underlying
mechanisms regulating GBM progression. There are certain
limitations in the present study. The fresh sample size was small
and needs to be increased to be of statistical value. The only cell
model used in the present study was U87, to verify the results and
conclusions, more cells need to be used in the future. Detailed
underlying mechanisms require further investigation, which will be
the focus of future studies by the authors.
Acknowledgements
The authors would like to thank the central lab for
providing technical instruction and assistance. The authors would
also like to thank Dr Chen Dewei for his assistance with
bioinformatics data mining. The present study was supported by the
National Natural Science Foundation of China (grant no.
81502283).
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Berindan-Neagoe I, Pdel C Monroig,
Pasculli B and Calin GA: MicroRNAome genome: A treasure for cancer
diagnosis and therapy. CA Cancer J Clin. 64:311–336. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen CZ: MicroRNAs as oncogenes and tumor
suppressors. N Engl J Med. 353:1768–1771. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lei CJ, Li L, Gao X, Zhang J, Pan QY, Long
HC, Chen CZ, Ren DF and Zheng G: hsa-miR-132 inhibits proliferation
of hepatic carcinoma cells by targeting YAP. Cell Biochem Funct.
33:326–333. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li W, Zhang J, Chen T, Yin P, Yang J and
Cao Y: miR-132 upregulation promotes gastric cancer cell growth
through suppression of FoxO1 translation. Tumour Biol. Aug
23–2015.(Epub ahead of print).
|
|
8
|
Mokutani Y, Uemura M, Munakata K, Okuzaki
D, Haraguchi N, Takahashi H, Nishimura J, Hata T, Murata K,
Takemasa I, et al: Down-Regulation of microRNA-132 is associated
with poor prognosis of colorectal cancer. Ann Surg Oncol. 23 Suppl
5:S599–S608. 2016. View Article : Google Scholar
|
|
9
|
Tavolaro S, Colombo T, Chiaretti S,
Peragine N, Fulci V, Ricciardi MR, Messina M, Bonina S, Brugnoletti
F, Marinelli M, et al: Increased chronic lymphocytic leukemia
proliferation upon IgM stimulation is sustained by the upregulation
of miR-132 and miR-212. Genes Chromosomes Cancer. 54:222–234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Makeyev EV, Zhang J, Carrasco MA and
Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation
by triggering brain-specific alternative pre-mRNA splicing. Mol
Cell. 27:435–448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Magill ST, Cambronne XA, Luikart BW, Lioy
DT, Leighton BH, Westbrook GL, Mandel G, Goodman RH, et al:
microRNA-132 regulates dendritic growth and arborization of newborn
neurons in the adult hippocampus. Proc Natl Acad Sci USA.
107:20382–20387. 2010; View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kawashima H, Numakawa T, Kumamaru E,
Adachi N, Mizuno H, Ninomiya M, Kunugi H and Hashido K:
Glucocorticoid attenuates brain-derived neurotrophic
factor-dependent upregulation of glutamate receptors via the
suppression of microRNA-132 expression. Neuroscience.
165:1301–1311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shaked I, Meerson A, Wolf Y, Avni R,
Greenberg D, Gilboa-Geffen A and Soreq H: MicroRNA-132 potentiates
cholinergic anti-inflammatory signaling by targeting
acetylcholinesterase. Immunity. 31:965–973. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miller BH, Zeier Z, Xi L, Lanz TA, Deng S,
Strathmann J, Willoughby D, Kenny PJ, Elsworth JD, Lawrence MS, et
al: MicroRNA-132 dysregulation in schizophrenia has implications
for both neurodevelopment and adult brain function. Proc Natl Acad
Sci USA. 109:3125–3130. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee ST, Chu K, Im WS, Yoon HJ, Im JY, Park
JE, Park KH, Jung KH, Lee SK, Kim M and Roh JK: Altered microRNA
regulation in Huntington's disease models. Exp Neurol. 227:172–179.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Q, Liao F, Wu H, Cai T, Yang L, Wang
ZF and Zou R: Upregulation of miR-132 expression in glioma and its
clinical significance. Tumour Biol. 35:12299–12304. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ruan J, Lou S, Dai Q, Mao D, Ji J and Sun
X: Tumor suppressor miR-181c attenuates proliferation, invasion,
and self-renewal abilities in glioblastoma. Neuroreport. 26:66–73.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Setty M, Helmy K, Khan AA, Silber J, Arvey
A, Neezen F, Agius P, Huse JT, Holland EC and Leslie CS: Inferring
transcriptional and microRNA-mediated regulatory programs in
glioblastoma. Mol Syst Biol. 8:6052012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
An F, Zhan Q, Xia M, Jiang L, Lu G, Huang
M, Guo J and Liu S: From moderately severe to severe
hypertriglyceridemia induced acute pancreatitis: Circulating miRNAs
play role as potential biomarkers. PLoS One. 9:e1110582014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Verhaak RG, Hoadley KA, Purdom E, Wang V,
Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, et al:
Integrated genomic analysis identifies clinically relevant subtypes
of glioblastoma characterized by abnormalities in PDGFRA, IDH1,
EGFR, and NF1. Cancer Cell. 17:98–110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang H, Li XT, Wu C, Wu ZW, Li YY, Yang
TQ, Chen GL, Xie XS, Huang YL, Du ZW and Zhou YX: miR-132 can
inhibit glioma cells invasion and migration by target MMP16 in
vitro. Onco Targets Ther. 8:3211–3218. 2015.PubMed/NCBI
|
|
24
|
Yang Z, Chen Y, Fu Y, Yang Y, Zhang Y,
Chen Y and Li D: Meta-analysis of differentially expressed genes in
osteosarcoma based on gene expression data. BMC Med Genet.
15:802014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tian H, Hou L, Xiong YM, Huang JX, Zhang
WH, Pan YY and Song XR: miR-132 targeting E2F5 suppresses cell
proliferation, invasion, migration in ovarian cancer cells. Am J
Transl Res. 8:1492–1501. 2016.PubMed/NCBI
|
|
26
|
Phillips HS, Kharbanda S, Chen R, Forrest
WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, et
al: Molecular subclasses of high-grade glioma predict prognosis,
delineate a pattern of disease progression, and resemble stages in
neurogenesis. Cancer Cell. 9:157–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zagore LL, Grabinski SE, Sweet TJ,
Hannigan MM, Sramkoski RM, Li Q and Licatalosi DD: RNA binding
protein Ptbp2 is essential for male germ cell development. Mol Cell
Biol. 35:4030–4042. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang D, Yang H, Lin J, Teng Y, Jiang Y,
Chen J and Li Y: 17β-estradiol regulates cell proliferation, colony
formation, migration, invasion and promotes apoptosis by
upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell
MG-63 in an estrogen receptor-independent manner. Biochem Biophys
Res Commun. 457:500–506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zheng S, Gray EE, Chawla G, Porse BT,
O'Dell TJ and Black DL: PSD-95 is post-transcriptionally repressed
during early neural development by PTBP1 and PTBP2. Nat Neurosci.
15:381–388. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheung HC, Hai T, Zhu W, Baggerly KA,
Tsavachidis S, Krahe R and Cote GJ: Splicing factors PTBP1 and
PTBP2 promote proliferation and migration of glioma cell lines.
Brain. 132:2277–2288. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han W, Wang L, Yin B and Peng X:
Characterization of a novel posttranslational modification in
polypyrimidine tract-binding proteins by SUMO1. BMB Rep.
47:233–238. 2014. View Article : Google Scholar : PubMed/NCBI
|