Introduction
The occurrence of tissue and organ fibrosis is a
hallmark of the aging process, and pulmonary fibrosis is no
exception. The replicative senescence of alveolar epithelial cells
is a prominent pathological finding in idiopathic pulmonary
fibrosis (IPF) (1,2). IPF occurs primarily in the elderly,
and the incidence of IPF is low in younger people (3). A previous study demonstrated that the
occurrence of IPF may be associated with the epigenetic
deregulation mechanisms, which are associated with aging (4). It has been previously reported that
long non-coding RNAs (lncRNAs) may be associated with the
occurrence of pulmonary fibrosis. A previous study observed that
lncRNAs may regulate epithelial-mesenchymal transition in a
bleomycin-induced rat model (5).
LncRNAs may be involved in aging (6) and the regulation of aging-associated
diseases (7). It has been
previously reported that the protein kinase mTOR (mTOR) signaling
pathway may have an important role in the aging process (8,9). A
number of factors are closely associated with aging, including
stress, growth factors, nutritional status and energy supply, which
affect the mTOR signaling pathway. Therefore, the mTOR signaling
pathway may respond to various stimuli and alterations in the body
during the aging process (10).
Studies have demonstrated that lncRNAs and the mTOR signaling
pathway are involved in the process of aging and IPF; therefore,
the present study hypothesized that if there were alterations in
the lncRNAs associated with mTOR, this may provide further evidence
of the association between IPF and aging.
Transforming growth factor-β1 (TGF-β1) has been
demonstrated to have a multifunctional role in IPF. It has been
previously reported that TGF-β1 is able to induce rapid senescence
in A549 cells without significantly inhibiting cell growth, and may
further direct A549 cells to enter a replicative senescent state
(11). TGF-β1 has an important
function in the pathogenesis of IPF (12). Pulmonary fibrosis has been observed
to occur in the elderly in clinical setting; however, the
underlying mechanism remains to be elucidated.
Therefore, the present study aimed to investigate
the differential expression of lncRNAs using high-throughput
sequencing and bioinformatics analysis.
Materials and methods
Study population
According to diagnostic criteria established by the
American Thoracic Society (13),
24 patients with IPF were selected. Simultaneously, 24 healthy
controls were selected (both female:male, 2:18; age, 65±2.2 vs.
65±3.8 years). A total of 4 patients with IPF and 4 healthy
controls (all males; age, 67±3.2 vs. 64±2.8 years; P>0.05) were
selected at random and 3 ml venous blood was extracted for
transcriptome sequencing and bioinformatics analysis. All the
peripheral blood was snap-frozen in liquid nitrogen immediately
following collection and stored at −80°C prior to RNA extraction.
Blood samples were obtained from patients and health controls at
The First Affiliated Hospital of Shanxi Medical University from
January 2016 to July 2016 (Taiyuan, China). All the methods were
performed in accordance with the approved guidelines. The present
study was approved by the Shanxi Medical University Ethics
Committee. Written informed consent was obtained from patients and
healthy individuals.
RNA extraction and sequencing
Total RNA was extracted from blood samples using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's protocol. The
integrity of the RNA was evaluated using a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Wilmington, DE, USA) and standard denaturing 1% agarose gel
electrophoresis. RNA libraries were constructed using rRNA-depleted
RNAs with the TruSeq Stranded Total RNA Library Prep kit (Illumina,
Inc., San Diego, CA, USA), according to the manufacturer's
protocol.
Total RNA (1 µg) was used to remove the rRNAs using
the Ribo-Zero Gold kit (Illumina, Inc.). Libraries of 10 pM were
denatured as single-stranded DNA molecules, captured on Illumina
flow cells, amplified in situ as clusters and finally sequenced for
150 cycles on an Illumina HiSeq Sequencer (Illumina, Inc.)
according to the manufacturer's protocol. Cutadapt version 1.9
(14) and TopHat software (version
2.0.13) (15) were used to further
analyze the results. Subsequently, cuffdiff (version 2.2.1)
(cole-trapnell-lab.github.io/cufflinks/cuffdiff) was used to obtain
the gene level in fragments per kilobase of transcript per million
mapped reads (FPKM), as the expression profiles of lncRNAs and
mRNAs, and fold-changes and q-values were calculated based on the
FPKM. Differentially-expressed lncRNAs and mRNAs were
identified.
Gene ontology (GO) analysis and
pathway analysis
GO and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway analyses were performed to facilitate the
elucidation of the biological implications of unique genes, and to
identify significant pathways in the present study (16). In order to elucidate the potential
roles of differentially-expressed lncRNAs, the KEGG database
(www.genome.jp/kegg) was used to identify
significant pathways for predicted target genes. GO term enrichment
and the biological pathways used significant P-values (P<0.05)
associated with the target genes of differentially-expressed
lncRNAs. Pathway analysis is a functional analysis which
facilitates the mapping of genes to KEGG pathways. The NCBI
database (www.ncbi.nlm.nih.gov/) records gene-specific
information, including gene naming, map location, gene product and
its properties, markers, phenotype and sequences, and this
information was available to the present study. Ensembl database
(www.ensembl.org/index.html) has been
used for the annotation, analysis and display of vertebrate genomes
in the present study. Differentially-expressed mRNA pathway
analysis may be used to infer their biological functions. The
Fisher P-value denotes the significance of the correlation of the
pathway to the condition. P<0.05 was considered to be the
threshold for significance.
A549 cells in culture and
β-galactosidase activity
A549 (American Type Culture Collection, Manassas,
VA, USA) cells are adenocarcinomic human alveolar basal epithelial
cells. The A549 cells used in the present study were cultured in
RPMI 1640 (Gibco; Thermo Fisher Scientific, Inc.) medium. All
medium was supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), in a humidified air
atmosphere of 5% CO2 at 37°C. Trypsin (0.25% with 1 mM EDTA;
Invitrogen; Thermo Fisher Scientific, Inc.) was used to harvest the
cells for further experiments.
A549 cells were seeded in 6-well plates, incubated
for 7 day at 37°C and the cell culture medium was replaced every
other day. TGF-β1 (Austral Biologicals, San Ramon, CA, USA) was
added every other day to the culture at a final concentration of 10
ng/ml (11). There were two groups
in the present study: A549 cells and A549 cells treated with TGF-β1
for 7 days.
X-gal (Wuhan Boster Biological Technology, Ltd.,
Wuhan, China) is a chromogenic substrate of β-galactosidase, which
produces a blue product under the catalysis by β-galactosidase.
Cells at a density of 1×106 cells per well in 6-well
plates were fixed in 3% formaldehyde at 4°C for 15 min.
Subsequently, the cells were then incubated with staining solution
(composed of 40 mM citric acid sodium phosphate, 1 mg/ml X-gal, 5
mM potassium ferricyanide, 150 mM NaCl and 2 mM MgCl2)
at 37°C for 12 h. β-galactosidase positive cells were enumerated by
counting over 400 cells in three in dependent fields.
Validation of differential expression
of lncRNA and mRNA with reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
In order to validate the sequencing results, the
remaining twenty samples were analyzed to exclude the individual
differences. A549 cells were used to validate the hypothesis that
AP003419.16 and gene ribosomal protein S6 kinase B-2 (RPS6KB2) may
be associated with aging and IPF.
Total RNA was prepared using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) and reverse-transcribed using an
RNA-to-cDNA kit (Takara Biotechnology Co., Ltd., Dalian, China).
Reverse transcription was performed to convert the extracted RNA
into cDNA with an oligo (dT) primer (Takara Biotechnology Co.,
Ltd.) and reverse transcriptase Master mix (Takara Biotechnology
Co., Ltd.) and incubated at 37°C for 15 min and 85°C for 5 sec.
RT-qPCR analysis is the gold standard for data verification. The
expression levels of lncRNAs and mRNAs were determined using SYBR
Green I-based qPCR (Takara Biotechnology Co., Ltd.). The reaction
conditions were 95°C for 10 min, followed by 40 cycles of 95°C for
10 sec and 60°C for 60 sec. β-actin was used as an endogenous
control. Primer Premier version 5.0 (Primer Biosoft International,
Palo Alto, CA, USA) was used to design the primers. The primers
were as follows: AP003419.16 forward, TTAATCTTCCACGGGAGCAG and
reverse, GCTGTGAGAGCAGCAGGAC; RPS6KB2 forward, TCCACTCCTGCCACCGC
and reverse, TCGCCACCTGCCTCACA; and β-actin forward,
GTGGCCGAGGACTTTGATTG and reverse, CCTGTAACAACGCATCTCATATT. The
relative expression levels of AP003419.16 and RPS6KB2 were
calculated using the comparative 2−∆∆Cq method (17).
Statistical analysis
All statistical data were analyzed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Results are expressed as
mean ± standard deviation. Student's t-test were used for
inter-group comparisons. P<0.05 was considered to indicate a
statistically significant difference.
Results
lncRNA and mRNA expression
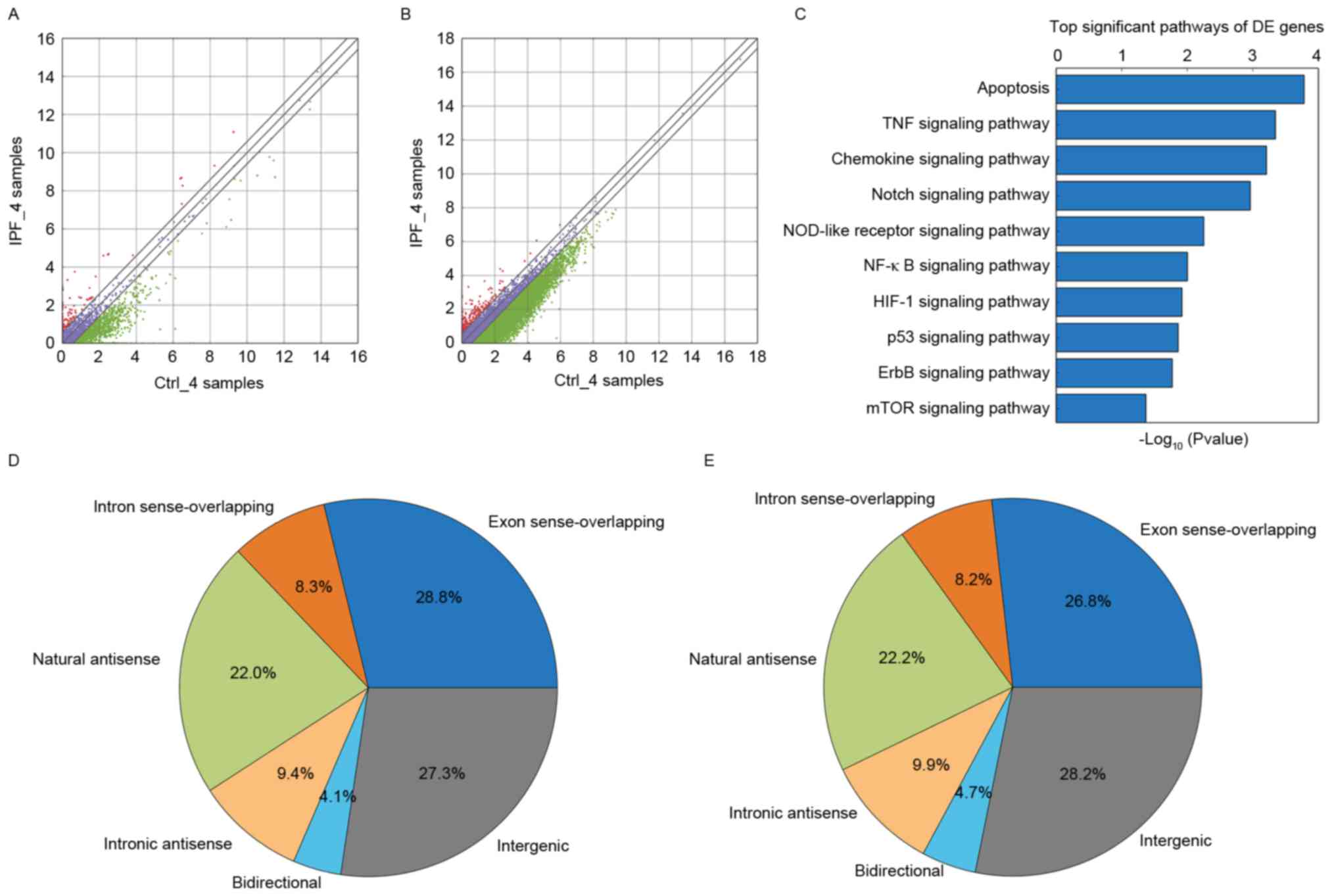
Compared with the control group, it was observed
that there were 440 lncRNAs that were upregulated and there were
1,376 lncRNAs that were downregulated in the IPF group. There were
1,816 differentially-expressed lncRNAs identified by this
screening, as presented in Fig.
1A. A total of 12 differentially expressed lncRNAs were
randomly selected (upregulated lncRNAs: AP003419.16, AB586698,
ADSS, AF080092, AF520792, AK126278; downregulated lncRNAs:
AB062083, ADAMTSL4-AS1, ADRM1, ADSL, AF063596, AJ276246) from the
aberrantly-expressed lncRNAs for further analysis. The present
study examined AP003419.16, which was upregulated in the
transcriptome sequencing results. Compared with the control group,
it was observed that there were 361 mRNAs that were upregulated and
there were 1,124 mRNAs that were downregulated in the IPF group.
There were 1,485 differentially-expressed mRNAs identified by this
screening in Fig. 1B.
Pathway analysis
GO and KEGG pathway analysis of
differentially-expressed mRNAs provided a measure of the critical
function. The most enriched GO term was associated with regulation
of chromosome segregation in the GO biological process analysis
(Table I). Significant pathways
(Table II; Fig. 1C) were identified for predicting
the target lncRNA genes according to the KEGG database. The mTOR
signaling pathway was observed to be associated with aging, in
addition to with the IPF. According to the KEGG database, the
Fisher P-value was 0.04 (P<0.05) and the enrichment score was
1.37. According to their position in the genome, which is
associated with protein-coding genes, lncRNAs may be divided into
six types: Exon sense-overlapping; intronic antisense; natural
antisense; intergenic; bidirectional; and intron sense-overlapping
(18,19). According to the result of
transcriptome sequencing and bioinformatics analysis in the the IPF
group and the control group, it was found that natural antisense
represented 22.0% of the total lncRNAs in the IPF group (Fig. 1D). Natural antisense accounted for
22.2% in the control group (Fig.
1E).
 | Table I.Target gene-associated GO
analysis. |
Table I.
Target gene-associated GO
analysis.
| A, Biological
processes |
|---|
|
|---|
| GO ID | Term | P-value | Enrichment
score | List total | Genes |
|---|
| GO:0000086 | G2/M transition of
mitotic cell cycle | 0.000579 | 3.23 | 317 | RAD51B, CCND1,
AURKB, PLK4, CIT, NEDD1, PLK1, HAUS2, CCNB2, LIN52, MELK |
| GO:0051983 | Regulation of
chromosome segregation | 0.00027 | 3.566 | 317 | BUB1, ESPL1,
KNSTRN, CDCA5, AURKB |
| GO:0007063 | Regulation of
sister chromatid cohesion | 0.001 | 2.95 | 317 | ESPL1, CDCA5,
BUB1 |
|
| B, Cellular
components |
|
| GO ID | Term | P-value | Enrichment
score | List total | Genes |
|
| GO:0051233 | Spindle
midzone | 0.0092 | 2.034 | 346 | CENPE, PLK1,
BUB1B |
| GO:0005763 | Mitochondrial small
ribosomal subunit | 0.005 | 2.284 | 346 | MRPS31, MRPS18C,
MRPS33 |
| GO:0005758 | Mitochondrial
intermembrane space | 0.009 | 2.01 | 346 | TIMM10, TIMM8B,
SLMO2, TRIAP1, ACN9 |
 | Table II.Target gene-associated pathways. |
Table II.
Target gene-associated pathways.
| Pathway ID | Definition | P-value | Selection
counts | Size | Enrichment
score | Genes |
|---|
| hsa04150 | mTOR signaling
pathway | 0.04 | 20 | 6,890 | 1.37 | AKT1, AKT2, EIF4E2,
IKBKB, IRS1, MAPK3, PDPK1, PIK3CD, PIK3R5, PRKCA, PRKCB, RPS6KA2,
RPS6KB2, RRAGA, STK11, STRADA, TNF, TSC2, ULK1, ULK3 |
| hsa04923 | Regulation of
lipolysis in adipocytes | 0.033 | 4 | 6,890 | 1.474 | PIK3R3, PLA2G16,
PRKG1, TSHR |
| hsa04012 | ErbB signaling
pathway | 0.02 | 29 | 6,890 | 1.77 | AKT1, AKT2, ARAF,
CAMK2G, CDKN1A, CDKN1B, CRK, CRKL, ELK1, GRB2, GSK3B, HRAS, MAP2K7,
MAPK3, MYC, NCK1, NCK2, PAK2, PIK3CD, PIK3R5, PLCG1, PRKCA, PRKCB,
RPS6KB2, SHC1, SRC, STAT5A, STAT5B, TGFA |
| hsa04621 | NOD-like receptor
signaling pathway | 0.005 | 22 | 6,890 | 2.252 | CARD6, CARD8,
CARD9, CASP5, CASP8, CHUK, CXCL1, IKBKB, MAPK13, MAPK14, MAPK3,
MEFV, NFKB1, NLRP1, NLRP3, NOD1, NOD2, PSTPIP1, PYCARD, RELA,
SUGT1, TNF |
| hsa05133 | Pertussis | 0.02 | 5 | 6,890 | 1.65 | C4BPA, C5, CASP7,
IL10, IL12A |
| hsa04330 | Notch signaling
pathway | 0.001 | 21 | 6,890 | 2.96 | APH1A, CREBBP,
CTBP2, DTX1, DTX3, DVL1, DVL2, DVL3, HDAC1, HDAC2, JAG2, KAT2A,
LFNG, MFNG, NCOR2, NCSTN, NOTCH1, NOTCH2, NUMB, PSENEN, RFNG |
Target lncRNA
RPS6KB2 is involved in the process of aging and
pulmonary fibrosis, given its activation by growth factors and its
regulation by the mTOR signaling pathway. RPS6KB2 is an important
molecule in the mTOR signaling pathway (Table II). It was demonstrated that the
adjacent gene mRNA to RPS6KB2 was AP003419.16, using bioinformatics
analysis. AP003419.16 is located on chromosome 11 at approximate
locations (chr11: 67195930-67202872), and the adjacent gene is
RPS6KB2. According to the result of our transcriptome sequencing
and bioinformatics analysis in the the IPF group and the control
group, it was found that the lncRNA AP003419.16 is one of the
differentially expressed lncRNAs, and the fold change between two
groups was 1.92. According to the Ensembl database, the gene ID of
AP003419.16 is ENSG00000255949, which is 470 bp in length and the
biotype is antisense.
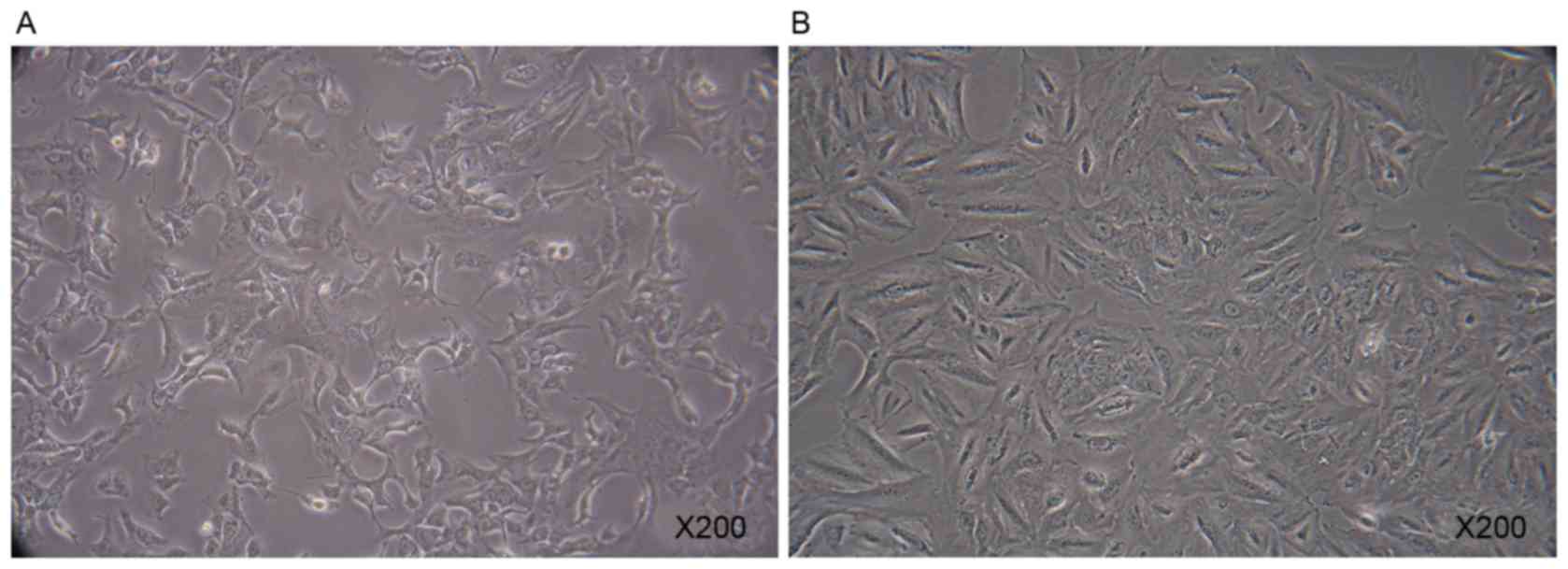
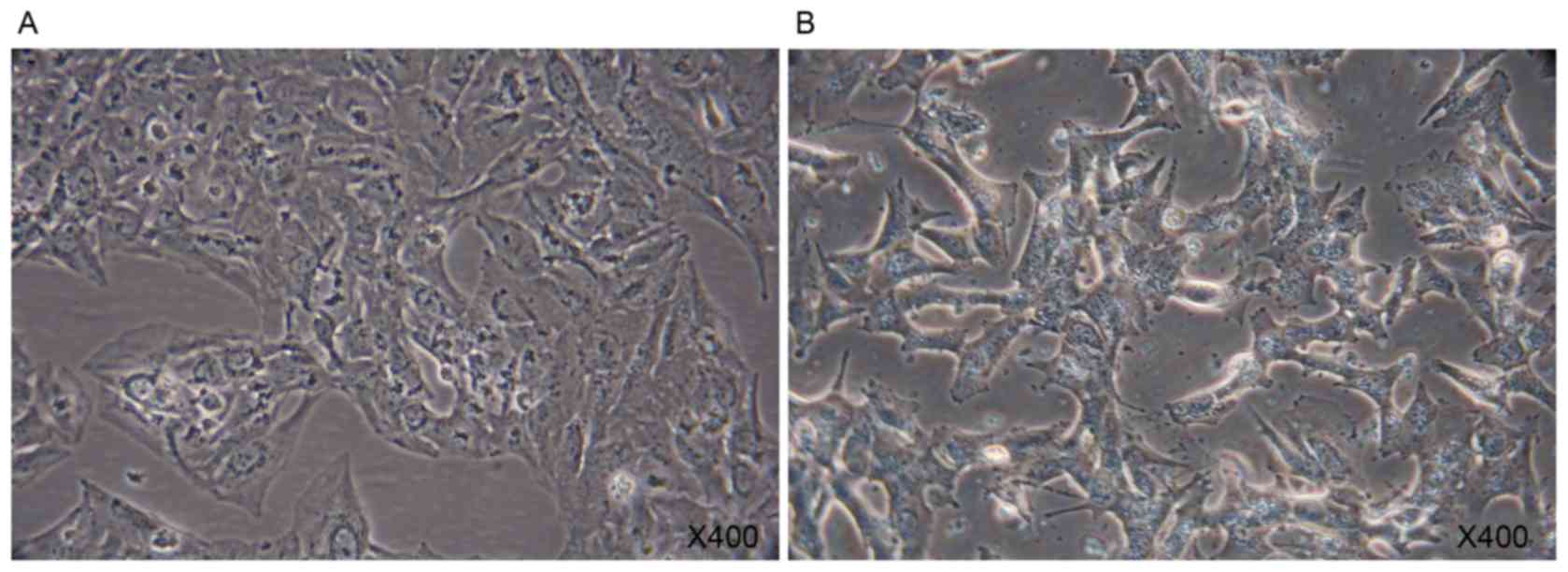
A549 cells
A549 cells were polygonal in the normal state
(Fig. 2A). When cultured in the
presence of 10 ng/ml TGF-β1 for 7 days, A549 cells exhibited rapid
morphological change (Fig. 2B). A
marked increase in the X-gal positive A549 cells was observed
following the addition of TGF-β1 for 7 days. TGF-β1 was able to
direct A549 cells to a replicative senescent state (Fig. 3).
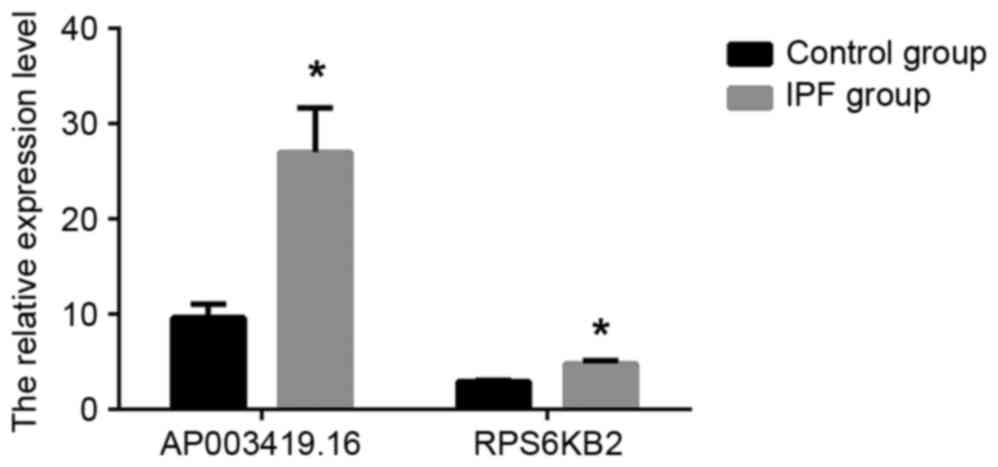
RT-qPCR validation
In order to validate the results independently and
to determine the role of lncRNAs in patients with IPF, RT-qPCR
analysis was used. AP003419.16 and RPS6KB2 were highly expressed in
the 20 patients with IPF and exhibited reduced expression in the
control group, according to the results of the RT-qPCR (Fig. 4; P<0.05). Similarly, AP003419.16
and RPS6KB2 were recorded at higher expression levels in
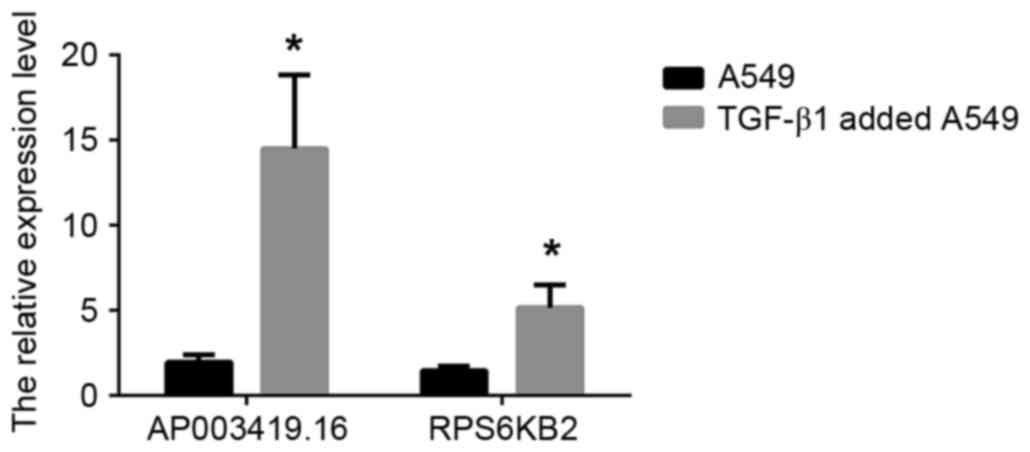
TGF-β1-treated A549 cells compared with the control group (Fig. 5; P<0.05).
Discussion
The clinical treatment for IPF predominantly relies
on ventilator maintenance, although outcomes are poor. According to
the American Thoracic Society treatment principles, the treatment
of choice for IPF is lung transplantation (20). However, due to high cost of
treatment and short survival time, lung transplantation represents
a psychological and economic burden on patients with IPF and their
families. Understanding the regulatory functions of lncRNAs in lung
fibroblasts may provide novel insights for IPF therapy.
LncRNAs are transcribed RNAs that are >200
nucleotides long. Previous studies have suggested that lncRNAs may
act as competitive endogenous RNAs and are involved in
physiological and pathological processes (21–23)
According to their position in the genome, which is associated with
protein-coding genes, lncRNAs may be divided into six types
(18,19): Exon sense-overlapping; intronic
antisense; natural antisense; intergenic; bidirectional; and intron
sense-overlapping. LncRNAs are able to regulate gene expression
through various mechanisms as follows (24): LncRNA transcription may interfere
with the expression of adjacent genes; or, it may act via the
transcription of protein-coding genes to form complementary
double-stranded regulatory gene expression. The expression levels
of specific lncRNAs may be altered during the progression of a
disease; therefore, they may be used as a marker for its diagnosis.
LncRNAs are able to regulate adjacent genes and thus contribute to
the incidence of associated diseases (25). Antisense lncRNAs may form stable
double-stranded structures with adjacent mRNAs and regulate their
expression to facilitate the development of disease (26). Antisense lncRNAs are able to
regulate the inhibitory effect of microRNAs on adjacent mRNAs in
the development of disease (27).
Antisense lncRNAs are involved in the regulation of
aging-associated diseases (28,29)
and aging-associated pathways (30). A previous study observed that
lncRNAs may regulate pulmonary fibrosis during pulmonary epithelial
cell transformation in a pulmonary fibrosis animal model (5). It has additionally been reported that
the silencing of lncRNAs may inhibit cell proliferation and
α-smooth muscle actin expression, whilst increasing collagen
expression, in pulmonary fibroblasts (31). These previous findings suggested
that lncRNAs may be involved in the occurrence of aging and
IPF.
Rapamycin, an anti-aging drug, is effective in
extending mammalian life and in the treatment of diseases,
including premature aging (32).
In a bleomycin-established IPF animal model, lung epithelial cells
were protected by mTOR signaling pathway inhibitors, which may have
therapeutic implications in pulmonary fibrosis (33,34).
mTOR has been previously reported to be associated with collagen
production in IPF and promote the pathogenesis of IPF (35). mTOR expression has been observed to
be associated with the degree of pulmonary fibrosis in IPF
(36).
According to the KEGG pathway analysis performed by
the present study, RPS6KB2 is an important gene in the mTOR
signaling pathway. According to the NCBI database, the protein
encoded by the mRNA is a ribosomal S6 kinase (S6K), which contains
a kinase catalytic domain and may phosphorylate the S6 ribosomal
protein. RPS6KB2 primarily regulates cell senescence and the cell
cycle through phosphorylation of S6K. S6K is involved in regulating
the physiological processes of DNA replication and translation.
RPS6KB2 is associated with aging and pulmonary fibrosis. The S6K
pathway is involved in the progression of pulmonary fibrosis
(37).
High-throughput sequencing and bioinformatics
analysis performed in the present study identified AP003419.16 to
be adjacent to the protein-coding gene RPS6KB2. RPS6KB2 is believed
to be involved in the process of aging and pulmonary fibrosis, due
to its activation by growth factors and regulation by the mTOR
signaling pathway. The expression levels of AP003419.16 and RPS6KB2
were higher in the 20 patients with IPF compared with the control
group, as demonstrated by RT-qPCR analysis.
TGF-β1 has an important role in the pathogenesis of
IPF, and is considered to be an important profibrotic factor
(38,39). The results of the present study
demonstrated a marked increase in the number of X-gal-positive A549
cells following treatment with TGF-β1 for 7 days. TGF-β1 was
additionally demonstrated to direct A549 cells to a replicative
senescent state. AP003419.16 and RPS6KB2 were highly expressed in
TGF-β1-treated A549 cells compared with the control group.
Therefore, IPF may be associated with senescence at the cellular
level.
The present study demonstrated that the expression
of AP003419.16 increased significantly in patients with IPF, while
its adjacent gene RPS6KB2 increased simultaneously. Analysis of the
expression of AP003419.16 may predict an increased risk of
aging-associated IPF. The findings of the present study elucidated
a molecular hypothesis of IPF progression in the aging process and
provided novel molecular targets for clinical treatment of IPF.
Glossary
Abbreviations
Abbreviations:
|
IPF
|
idiopathic pulmonary fibrosis
|
|
lncRNA
|
long noncoding RNA
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
RPS6KB2
|
ribosomal protein S6 kinase B-2
|
|
S6K
|
S6 kinase
|
|
TGF-β1
|
transforming growth factor-β1
|
References
|
1
|
Cookson WO and Moffatt MF: Bedside to gene
and back in idiopathic pulmonary fibrosis. N Engl J Med.
368:2228–2230. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fernandez IE and Eickelberg O: New
cellular and molecular mechanisms of lung injury and fibrosis in
idiopathic pulmonary fibrosis. Lancet. 380:680–688. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Puglisi S, Torrisi S, Giuliano R, Vindigni
V and Vancheri C: What we know about the pathogenesis of idiopathic
pulmonary fibrosis. Semin Respir Crit Care Med. 37:358–367. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
King TE, Pardo A and Selman M: Idiopathic
pulmonary fibrosis. Lancet. 378:1949–1961. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun H, Chen J, Qian W, Kang J, Wang J,
Jiang L, Qiao L, Chen W and Zhang J: Integrated long non-coding RNA
analyses identify novel regulators of epithelial-mesenchymal
transition in the mouse model of pulmonary fibrosis. J Cell Mol
Med. 20:1234–1246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Puvvula PK, Desetty RD, Pineau P, Marchio
A, Moon A, Dejean A and Bischof O: Long noncoding RNA PANDA and
scaffold-attachment-factor SAFA control senescence entry and exit.
Nat Commun. 5:53232014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Szafranski K, Abraham KJ and Mekhail K:
Non-coding RNA in neural function, disease and aging. Front Genet.
6:872015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Magnuson B, Ekim B and Fingar DC:
Regulation and function of ribosomal protein S6 kinase (S6K) within
mTOR signalling networks. Biochem J. 441:1–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Selman C, Tullet JM, Wieser D, Irvine E,
Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D,
Ramadani F, et al: Ribosomal protein S6 kinase 1 signaling
regulates mammalian life span. Science. 326:140–144. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharp ZD: Aging and TOR: Interwoven in the
fabric of life. Cell Mol Life Sci. 68:587–597. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Katakura Y, Nakata E, Miura T and
Shirahata S: Transforming growth factor β triggers two
independent-senescence programs in cancer cells. Biochem Biophys
Res Commun. 255:110–115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qu Y, Zhang L, Kang Z, Jiang W and Lv C:
Ponatinib ameliorates pulmonary fibrosis by suppressing
TGF-β1/Smad3 pathway. Pulm Pharmacol Ther. 34:1–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Raghu G, Rochwerg B, Zhang Y, Garcia CA,
Azuma A, Behr J, Brozek JL, Collard HR, Cunningham W, Homma S, et
al: An official ATS/ERS/JRS/ALAT clinical practice guideline:
Treatment of idiopathic pulmonary fibrosis. Am J Respir Crit Care
Med. 192:e3–e19. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet J.
17:10–12. 2011. View Article : Google Scholar
|
|
15
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Bio. 14:R362013. View Article : Google Scholar
|
|
16
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. Nature
genetics. 25:25–29. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
De Oliveira NC, Julliard W, Osaki S,
Maloney JD, Cornwell RD, Sonetti DA and Meyer KC: Lung
transplantation for high-risk patients with idiopathic pulmonary
fibrosis. Sarcoidosis Vasc Diffuse Lung Dis. 33:235–241.
2016.PubMed/NCBI
|
|
21
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng EC and Lin H: Repressing the
repressor: A lincRNA as a MicroRNA sponge in embryonic stem cell
self-renewal. Dev Cell. 25:1–2. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song X, Cao G, Jing L, Lin S, Wang X,
Zhang J, Wang M, Liu W and Lv C: Analysing the relationship between
lncRNA and protein-coding gene and the role of lncRNA as ceRNA in
pulmonary fibrosis. J Cell Mol Med. 18:991–1003. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee DY, Moon J, Lee S-T, Jung KH, Park DK,
Yoo JS, Sunwoo JS, Byun JI, Shin JW, Jeon D, et al: Distinct
expression of long non-coding RNAs in an Alzheimer's disease model.
J Alzheimers Dis. 45:837–849. 2015.PubMed/NCBI
|
|
26
|
Faghihi MA, Modarresi F, Khalil AM, Wood
DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ and
Wahlestedt C: Expression of a noncoding RNA is elevated in
Alzheimer's disease and drives rapid feed-forward regulation of
β-secretase. Nat Med. 14:723–730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Faghihi MA, Zhang M, Huang J, Modarresi F,
Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G III and
Wahlestedt C: Evidence for natural antisense transcript-mediated
inhibition of microRNA function. Genome Biol. 11:R562010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Carrieri C, Cimatti L, Biagioli M, Beugnet
A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C,
et al: Long non-coding antisense RNA controls Uchl1 translation
through an embedded SINEB2 repeat. Nature. 491:454–457. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Wu Z, Fu X and Han W: lncRNAs:
Insights into their function and mechanics in underlying disorders.
Mutat Res Rev Mutat Res. 762:1–21. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang D, Lian T, Tu J, Gaur U, Mao X, Fan
X, Li D, Li Y and Yang M: LncRNA mediated regulation of aging
pathways in Drosophila melanogaster during dietary restriction.
Aging (Albany NY). 8:2182–2203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang C, Yang Y and Liu L: Interaction of
long noncoding RNAs and microRNAs in the pathogenesis of idiopathic
pulmonary fibrosis. Physiol Genomics. 47:463–469. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mendelsohn AR and Larrick JW: Rapamycin as
an antiaging therapeutic? Targeting mammalian target of rapamycin
to treat Hutchinson-Gilford Progeria and neurodegenerative
diseases. Rejuvenation Res. 14:437–441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yoshizaki A, Yanaba K, Yoshizaki A, Iwata
Y, Komura K, Ogawa F, Takenaka M, Shimizu K, Asano Y, Hasegawa M,
et al: Treatment with rapamycin prevents fibrosis in tight-skin and
bleomycin-induced mouse models of systemic sclerosis. Arthritis
Rheum. 62:2476–2487. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang W, Wei K, Ho L, Berry GJ, Jacobs SS,
Chang CH and Rosen GD: A critical role for the mTORC2 pathway in
lung fibrosis. PLoS One. 9:e1061552014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Nho RS and Hergert P: IPF fibroblasts are
desensitized to type I collagen matrix-induced cell death by
suppressing low autophagy via aberrant Akt/mTOR kinases. PLoS One.
9:e946162014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park JS, Park HJ, Park YS, Lee SM, Yim JJ,
Yoo CG, Han SK and Kim YW: Clinical significance of mTOR, ZEB1,
ROCK1 expression in lung tissues of pulmonary fibrosis patients.
BMC Pulm Med. 14:1682014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Madala SK, Thomas G, Edukulla R, Davidson
C, Schmidt S, Schehr A and Hardie WD: p70 ribosomal S6 kinase
regulates subpleural fibrosis following transforming growth
factor-α expression in the lung. Am J Physiol Lung Cell Mol
Physiol. 310:L175–L186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li FZ, Cai PC, Song LJ, Zhou LL, Zhang Q,
Rao SS, Xia Y, Xiang F, Xin JB, Greer PA, et al: Crosstalk between
calpain activation and TGF-β1 augments collagen-I synthesis in
pulmonary fibrosis. Biochim Biophys Acta. 1852:1796–1804. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Strieter RM and Mehrad B: New mechanisms
of pulmonary fibrosis. Chest. 136:1364–1370. 2009. View Article : Google Scholar : PubMed/NCBI
|