Introduction
DNA damage occurs constantly due to intrinsic DNA
metabolic activities and extrinsic agents, including irradiation
and various DNA-damaging chemicals. A double-strand break (DSB) is
one of the most serious lesions as it can lead to genome
rearrangements if not properly sensed and repaired (1). To respond to this challenge, cells
elicit a coordinated DNA-damage response, which includes DNA
repair, checkpoint activation, and consequent cell cycle arrest
and/or apoptosis to maintain genomic integrity (2,3).
The involvement of ring finger protein 8 (RNF8), an
FHA domain and RING finger motif-containing protein, is important
in the cellular response to DNA damage. RNF8 is rapidly accumulated
at DSBs via the interaction of its N-terminal FHA domain with the
phosphorylated mediator of DNA-damage checkpoint 1 (MDC1), which
binds to the phosphorylated H2AX (γ-H2AX) accumulating at the DNA
lesions, where it functions as an E3 ligase to ubiquitinate H2A,
H2AX and other unknown proteins (4–8). The
ubiquitination of H2A and H2AX recruits RNF168, another E3 ligase,
to sites of DSBs to amplify the RNF8-dependent H2A ubiquitination
and promote the formation of Lys63-linked ubiquitin chains for the
recruitment of p53-binding protein 1 (53BP1), breast cancer 1
(BRCA1) and other damage repair proteins (9). In this manner, RNF8 serves as a
molecular link between the protein phosphorylation and protein
ubiquitination pathways, which are essential in the DNA damage
response.
Small ubiquitin-like modifier (SUMO) modification is
another post-translational modification known to regulate the DNA
damage response. In mammals, three SUMO paralogues, SUMO1, SUMO2
and SUMO3, are expressed. SUMO2 and SUMO3 differ by three
N-terminal residues, whereas SUMO1 is 45% identical to either SUMO2
or SUMO3. All three SUMO species can be covalently attached to
target proteins by forming an isopeptide bond between the target
lysine and the activated SUMO c-terminal glycine residue by a
mechanism similar to that of ubiquitination. SUMO modification has
been shown to be required for complete accumulation of DNA
damage-repair proteins, including RNF8, RNF168, BRCA1 and 53BP1, at
sites of DNA lesions (10).
Furthermore, sumoylation of DSB response-associated ubiquitin
ligases, for example BRCA1, enhances ubiquitin ligase activity
(11).
In addition to modification by SUMO, target proteins
can bind noncovalently to SUMO via its short hydrophobic region,
known as SUMO interaction motif (SIM), to alter its cellular
localization and/or activity (12–14).
In the majority of cases, however, SUMO-mediated protein-protein
interactions during the DNA damage response and the consequent
effect on genomic integrity remain to be elucidated.
In the present study, a yeast two-hybrid assay was
performed to identify RNF8-binding proteins, which identified
SUMO2/3 as its binding partner. The data presented showed that RNF8
bound directly and noncovalently to SUMO2/3, but not to SUMO1. The
accumulation of RNF8 and SUMO2/3 at sites of DNA lesions
contributed to DSB repair following ionizing radiation.
Materials and methods
Plasmids
The full-length human RNF8 gene was obtained by
polymerase chain reaction (PCR) from the p-RNF8 plasmid (provided
by Professor Tiebang Kang of Sun Yat-sen University Cancer Center,
Guangzhou, China) and subcloned into the pBridge vector (Clontech
Laboratories, Inc., Mountainview, CA, USA) carrying the Gal4
DNA-binding domain, pcDNA3.0-Flag vector and pEGFP-N1 vector
(Clontech Laboratories, Inc.). Human SUMO1, SUMO2 and SUMO3 were
PCR-amplified from the HeLa cDNA library (Clontech Laboratories,
Inc.) and cloned into the pGEX4T-1, pDsRed2-N1, pGAD-T7 or pXJ40-HA
vector. The SUMO3
(G91G92→A91A92)
mutation was generated by PCR with mutation-specific primers and
cloned into the pXJ40-HA vector (provided by Professor Xuemin Zhang
of National Center of Biomedical Analysis, Beijing, China). The
primer sequences used for PCR amplification were as follows: SUMO1
forward, 5′-ATGTCTGACCAGGAGGCAAAACCTTC-3 and reverse,
5′-CTAAACTGTTGAATGACCCCCCGTT-3; SUMO2 forward,
5′-ATGGCCGACGAAAAGCCCAAG-3 and reverse,
5′-TCAGTAGACACCTCCCGTCTGCTG-3; SUMO3 forward,
5′-ATGTCCGAGGAGAAGCCCAAG-3 and reverse,
5′-CTAGAAACTGTGCCCTGCCAG-3′. The cycle conditions included an
initial denaturation step at 94°C for 5 min followed by 35 cycles
of 94°C for 30 sec, 58°C for 30 sec and 72°C for 1 min and then a
final extension at 72°C for 5 min.
Antibodies and reagents
The mouse anti-Flag antibody (cat. no. F3165) was
purchased from Sigma-Aldrich; Merck Millipore (Darmstadt, Germany).
Mouse anti-hemagglutinin (HA; cat. no. sc-7392) antibody was
provided by Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA)
and goat anti-HA antibody was from GenScript (Nanjing, China).
Rabbit anti-phospho-H2A.X (cat.no. 9718) was from Cell Signaling
Technology, Inc. (Danvers, MA, USA). Fluorescein isothiocyanate
(FITC)-conjugated anti-mouse IgG (cat. no. 715-095-151) and
tetramethyl rhodamine isocyanate (TRITC)-conjugated anti-goat IgG
(cat. no. 305-025-003) were from Jackson ImmunoResearch
Laboratories, Inc. (West Grove, PA, USA). Glutathione-Sepharose 4B
beads were purchased from GE Healthcare Life Sciences (Uppsala,
Sweden) and Protein G beads were from Santa Cruz Biotechnology,
Inc. (Santa Cruz, CA, USA). The small interfering (si)RNA
oligonucleotides against RNF8, SUMO1 and SUMO2/3 were chemically
synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
siRNA against RNF8 was 5′AGAAUGAGCUCCAAUGUAUU3′; siRNA against
SUMO1 was 5′GGACAGGAUAGCAGUGAGA3′; and siRNA against SUMO2/3 was
5′CAAUGAGGCAGAUCAGAUU3′.
Yeast two-hybrid screening
Full length cDNA from human RNF8 was cloned in frame
with the Gal4 DNA-binding domain in pBridge. pBD-RNF8 was
co-transfected into the Saccharomyces cerevisiae strain
CG1945 together with a human fetal brain cDNA library (Clontech
Laboratories, Inc., CA, USA) using the lithium acetate.
Transformants(~107) were selected on 100-mm Sabouraud
Dextrose/-Trp-Leu-His dropout plates (30°C; 4–5 days) for the
primary screening and were analyzed using a LacZ assay for the
second screening. Following rescue, the potential positive plasmids
were isolated and re-transformed into the CG1945 yeast strain
containing the RNF8 bait plasmid. Only clones, which were confirmed
to be positive by at least two independent tests were selected for
specific interactions and sequenced.
Cell culture and transfection
HeLa human epithelial carcinoma cells and U2OS human
osteosarcoma cells were obtained from Peking Union Medical College
(Beijing, China) and grown in DMEM supplemented with 8% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), 100 U/ml penicillin and 100 µ/ml streptomycin, at 37°C in a
humidified atmosphere containing 5% CO2 and 95% air.
Cell transfection was performed using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol.
Ionizing radiation
The U2OS cells were plated into a 6-well plate at a
density of 6×105 cells/well and irradiated using a
Biological X-ray irradiator (RS 2000; Rad Source Technologies,
Inc., Suwanee, GA, USA) at 160 kV and 25 mA, with a dose of 10 Gy
and a dose rate of 2 Gy/min.
Western blot analysis
Total proteins were extracted using cell lysis
buffer (Cell Signaling Technology, Inc.). Whole cell lysates were
quantified using a BCA Protein assay kit (Beyotime Institute of
Biotechnology, Shanghai, China) and 20 µg of protein was loaded per
lane to separate on 10 or 12% sodium dodecyl sulfate polyacrylamide
gels. Subsequently, the separated proteins were transferred onto
polyvinylidene difluoride membranes. The membrane was then blocked
with 5% non-fat milk in Tris-buffered saline (TBS) for 2 h at room
temperature and incubated overnight at 4°C anti-Flag (1:1,000) and
anti-HA (1:1,000) antibodies. After washing three times for 10 min
each with TBST, the membrane was incubated for 2 h at room
temperature with a horseradish peroxidase-conjugated secondary
antibody (1:5,000). The bands were visualized using an ECL
detection kit (Applygen Technologies, Inc., Beijing, China).
Glutathione S-transferase (GST)
pull-down assay
Recombinant GST and GST-fused human SUMO2 or SUMO3
proteins were purified from Escherichia coli BL21 (DE3) (New
England BioLabs, Inc., Ipswich, MA, USA) cells using
glutathione-sepharose 4B beads. The cell lysates from the HeLa
cells transiently transfected with Flag-RNF8, Flag-RNF8 (ΔFHA) and
Flag-RNF8 (ΔRING) were incubated with purified GST, GST-SUMO2 and
GST-SUMO3 at 4°C overnight. The beads were then washed four times
with lysis buffer. The bound proteins were eluted by boiling in 2X
SDS loading buffer and analyzed using western blot analysis with
the anti-Flag antibody.
Immunoprecipitation
The HeLa cells were seeded into 10 cm diameter
dishes (1×106 cells/dish) and co-transfected with
Flag-RNF8 and HA-SUMO1, HA-SUMO2, HA-SUMO3 or HA-SUMO3 M
(G91G92→A91A92). At 36
h post-transfection, the cells were harvested and lysed using cell
lysis buffer (Cell Signaling Technology, Inc.). The cell lysates
were centrifuged at 12,000 × g for 10 min at 4°C and the
supernatants were incubated with anti-HA antibody at 4°C for 2 h.
Protein G agarose beads were then added, and the mixture was
incubated at 4°C overnight. The beads were washed three times with
cell lysis buffer, boiled in 2X SDS loading buffer for 6 min, and
the precipitated proteins were subjected to western blot
analysis.
Immunostaining procedure
To observe the cellular co-localization of RNF8 and
SUMO2/3, the U2OS cells were co-transfected with RNF8-GFP and
SUMO2-Red or SUMO3-Red for 28 h. The cells were then washed with
PBS and incubated with 4% paraformaldehyde for 20 min at room
temperature. Following washing with PBS, the coverslips were
mounted onto glass slides and observed under a Leica confocal
microscope (Leica Microsystems GmbH, Wetzlar, Germany).
To detect whether RNF8 and SUMO2/3 accumulated at
DNA-damage sites, the U2OS cells were seeded at a density of
6×105 cells/well on cover slips in a 6-well plated and
transfected with Flag-RNF8 and HA-SUMO2 for 28 h. The cells were
irradiated with a 10 Gy dose, followed by recovery for 6 h. The
cells were then washed three times with PBS, fixed with 4%
paraformaldehyde and permeabilized with 0.2% Triton X-100 for 4 min
at room temperature. The samples were blocked with 5% goat serum
(cat. no. 5425, Cell Signaling Technology, Inc.) followed by
incubation with anti-Flag, anti-HA and anti-phospho-H2AX antibodies
at 37°C for 45 min. Following incubation with TRITC- and
FITC-conjugated secondary antibodies, the cells were visualized
under a Leica confocal microscope.
Survival assay
The U2OS cells were seeded in 12-well plate at a
density of 2×105 cells/well and transfected with either
control siRNA or siRNA against RNF8, SUMO1 or SUMO2/3 using
Lipofectamine 2000 reagent according to the manufacturer's
instructions. At 48 h post-transfection, the cells were exposed to
different dose of radiation (1–3 Gy) and left to grow for 10 days.
The cells were then fixed and stained with crystal violet, and the
numbers of colonies were counted using a light microscope. The
experiment was performed three times.
Statistical analysis
The samples were assayed in triplicate. Data are
presented as the mean ± standard deviation. The results shown are
representative of at least three experiments. Differences were
determined using one-way analysis of variance followed by Student's
t-test. P<0.05 was considered to indicate a statistically
significant difference.
Results
RNF8 binds directly and noncovalently
to SUMO2/3
To better understand the roles of RNF8 in DNA damage
signaling, the present study performed yeast-two hybrid assays to
identify RNF8-binding proteins. Among the proteins recognized by
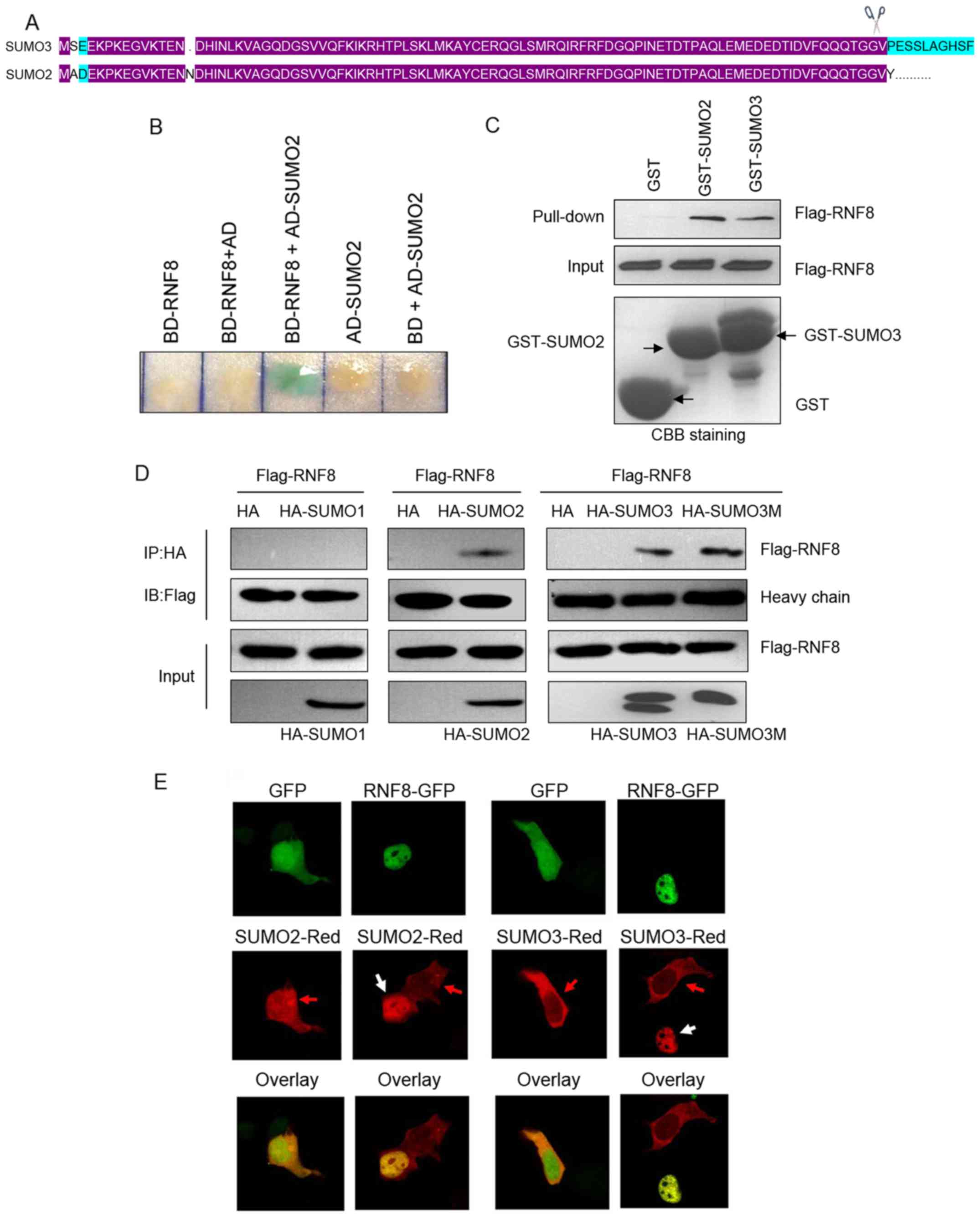
RNF8, SUMO2/3 was of particular interested (Fig. 1A and B). The interaction between
RNF8 and SUMO2/3 was further confirmed in the in vitro
GST-pull down assay (Fig. 1C).
Mammalian cells express three paralogs: SUMO2 and SUMO3, which are
96% identical, and SUMO1, which is 45% identical to SUMO2/3.
Therefore, the present study investigated whether RNF8 binds to
SUMO1. Using an in vivo co-immunoprecipitation assay in HeLa
cells overexpressing Flag-tagged RNF8 and HA-tagged SUMO1, SUMO2 or
SUMO3, it was found that RNF8 bound efficiently to SUMO2 and SUMO3,
but almost no binding was observed to SUMO1 (Fig. 1D). It is known that SUMO can
interact covalently and/or noncovalently with target proteins. To
distinguish whether the association of RNF8 with SUMO2/3 was
covalent or noncovalent, the present study constructed a
nonconjugatable form of SUMO3
(G91G92→A91A92) and
contransfected it with Flag-RNF8 into HeLa cells. As shown in
Fig. 1D, mutation of SUMO3 did not
affect binding to RNF8, indicating that RNF8 binds noncovalently to
SUMO3. Finally, a Leica confocal microscope was used to detect the
cellular co-localization of RNF8 with SUMO2/3 in HeLa cells. As
shown in Fig. 1E, SUMO2-Red and
SUMO3-Red co-localized specifically with RNF8-GFP in the nucleus,
but not with GFP. Together, these data suggested that RNF8 was able
to bind directly and noncovalently to SUMO2/3.
Mapping the SUMO2/3 binding domain in
RNF8
In order to map the SUMO2/3 binding domain in RNF8
in the present study, a series of RNF8 truncation mutants were
generated (Fig. 2A). The results
from one-to-one yeast two-hybrid assays performed with RNF8
deletions and SUMO2 indicated that the region between amino acids
17 and 120 of RNF8, which includes the FHA domain, is important for
binding to SUMO2 (Fig. 2B). This
result was further confirmed in the in vitro GST-pull down
assay. As shown in Fig. 2C,
deletion of the RING domain bound SUMO2 and wild-type RNF8, whereas
truncation of the FHA domain did not bind to GST-SUMO2. Thus,
deletion analysis revealed that the RNF8 FHA domain is required for
binding to SUMO2/3. The FHA domain-mediated interaction between
RNF8 and SUMO2/3 was also confirmed using a co-immunoprecipitation
assay with FHA domain deletion mutants of RNF8. As shown in
Fig. 2D, the FHA domain was found
to bind efficiently to SUMO2. Furthermore, SUMO2 co-localized
specifically with RNF8 in the nucleus, but not with the RNF8 FHA
deletion mutant (Fig. 2E).
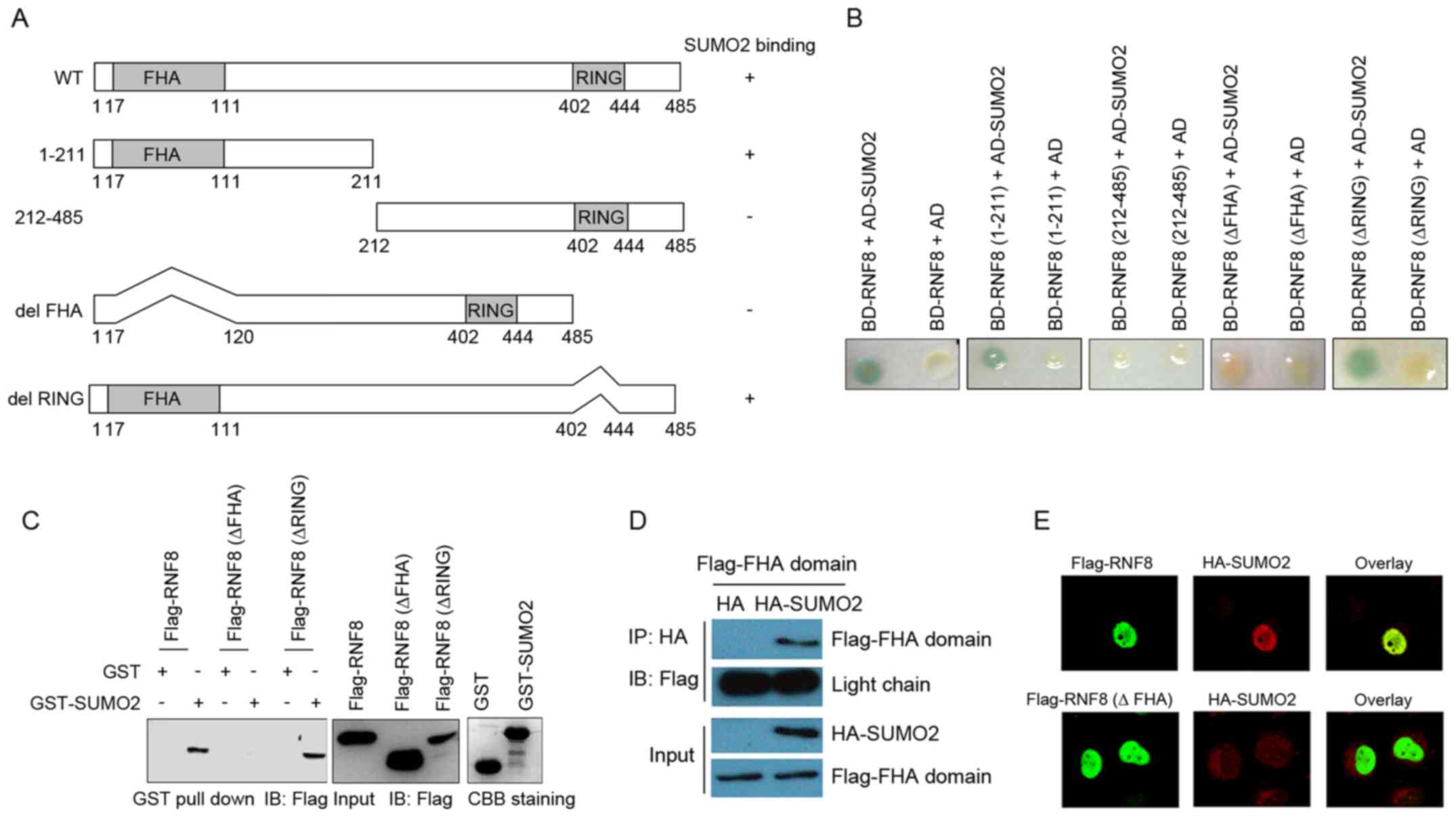 | Figure 2.Mapping the SUMO2/3 interaction domain
in RNF8. (A) Schematic representation of the deletion mutants of
RNF8. The binding of SUMO2 by RNF8 deletion mutants is indicated on
the right based on the one-to-one yeast two-hybrid assays. (B)
Yeast strain CG1945 was co-transformed with the bait and prey
plasmids as indicated. A single yeast colony was subjected to a
liquid β-gal assay, with the presence of a blue product indicating
a positive interaction. (C) FHA domain of RNF8 was required for
binding to SUMO2 based on the in vitro GST pull-down assay.
Lysates of HeLa cells transfected with Flag-RNF8, Flag-RNF8 (ΔFHA)
and Flag-RNF8 (ΔRING) were incubated with GST or GST-SUMO2 at 4°C
overnight. The bound proteins were analyzed using western blot
analysis with an anti-Flag antibody. (D) Co-immunoprecipitation
showing the interaction of HA-SUMO2 and Flag-FHA domain of RNF8 in
HeLa cells. (E) Immunofluorescence assay showing nuclear
co-localization of SUMO2 and WT RNF8, but not with the RNF8 FHA
deletion mutant. The cells were observed under a Leica confocal
microscope. Magnification, ×630. GST, glutathione S-transferase;
RNF8, ring finger protein 8; SUMO, small ubiquitin-like modifier;
CBB, coomassie brilliant blue; HA, hemaglutinin; WT, wild-type. |
Interaction of RNF8 with SUMO2/3
increases cellular resistance to genotoxic stress
Upon DNA damage, checkpoint proteins localize to
chromatin structures at the vicinity of DNA breaks. Thus, focus
formation following DNA damage can be observed using
immunostaining. RNF8 is well known to accumulate at the chromatin
following DNA damage. The present study also observed RNF8 focus
formation following ionizing radiation and theses foci co-localized
with the DNA damage marker, γ-H2AX. Furthermore, SUMO2 foci were
readily visualized following DNA damage and the SUMO2 foci
overlapped with those of RNF8 (Fig.
3A). Thus, it appeared that the binding of RNF8 to SUMO2/3 was
involved in the RNF8 response to genotoxic stress. Therefore, the
present study examined whether the binding of RNF8 to SUMO2/3 was
required for cell survival following DNA damage. As shown in
Fig. 3B, consistent with the
expected results, the transient knockdown of RNF8, SUMO1 or SUMO2/3
resulted in a significant increase in cellular radiation
sensitivity. Of note, it was found that the knockdown of RNF8
together with SUMO2/3 had an additive effect on increased cellular
radiation sensitivity, beyond the effect of either RNF8 or SUMO2/3
alone. However, this was not observed in the cells co-transfected
with siRNAs targeting RNF8 and SUMO1. Together, these results
suggested that binding of RNF8 to SUMO2/3 promoted DNA repair.
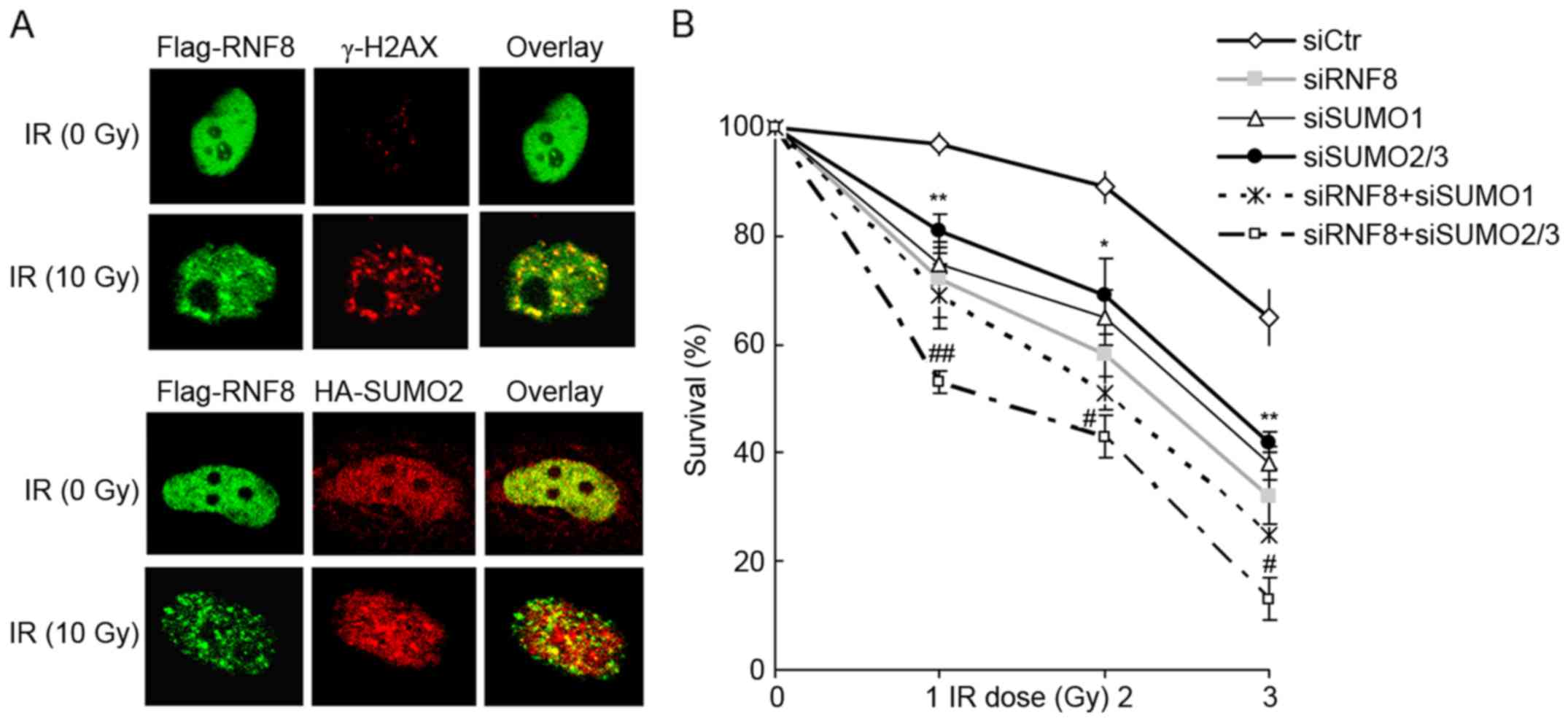 | Figure 3.RNF8 and SUMO2/3 are localized to
sites of DNA lesions and required for cell survival following DNA
damage. (A) Co-localization of RNF8 and SUMO2/3 at sites of DNA
lesions in U2OS cells. U2OS cells were transfected with Flag-RNF8
or co-transfected with Flag-RNF8 and HA-SUMO2 for 28 h. The cells
were irradiated with 10 Gy or left untreated and immunostained with
anti-Flag, anti-γ-H2AX and anti-HA antibodies. The cells were
observed under a Leica confocal microscope. Magnification, ×630.
(B) Survival of siRNA-transfected U2OS cells exposed to
irradiation. U2OS cells were transfected with siRNAs for 48 h,
followed by exposure to different doses of irradiation and left to
grow for 10 days. The cells were then fixed and stained with
crystal violet, and the number of colonies was counted. The
experiment was performed in triplicate and data are presented as
the mean ± standard deviation of three independent experiments.
**P<0.01 and *P<0.05, vs. siCtr group; ##P<0.01
and #P<0.05, vs. siRNF8 group. RNF8, ring finger
protein 8; SUMO, small ubiquitin-like modifier; si/siRNA, small
interfering RNA; Ctr, control; HA, hemaglutinin; IR
irradiation. |
Discussion
Although the sumoylation pathway has been well
characterized to have a significant role in the mammalian DNA
damage response, the functional properties of SUMO paralogues,
which mediate noncovalent binding to targeting proteins in the
cellular response to genotoxic stress, remain to be fully
elucidated. In the present study, a direct, noncovalent interaction
was identified between RNF8 and SUMO2/3. RNF8 and SUMO2/3
co-localize at the sites of DNA damage following genotoxic stress.
The direct binding of RNF8 to SUMO2/3 is required for cell survival
upon DNA damage. The results of the present study suggested that
RNF8 mediated the cooperation between the ubiquitin and SUMO
systems in the cellular response to DNA damage.
Sumoylation has been well characterized as a
pervasive mechanism for controlling the cellular response to DSBs.
A direct link between sumoylation and DSB responses in mammalian
cells was identified through observation that SUMO1, SUMO2/3 and
the E3 ligase enzymes, PIAS4 and PIAS1, accumulated at DSB sites
(10). In addition, in the absence
of PIAS proteins, DNA damage response factors, including RNF168,
53BP1, BRCA1 and RAP80, do not co-localize to the DSB, and DNA
repair is impeded (10,11). In this context, SUMO often provides
a binding platform for attracting coregulatory proteins, which
contain a SIM to a sumoylated protein. The SIM-mediated
protein-protein interaction drives key processes in the DNA damage
repair response by the regulation of ubiquitin ligase activity and
the promotion of SUMO-targeted E3 ubiquitin ligase
(STUbL)-dependent ubiquitination of SUMO-conjugated target protein.
The sumoylation of BRCA1 increases the activity of BRCA1 ubiquitin
ligase, possibly via a productive association with SIM-containing
target proteins (11). The STUbL
RNF4 is recruited to DNA lesions via binding of its N-terminal SIMs
to sumoylated DNA-damage response proteins, including 53BP1, MDC1
and replication protein A, where it mediates the accumulation of
ubiquitin adducts, which may be coupled with target degradation
(13,15). In the present study, data revealed
that the E3 ubiquitin ligase, RNF8, bound directly to SUMO2/3 and
that this interaction was important for DSB repair. Whether the
binding of RNF8 to SUMO2/3 alters its ubiquitin ligase activity and
whether RNF8 is an STUbL remains to be elucidated.
SIM is characterized by a loose consensus sequence
with multiple variants. The first SIM was identified as a common
Ser-X-Ser sequence surrounded by a hydrophobic core and acidic
amino acids (16), whereas a study
by Song et al suggested that SIM was the hydrophobic core
with the consensus Val/Ile-X-Val/Ile-Val/Ile (17). Subsequently, Hannich et al
defined SIM as a hydrophobic core flanked by acidic residues
(18). It has also been suggested
that hydrophobic and acidic residues in SIMs may determine its
specificity in binding to distinct SUMO isoforms (19). Hecker et al showed that the
presence of acidic amino acids or phosphorylated residues are
necessary for the binding to SUMO1, but not to SUMO2/3 (12). Certain SUMO-binding partners do not
contain one of the universal SIMs, including the I-V-D-V SIM motif
in TTRAP and V-Q-E-V in thymine DNA glycosylase (12,20).
In the present study, it was found that the FHA domain of RNF8 was
responsible for the binding to SUMO2/3. The FHA domain does not
contain the universal SIM motif, and characterization of the
residues in the FHA domain responsible for the binding to SUMO2/3
is required.
In conclusion, the present study described the
noncovalent interaction between the E3 ubiquitin ligase RNF8 and
SUMO2/3 and indicated that this interaction promoted DSB
repair.
Acknowledgements
The present study was supported by the China
Postdoctoral Science Foundation (grant no. 2015T80145).
References
|
1
|
Jackson SP and Bartek J: The DNA-damage
response in human biology and disease. Nature. 461:1071–1078. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Park JM, Choi JY, Yi JM, Chung JW, Leem
SH, Koh SS and Kang TH: NDR1 modulates the UV-induced DNA-damage
checkpoint and nucleotide excision repair. Biochem Biophys Res
Commun. 461:543–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jackson SP and Durocher D: Regulation of
DNA damage responses by ubiquitin and SUMO. Mol Cell. 49:795–807.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mailand N, Bekker-Jensen S, Faustrup H,
Melander F, Bartek J, Lukas C and Lukas J: RNF8 ubiquitylates
histones at DNA double-strand breaks and promotes assembly of
repair proteins. Cell. 131:887–900. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huen MS, Grant R, Manke I, Minn K, Yu X,
Yaffe MB and Chen J: RNF8 transduces the DNA-damage signal via
histone ubiquitylation and checkpoint protein assembly. Cell.
131:901–914. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stucki M, Clapperton JA, Mohammad D, Yaffe
MB, Smerdon SJ and Jackson SP: MDC1 directly binds phosphorylated
histone H2AX to regulate cellular responses to DNA double-strand
breaks. Cell. 123:1213–1226. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yoshioka T, Kimura M, Saio M, Era S and
Okano Y: Plk1 is negatively regulated by RNF8. Biochem Biophys Res
Commun. 410:57–61. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu G, Chapman JR, Brandsma I, Yuan J,
Mistrik M, Bouwman P, Bartkova J, Gogola E, Warmerdam D, Barazas M,
et al: REV7 counteracts DNA double-strand break resection and
affects PARP inhibition. Nature. 521:541–544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mattiroli F, Vissers JH, van Dijk WJ, Ikpa
P, Citterio E, Vermeulen W, Marteijn JA and Sixma TK: RNF168
ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling.
Cell. 150:1182–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Galanty Y, Belotserkovskaya R, Coates J,
Polo S, Miller KM and Jackson SP: Mammalian SUMO E3-ligases PIAS1
and PIAS4 promote responses to DNA double-strand breaks. Nature.
462:935–939. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Morris JR, Boutell C, Keppler M, Densham
R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T,
et al: The SUMO modification pathway is involved in the BRCA1
response to genotoxic stress. Nature. 462:886–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hecker CM, Rabiller M, Haglund K, Bayer P
and Dikic I: Specification of SUMO1- and SUMO2-interacting motifs.
J Biol Chem. 281:16117–16127. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin Y, Seifert A, Chua JS, Maure JF,
Golebiowski F and Hay RT: SUMO-targeted ubiquitin E3 ligase RNF4 is
required for the response of human cells to DNA damage. Genes Dev.
26:1196–1208. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim ET, Kim KK, Matunis MJ and Ahn JH:
Enhanced SUMOylation of proteins containing a SUMO-interacting
motif by SUMO-Ubc9 fusion. Biochem Biophys Res Commun. 388:41–45.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Galanty Y, Belotserkovskaya R, Coates J
and Jackson SP: RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes
DNA double-strand break repair. Genes Dev. 26:1179–1195. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Minty A, Dumont X, Kaghad M and Caput D:
Covalent modification of p73alpha by SUMO-1. Two-hybrid screening
with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1
interaction motif. J Biol Chem. 275:36316–36323. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song J, Durrin LK, Wilkinson TA, Krontiris
TG and Chen Y: Identification of a SUMO-binding motif that
recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 101:pp.
14373–14378. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hannich JT, Lewis A, Kroetz MB, Li SJ,
Heide H, Emili A and Hochstrasser M: Defining the SUMO-modified
proteome by multiple approaches in Saccharomyces cerevisiae. J Biol
Chem. 280:4102–4110. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kerscher O: SUMO junction-what's your
function? New insights through SUMO-interacting motifs. EMBO Rep.
8:550–555. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baba D, Maita N, Jee JG, Uchimura Y,
Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H and Shirakawa
M: Crystal structure of thymine DNA glycosylase conjugated to
SUMO-1. Nature. 435:979–982. 2005. View Article : Google Scholar : PubMed/NCBI
|