Introduction
Human bladder cancer (BCa), one of the most common
genitourinary malignancies arising from mucous membrane, accounts
for >3% of all malignant tumors around the world and more and
more people are diagnosed each year (1). Early stage of bladder cancers do not
usually cause symptoms and, with regard to those of late stage, it
still frequently recurs and gradually progresses into muscle
invasive BCa (2). The gold
standard of BCa diagnosis is based on cystoscopy, which is invasive
and relatively expensive. Currently, non-invasive and specific
markers are used for urinary cytology, however, this method is not
sensitive to detection of low-grade BCa. Thus, new, highly
sensitive and specific urine-based diagnostic tools are
particularly attractive because urine is a promising and readily
available source for molecular markers, including RNA.
As small nucleotides of RNA, by binding to
complementary sequences in the 3′-untranslated regions of specific
mRNAs, micro (mi)RNAs inhibit the translation of specific target
genes (3). In previous years, many
cardinal cellular and physiological processes have been
demonstrated to be associated with altered change in miRNAs
expression levels (4–6). miRNAs are involved in the regulation
of various cellular processes including cellular differentiation,
cell cycle progression and apoptosis. Moreover, miRNAs, as
important factors in tumorigenesis and metastasis and their
expression signatures are associated with the prognosis and
progression in a variety of cancers (7–9).
Therefore, the authors could suppose that those miRNAs of which the
expressions are changed significantly in tumors relative to normal
tissues may have influence on tumor progression (10). Moreover, identifying target genes
united with differentially expressed miRNAs might illustrate the
roles of miRNAs in cancer biology (11). Previous studies have reported that
we could distinguish malignant or normal tissue, as well as various
tumor entities by miRNA expression profiles (12–16).
As highly stable molecules, the authors could quantify miRNAs in
tissues and body fluids, which makes them considered as promising
cancer biomarkers. It has been reported that diagnosis, stage and
sub-classification of cancer can be explored by differentially
expressed miRNAs, which also can predict treatment efficacy and
prognosis (4). However, the
molecular mechanisms are not yet clearly elucidated, which makes it
necessary to identify novel miRNAs.
In the present study, the group performed an
MIBC-related miRNA expression profile, so as to address the
functional roles of miRNAs in MIBC. Several human MIBC tissues and
normal bladder tissues were conducted to conduct a microarray
analysis (GEO accession No. GSE76211), revealing several
differentially expressed miRNAs. The putative target genes of
dysregulated miRNAs were predicted using miRNA databases. Then the
networks among miRNAs and genes, gene ontologies (GOs) and pathways
were built. The purpose of the current study was to identify
candidate predictive tumor-associated miRNAs in MIBC patients.
Materials and methods
Preparation for human bladder
samples
Three MIBC tissues samples were collected from
patients after surgery at Zhongnan Hospital of Wuhan University
(Wuhan, China). Three normal bladder tissue samples were collected
from donors by accidental death. Information of the MIBC patients
and donors was listed in Table I.
Those samples used in the study have been described in previous
publication (17–19). Briefly, two pathologists confirmed
the histology diagnosis independently and all the tissues were
snap-frozen for total RNA isolation at liquid nitrogen after
excision from operation room. Informed consent was obtained for
surgery patients and normal donors from the patients and their
relatives, respectively. The Ethics Committee at Zhongnan Hospital
of Wuhan University (Wuhan, China) approved the experiments using
human bladder tissue samples for RNA isolation analysis (approval
number: 2015029). All methods used for human bladder tissue samples
were performed in accordance with the approved guidelines and
regulations.
 | Table I.Information of the patients and
donors. |
Table I.
Information of the patients and
donors.
|
Characteristics | MIBC patients | Donors |
|---|
| Number | 3 | 3 |
| Age, years (mean ±
SD) | 62±1.581 | 37±2.327 |
| Gender | Male | Male |
| BCa stage | Stage II | – |
| Surgical
method | Radical
resection | – |
RNA extraction
Based on the manufacturer's protocol, total RNA was
extracted from the frozen tissue block using RNeasy Mini kit (cat.
no. 74101, Qiagen GmbH, Hilden, Germany), combined with QIAshredder
(cat. no. 79654, Qiagen GmbH) using a centrifuge (cat. no. 5424,
Eppendorf, Hamburg, Germany). In order to remove genomic DNA, DNase
I digestion (cat. no. 79253, Qiagen GmbH), DNase I digestion (cat.
no. 79254, Qiagen GmbH) was used in each RNA preparation.
miRNA microarray
After assessing RNA quality and quantity, the miRNAs
microarray analysis (Affymetrix microRNA 4.0 Array, Affymetrix,
Inc., Santa Clara, CA, USA) was performed according to the
manufacturer's instructions. Briefly, 1 µg of total RNA was labeled
with Biotin using the FlashTag Biotin HSR RNA Labeling kit
(Genisphere LLC, Hatfield, PA, USA) and then hybridized overnight
with the array, which was washed, stained, and read by an GeneChip
Scanner 3000 7G (Affymetrix, Inc.). MiRNA microarray data GSE40355
used for validation were obtained from Gene Expression Omnibus
(GEO) database (http://www.ncbi.nlm.nih.gov/geo/). This dataset
included eight normal bladder tissues samples, eight low grade BCa
tissues samples and eight high grade BCa tissue samples. Among
them, significant expressed miRNAs were screened out from high
grade BCa tissues compared with normal bladder tissues.
Data analysis of miRNAs
microarray
CEL-files of the raw data were first exported by
Affymetrix GeneChip Command Console Software Version 4.0
(Affymetrix, Inc.) and then uploaded to the website of Gminix-Cloud
Biotechnology Information (GCBI) by Genminix Informatics Co., Ltd.
(Shanghai, China; http://www.gcbi.com.cn/gclib/html/index) for further
analysis, including difference analysis of miRNAs profiles,
prediction of miRNAs target genes, GO/pathway enrichment analysis,
miRNAs-gene-network and miRNAs-GO-network analysis. The miRNAs
array data used in the present paper has been uploaded to the NCBI
Gene Expression Omnibus and the GEO accession number is
GSE76211.
According to the GCBI online method description for
difference analysis, the procedure for candidate miRNAs selection
is as follows: When the number of samples in each group is no less
than 3, SAM method is used for difference analysis. The authors
implemented a series of steps to obtain the estimation of
significance of difference and false discovery rate for every
filtered gene:
i) Calculate the exchange factor s0:
Firstly, calculate the standard deviation for all genes, denote
sα as the α percentile for si. For the
percentile value q1 < q2 … q100
of the si, calculate the statistic:
vj=mad{diα=ri/(si+sα)|si=[qj,qj+1)}
Where mad denotes the mean absolute deviation. At
last, α (denote as ᾶ) was selected to make the CV (coefficient of
variation) of the vj achieve minimum. Then, the exchange
factor s0 is sᾶ used.
ii) Calculate the statistic value (d Score) for
every gene:
di=ri/(si+s0)
Where ri reflects the difference in
average level among different groups, si reflects the
variation of sample population. See details in references (20,21)
iii) Calculate the order statistic:
d(1)≤d(2)≤···≤d(i)≤···≤d(p)
iv) In order to get the above statistic's estimate,
the authors made a permutation method (a loop strategy through
every sample, the total number no less than 1,000; the detail are
omitted) the expected distribution of d score. The estimated
statistic values are denoted as follows:
d(1)*≤d(2)*≤···≤d(i)*≤···≤d(p)*
v) The authors obtain the order statistic value
under the permutation:
d¯(i)=∑i=11000d(i)*1000
vi) By calculating the maximum distance between the
order statistic d(i) and the expected order statistic
d¯(i), the authors constructed
a series of rejection regions for q-value. In fact, a grid of delta
values was obtained by dividing 50 equivalent delta value for the
above distance.
vii) For a fixed delta value, by computing the
difference ∆(i) = d(i)
-d¯(i) the authors identified
the nearest ∆(i) for gene i. The cut-up is marked as min
{∆(i) ≥ delta} for positive gene and the
cut-down max {∆(i) ≤ delta} as for negative
gene. The genes with differences above the cut-up value (we denote
the number of these genes as R(p)) were considered as
significantly positive genes. While the genes with differences
lower than the cut-down value were considered as significantly
negative genes.
viii) Under the above cut-up and cut-down
thresholds, the simulation of step VII was performed respectively
on the statistics obtained from step V, such that the number of
positive genes could be obtained under random state (≥1,000
permutations). The median of the 1,000 positive genes was estimated
as the number of false positive genes, to allow the false discovery
rate (FDR) to be estimated (the positive FDR=V(p)R(p) and the negative FDR was
similar), and thus the proportion of false positive genes in the
full set of positive genes.
ix) Finally, according to the definition of q-value
(22), the authors obtained the
q-value for the gene, i, by selecting the minimum of the FDR for
the 50 delta values determined in step VII (every delta as a
rejection region).
Results
Identification of differentially
expressed miRNAs (DE-miRNAs) in BCa tissues
The obtained miRNA expression profiles (GSE76211) of
BCa and normal bladder tissues were analyzed by the Affymetrix
microRNA 4.0 Array, which contains 2,578 probes and can interrogate
all mature miRNAs sequences in miRBase Release 20. The results
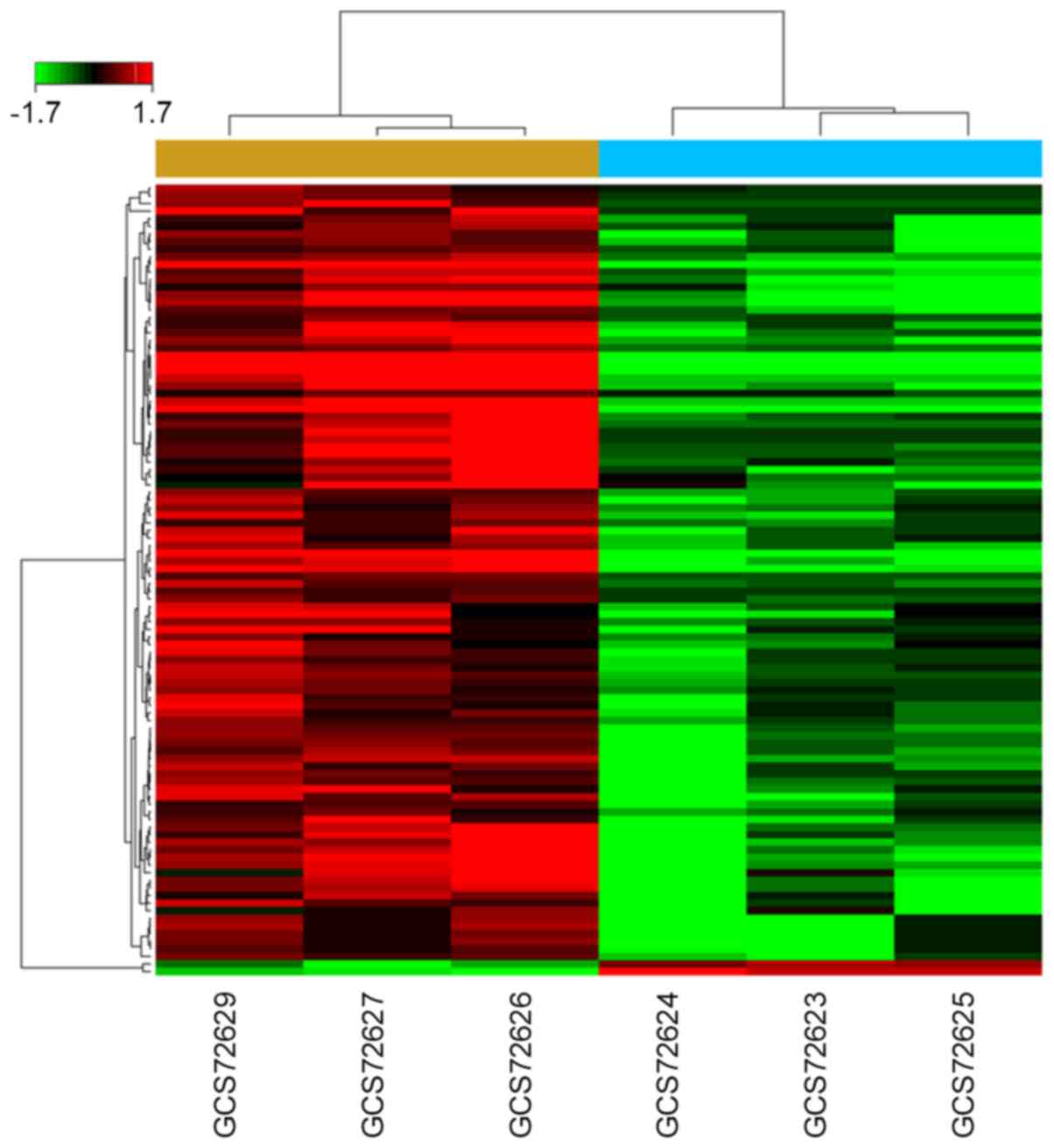
revealed that 104 miRNAs were dysregulated in BCa group under the
condition of ‘P<0.05 and fold change >1.5’, compared with
normal bladder group. Among them, 102 miRNAs were downregulated and
2 were upregulated (Fig. 1). All
of the dysregulated miRNA were listed in Table II.
 | Table II.Differently expressed miRNAs in human
MIBC tissues. |
Table II.
Differently expressed miRNAs in human
MIBC tissues.
| MIBC tissues vs.
normal bladder tissues. (P<0.05, Fold-change >1.5) |
|---|
|
|---|
| miRNA | Change | P-value |
Featurea | Rankb |
|---|
|
hsa-miR-4786-5p | −10.205618 | 0.00124 | Down |
1 |
| hsa-miR-490-3p | −35.773813 | 0.001358 | Down |
2 |
|
hsa-miR-3617-5p | −42.647285 | 0.001476 | Down |
3 |
| hsa-miR-490-5p | −75.331847 | 0.001594 | Down |
4 |
| hsa-miR-139-3p | −32.552806 | 0.001712 | Down |
5 |
| hsa-miR-133b | −121.234878 | 0.00183 | Down |
6 |
| hsa-miR-145-3p | −9.840205 | 0.001948 | Down |
7 |
|
hsa-miR-29b-1-5p | −10.581222 | 0.002066 | Down |
8 |
| hsa-miR-155-5p | 8.720112 | 0.002184 | UP |
9 |
| hsa-miR-1 | −40.712728 | 0.002302 | Down | 10 |
| hsa-miR-548q | −5.779384 | 0.00242 | Down | 11 |
|
hsa-miR-133a-5p | −17.075477 | 0.002774 | Down | 12 |
|
hsa-miR-146b-5p | 4.140864 | 0.002893 | UP | 13 |
| hsa-miR-28-3p | −3.834537 | 0.003129 | Down | 14 |
|
hsa-miR-30c-2-3p | −11.293849 | 0.003247 | Down | 15 |
| hsa-miR-143-5p | −15.820855 | 0.003365 | Down | 16 |
|
hsa-miR-6511b-3p | −6.432579 | 0.003601 | Down | 17 |
| hsa-miR-320e | −3.630028 | 0.003719 | Down | 18 |
| hsa-miR-99a-3p | −2.325859 | 0.003837 | Down | 19 |
| hsa-miR-30a-3p | −18.900354 | 0.003955 | Down | 20 |
|
hsa-miR-133a-3p | −63.82892 | 0.004073 | Down | 21 |
| hsa-miR-4510 | −2.805811 | 0.004545 | Down | 22 |
| hsa-miR-139-5p | −12.808018 | 0.004782 | Down | 23 |
| hsa-miR-4324 | −19.035251 | 0.005136 | Down | 24 |
| hsa-miR-145-5p | −3.499059 | 0.005372 | Down | 25 |
|
hsa-miR-125b-1-3p | −13.117689 | 0.005608 | Down | 26 |
| hsa-miR-143-3p | −3.894089 | 0.005962 | Down | 27 |
|
hsa-miR-193a-5p | −4.050292 | 0.006198 | Down | 28 |
| hsa-miR-5684 | −4.681531 | 0.007261 | Down | 29 |
| hsa-miR-328-3p | −8.31845 | 0.007379 | Down | 30 |
| hsa-miR-4429 | −2.76178 | 0.007497 | Down | 31 |
|
hsa-miR-1287-5p | −7.357196 | 0.007615 | Down | 32 |
|
hsa-miR-3605-5p | −2.739437 | 0.007969 | Down | 33 |
| hsa-miR-338-5p | −4.852428 | 0.008323 | Down | 34 |
|
hsa-miR-193a-3p | −9.317892 | 0.00856 | Down | 35 |
| hsa-miR-4257 | −3.696435 | 0.008796 | Down | 36 |
|
hsa-miR-6507-5p | −6.813232 | 0.010331 | Down | 37 |
|
hsa-miR-6722-3p | −2.904131 | 0.011275 | Down | 38 |
| hsa-miR-497-5p | −3.273151 | 0.011865 | Down | 39 |
|
hsa-miR-125b-2-3p | −7.613095 | 0.012574 | Down | 40 |
|
hsa-miR-29b-2-5p | −2.995467 | 0.013046 | Down | 41 |
|
hsa-miR-6511a-3p | −8.396761 | 0.013282 | Down | 42 |
| hsa-miR-628-3p | −4.613832 | 0.013636 | Down | 43 |
| hsa-miR-378b | −6.024561 | 0.014581 | Down | 44 |
|
hsa-miR-664a-5p | −6.119249 | 0.014699 | Down | 45 |
| hsa-miR-204-5p | −6.006885 | 0.015525 | Down | 46 |
| hsa-miR-7641 | −4.116459 | 0.015762 | Down | 47 |
| hsa-miR-320d | −2.149376 | 0.016706 | Down | 48 |
| hsa-miR-3656 | −2.093619 | 0.017414 | Down | 49 |
|
hsa-miR-1273g-3p | −2.156204 | 0.017769 | Down | 50 |
| hsa-miR-378g | −7.277976 | 0.018359 | Down | 51 |
|
hsa-miR-1225-5p | −2.552195 | 0.019067 | Down | 52 |
|
hsa-miR-3156-5p | −6.520017 | 0.019421 | Down | 53 |
| hsa-miR-383-5p | −3.70084 | 0.019658 | Down | 54 |
|
hsa-miR-3064-3p | −2.07381 | 0.020012 | Down | 55 |
| hsa-miR-378d | −5.5391 | 0.02013 | Down | 56 |
| hsa-miR-23b-5p | −9.113128 | 0.020366 | Down | 57 |
| hsa-miR-3195 | −10.384316 | 0.020484 | Down | 58 |
| hsa-miR-100-5p | −3.57042 | 0.020956 | Down | 59 |
| hsa-miR-378e | −14.003579 | 0.021547 | Down | 60 |
|
hsa-miR-4649-5p | −2.337674 | 0.022019 | Down | 61 |
|
hsa-miR-3622b-5p | −3.04368 | 0.022373 | Down | 62 |
|
hsa-miR-6840-3p | −6.817266 | 0.024616 | Down | 63 |
| hsa-miR-4322 | −6.233632 | 0.024852 | Down | 64 |
| hsa-miR-574-5p | −5.490031 | 0.025207 | Down | 65 |
| hsa-miR-4770 | −4.247774 | 0.025561 | Down | 66 |
|
hsa-miR-513a-5p | −3.907964 | 0.025679 | Down | 67 |
| hsa-miR-124-3p | −1.707013 | 0.025797 | Down | 68 |
|
hsa-miR-6819-5p | −2.385821 | 0.026151 | Down | 69 |
| hsa-miR-339-3p | −5.134485 | 0.027332 | Down | 70 |
| hsa-miR-26b-3p | −3.129563 | 0.02745 | Down | 71 |
| hsa-miR-29c-5p | −7.931159 | 0.027922 | Down | 72 |
| hsa-miR-29a-3p | −2.906434 | 0.02804 | Down | 73 |
|
hsa-miR-1227-5p | −2.287321 | 0.028276 | Down | 74 |
| hsa-miR-4327 | −2.710984 | 0.028512 | Down | 75 |
| hsa-miR-99a-5p | −4.947917 | 0.02863 | Down | 76 |
|
hsa-miR-1296-5p | −9.401257 | 0.029103 | Down | 77 |
| hsa-miR-378i | −4.649838 | 0.030283 | Down | 78 |
|
hsa-miR-6889-5p | −4.810854 | 0.030401 | Down | 79 |
| hsa-miR-186-5p | −3.276383 | 0.031464 | Down | 80 |
|
hsa-miR-6824-5p | −8.197661 | 0.031936 | Down | 81 |
| hsa-miR-195-3p | −4.796311 | 0.034416 | Down | 82 |
|
hsa-miR-3622a-5p | −4.145204 | 0.034888 | Down | 83 |
| hsa-miR-5572 | −6.053895 | 0.03536 | Down | 84 |
|
hsa-miR-6776-5p | −3.050977 | 0.035832 | Down | 85 |
|
hsa-miR-6790-5p | −2.868339 | 0.036895 | Down | 86 |
|
hsa-miR-6740-5p | −2.087867 | 0.037721 | Down | 87 |
| hsa-miR-4269 | −8.226931 | 0.038312 | Down | 88 |
|
hsa-miR-125b-5p | −2.694152 | 0.039138 | Down | 89 |
| hsa-miR-3188 | −8.008159 | 0.039256 | Down | 90 |
|
hsa-miR-6787-5p | −2.714687 | 0.03961 | Down | 91 |
| hsa-miR-99b-5p | −1.811603 | 0.039847 | Down | 92 |
| hsa-miR-30a-5p | −3.787576 | 0.040673 | Down | 93 |
|
hsa-miR-6765-5p | −2.365181 | 0.041027 | Down | 94 |
|
hsa-miR-1909-3p | −7.648533 | 0.041972 | Down | 95 |
| hsa-miR-4507 | −2.786848 | 0.042326 | Down | 96 |
| hsa-miR-28-5p | −2.529154 | 0.042916 | Down | 97 |
|
hsa-miR-6892-5p | −2.081233 | 0.046458 | Down | 98 |
|
hsa-miR-4433b-3p | −1.923165 | 0.046694 | Down | 99 |
|
hsa-miR-6737-5p | −3.166103 | 0.04693 | Down | 100 |
| hsa-miR-324-3p | −3.939764 | 0.048465 | Down | 101 |
| hsa-miR-211-3p | −3.140159 | 0.048819 | Down | 102 |
|
hsa-miR-6798-5p | −2.080154 | 0.049174 | Down | 103 |
| hsa-miR-4299 | −4.93377 | 0.049882 | Down | 104 |
Identification of putative target
genes
The current study has identified 104 miRNAs that
were significantly dysregulated in BCa tissues compared with normal
bladder tissues. As miRNAs play their functional roles by
regulating target genes expression at the posttranscriptional
level, the authors predicted the target genes of dysregulated
miRNAs using GCBI online tools, which were mainly based on the
algorithms of miRanda and TargetScan. A total of 11,884 genes were
predicted as putative target genes of dysregulated miRNAs.
GO/pathway enrichment analysis of
putative target genes of dysregulated miRNAs
To understand the role of miRNAs in cancer
development, GO and pathway enrichment analysis were performed. The
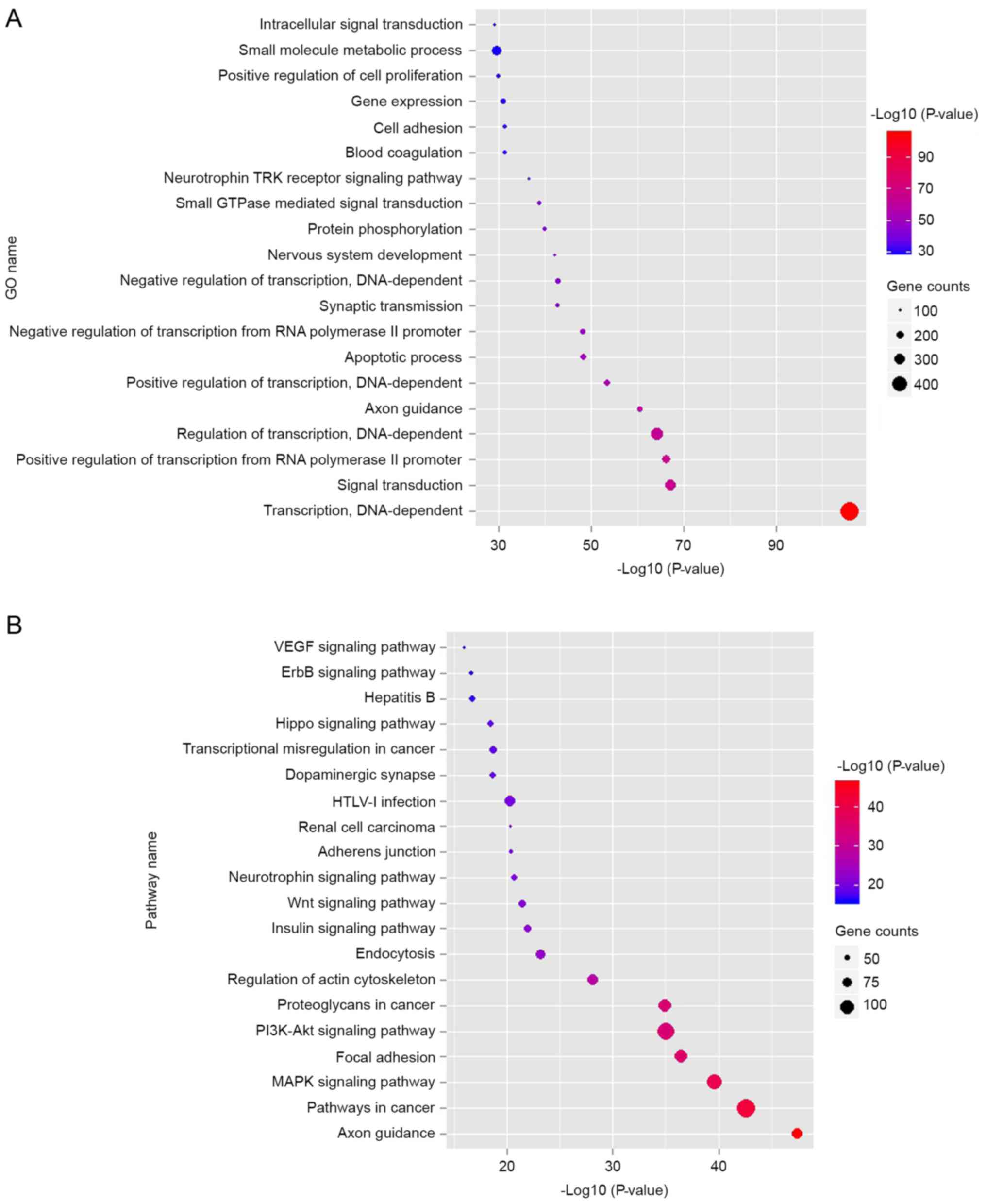
data in Fig. 2A indicated that the
top 10 dysregulated GOs were ‘transcription, DNA-dependent’,
‘signal transduction, positive regulation of transcription from RNA
polymerase II promoter’, ‘regulation of transcription,
DNA-dependent’, ‘axon guidance’, ‘positive regulation of
transcription, DNA-dependent’, ‘apoptotic process’, ‘negative
regulation of transcription from RNA polymerase II promoter’,
‘synaptic transmission’ and ‘negative regulation of transcription,
DNA-dependent’. GO analysis obviously suggests that many
dysregulated miRNAs may contribute to tumorigenesis of bladder
through many important functions such as transcription regulation,
signal transduction as well as apoptotic process. Combined with the
KEGG database, the authors analyzed the pathways involving the
putative target genes. As illustrated in Fig. 2B, the top ten dysregulated pathways
were the mitogen-associated protein kinase (MAPK) signaling
pathway, apoptosis, pathways in cancer, cell cycle, p53 signaling
pathway, calcium signaling pathway, Wnt signaling pathway, adherens
junction, focal adhesion and ErbB signaling pathway.
Pathway network analysis
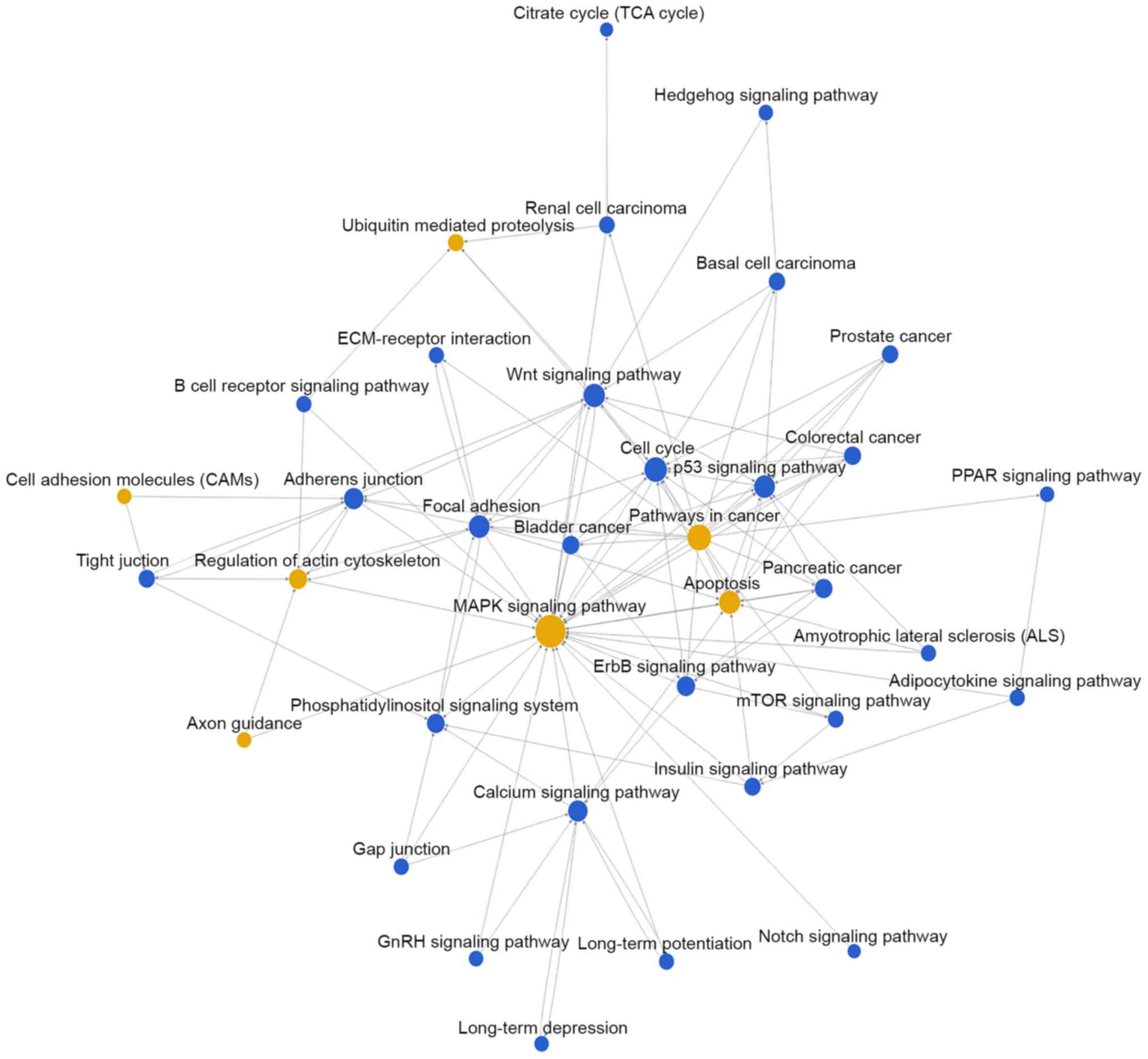
Then, pathway network (Path-net) analysis was
performed to draw an interaction network covering 36 significantly
changed pathways (Fig. 3). Among
them, the MAPK signaling pathway (degree=44), apoptosis
(degree=30), pathways in cancer (degree=29) and cell cycle
(degree=24) showed the highest degree, suggesting that these four
pathways might play a core role in regulation of bladder cancer
development.
miRNAs-gene-networks and
miRNAs-GO-networks
Based on the significantly regulated GOs and
pathways, the authors selected intersected genes and further
constructed miRNAs-gene-networks and miRNAs-GO-networks to screen
the key regulatory functions of the identified miRNAs and their
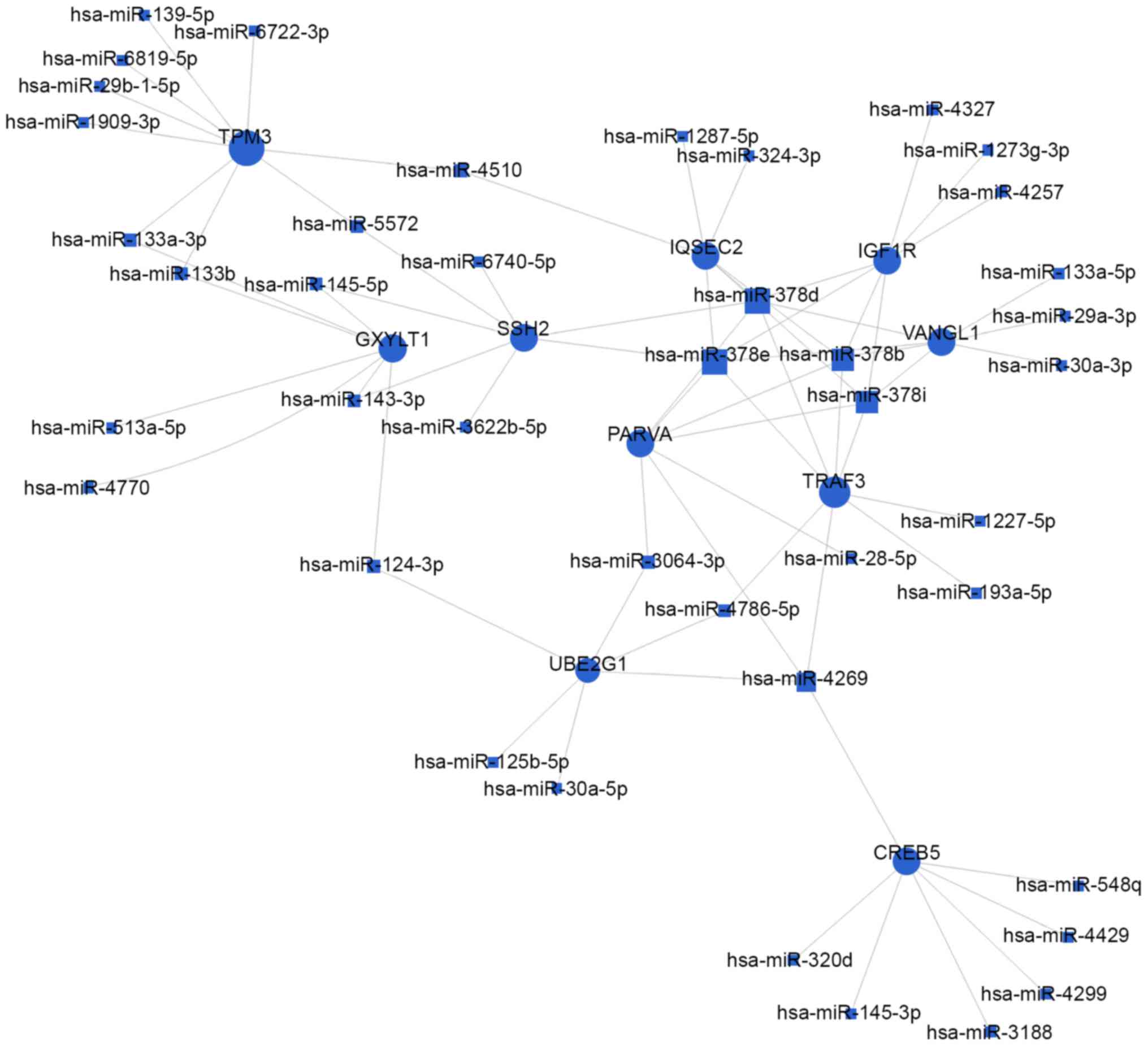
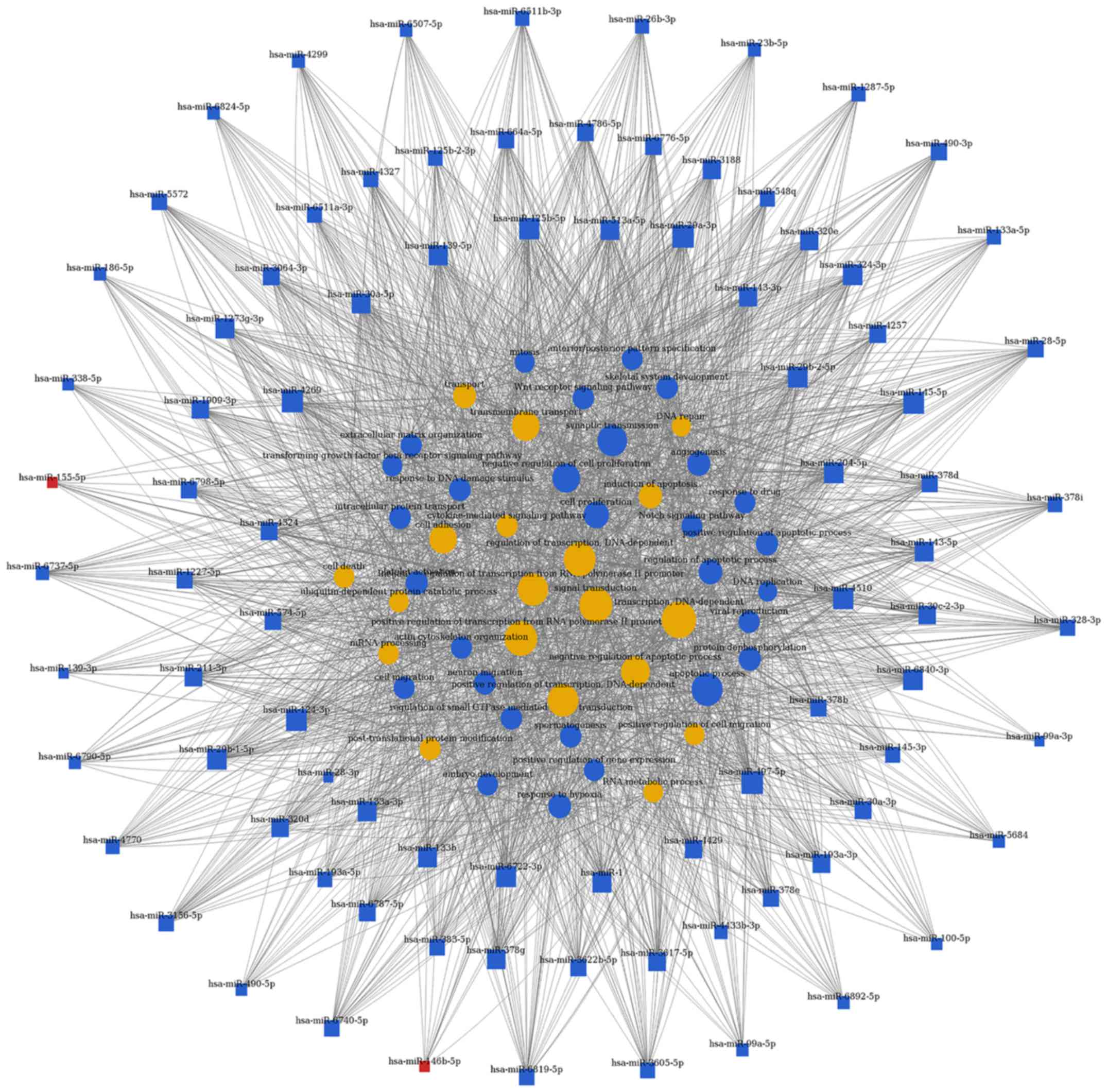
target genes, respectively. As shown in Figs. 4 and 5, and Table III, the top rated nine miRNAs
were hsa-miR-497-5p, hsa-miR-29a-3p, hsa-miR-124-3p, hsa-miR-4269,
hsa-miR-145-5p, hsa-miR-204-5p, hsa-miR-4510, hsa-miR-6840-3p and
hsa-miR-6722-3p. All of these miRNAs were downregulated in BCa
tissues compared with normal bladder tissues. The microarray
analysis demonstrated that the dysregulated miRNAs primarily play
vital roles in various biological processes, including
transcription regulation, apoptotic process, gene expression and
signal transduction. Taken together, deregulation of certain miRNAs
and several important pathways may be closely associated with human
bladder cancer development.
 | Table III.The top 10 miRNAs with high degrees
of miRNAs-gene-networks and miRNAs-GO-networks. |
Table III.
The top 10 miRNAs with high degrees
of miRNAs-gene-networks and miRNAs-GO-networks.
| Rank | miRNAs | miRNA-gene-networks
degreea |
Featureb | miRNAs | miRNA-GO-networks
degreea |
Featureb |
|---|
| 1 | hsa-miR-497-5p | 123 | Down | hsa-miR-497-5p | 637 | Down |
| 2 | hsa-miR-29a-3p | 88 | Down | hsa-miR-29a-3p | 518 | Down |
| 3 | hsa-miR-124-3p | 84 | Down | hsa-miR-124-3p | 516 | Down |
| 4 | hsa-miR-4269 | 64 | Down | hsa-miR-204-5p | 494 | Down |
| 5 | hsa-miR-145-5p | 63 | Down | hsa-miR-4269 | 458 | Down |
| 6 | hsa-miR-204-5p | 62 | Down | hsa-miR-145-5p | 430 | Down |
| 7 | hsa-miR-4510 | 61 | Down |
hsa-miR-6840-3p | 406 | Down |
| 8 |
hsa-miR-125b-5p | 57 | Down | hsa-miR-4510 | 404 | Down |
| 9 |
hsa-miR-6840-3p | 50 | Down |
hsa-miR-6722-3p | 387 | Down |
| 10 |
hsa-miR-6722-3p | 47 | Down | hsa-miR-1 | 383 | Down |
Validation of candidate miRNAs
To confirm that the top rated nine miRNAs identified
were indeed dysregulated in BCa tissues, the authors used another
miRNA expression profiling GSE40355 including 8 normal bladder
tissues samples, 8 low grade BCa tissues samples and 8 high grade
BCa tissue samples for validation. In the dataset of GSE40355, the
expression values of candidate miRNAs between normal bladder tissue
samples with high grade BCa tissue samples were extracted for t
test, and P<0.05 were considered statistically significant. In
addition, 157 dysregulated miRNAs including 69 upregulated and 88
downregulated miRNAs were obtained by analysis of GSE40355. As
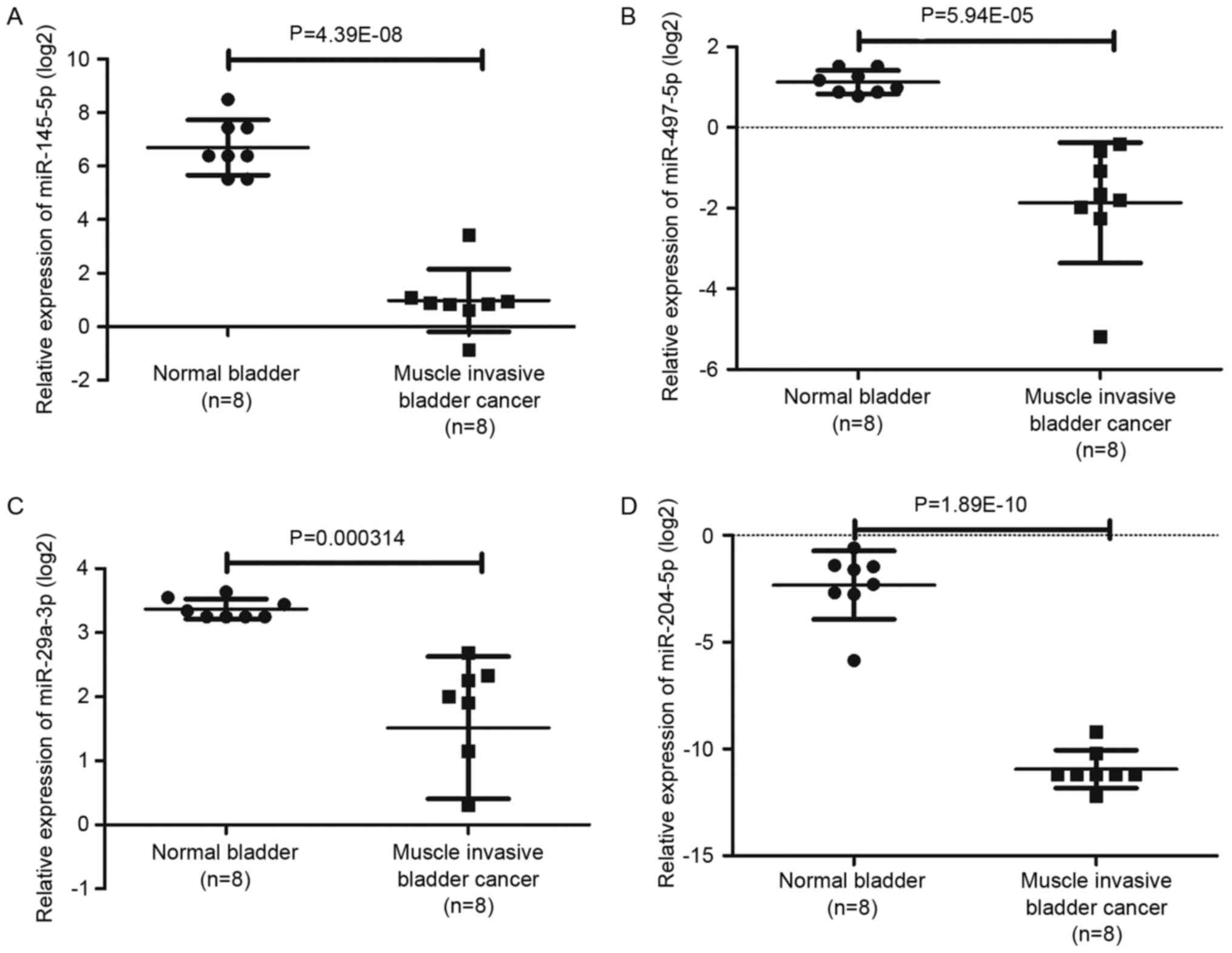
presented in Fig. 6 and Table IV, among the top rated nine miRNAs
screened from our miRNA microarray, four miRNAs involving
hsa-miR-497-5p, hsa-miR-29a-3p, hsa-miR-145-5p and hsa-miR-204-5p
were also significantly altered in GSE40355.
 | Table IV.The alteration of top rated nine
miRNAs in GSE76211 and GSE40355. |
Table IV.
The alteration of top rated nine
miRNAs in GSE76211 and GSE40355.
|
| GSE76211 | GSE40355 |
|---|
|
|
|
|
|---|
| miRNAs | Fold-change | P-value |
Featurea | Fold-change | P-value |
Featurea |
|---|
| hsa-miR-497-5p | −3.27 | 0.011865 | Down | −7.94 |
5.94×10−5 | Down |
| hsa-miR-29a-3p | −2.90 | 0.02804 | Down | −3.61 | 0.0003 | Down |
| hsa-miR-124-3p | −1.70 | 0.025797 | Down | – | – | Down |
| hsa-miR-4269 | −8.22 | 0.038312 | Down | – | – | Down |
| hsa-miR-145-5p | −3.49 | 0.005372 | Down | −52.69 |
4.39×10−8 | Down |
| hsa-miR-204-5p | −6.00 | 0.015525 | Down | −463.33 |
1.89×10−10 | Down |
| hsa-miR-4510 | −2.80 | 0.004545 | Down | – | – | Down |
|
hsa-miR-6840-3p | −6.81 | 0.024616 | Down | – | – | Down |
|
hsa-miR-6722-3p | −2.90 | 0.011275 | Down | – | – | Down |
Discussion
In the current study, by comparing array-based miRNA
expression profiling performed on three MIBC tissues and three
normal bladder tissues, the integrated bioinformatic analysis from
miRNA expression profiling identified that 104 miRNAs were
differentially expressed (P<0.05, fold change >1.5), of
which, 102 were downregulated and 2 were upregulated (Fig. 1 and Table II). Among the top 10 fold change
miRNAs, many of them were closely linked to occurrence and
development of bladder cancer and might play vital role as
oncogenes and tumor suppressor genes, which has been published in
several studies (23–27). Several studies have showed that
miR-490-5p was significantly downregulated in BCa tissue samples
compared with adjacent normal tissues (28). Low expression of miR-139-3p,
miR-133, miR-145 and miR-1 in MIBC tissue samples compared with
normal tissues was also reported by studies and played a functional
role in bladder cancer cell lines (29–32).
Then, 11,884 genes were predicted as putative target
genes of dysregulated miRNAs using target prediction method in GCBI
online tools. According to the GO analysis, the predicted target
genes were mainly involved in ‘transcription, DNA-dependent’,
signal transduction, positive regulation of transcription from RNA
polymerase II promoter, ‘regulation of transcription,
DNA-dependent’ and axon guidance (Fig.
2A). Interestingly, the authors noticed two pairs of opposite
GOs (Negative regulation of transcription, DNA-dependent vs.
Positive regulation of transcription, DNA-dependent and ‘Negative
regulation of transcription from RNA polymerase II promoter vs.
Positive regulation of transcription from RNA polymerase II
promoter’). miRNAs were involved in post-transcriptional regulation
of gene expression (33), which
played a key role in various cellular processes including cellular
differentiation, cell cycle progression and apoptosis. Among the
predictive target genes of miRNAs, there may be lots of oncogenes
and tumor suppressor genes. Oncogenes could promote BCa cell
proliferation by positive regulation of transcription, in contrast,
tumor suppressor genes could suppress BCa cell viability by
negatively regulation of transcription. Therefore, it can be
inferred that miRNA regulation disorders might account for these
biological behaviors. As for biological pathways, the MAPK
signaling pathway, apoptosis, pathways in cancer, cell cycle, p53
signaling pathway, calcium signaling pathway, Wnt signaling
pathway, adherens junction, focal adhesion and Erbb signaling
pathway were the top 10 enriched pathways of the predicted target
genes (Fig. 2B). Many studies
including our group (17–19) have ever reported that these
pathways such as the MAPK signaling pathway, Wnt signaling pathway,
p53 signaling pathway and Erbb signaling pathway play a functional
role in human bladder cancer cells (34,35).
Besides, the Path-net analysis covering 36 significant pathways
also showed that the MAPK signaling pathway, cell cycle, p53
signaling pathway, Wnt signaling pathway and calcium signaling
pathway have a close correlation with bladder cancer, indicating
that these pathways might play a key role in the development of
human bladder cancer. In order to find out the key miRNAs, the
authors conducted regulatory network analysis by overlapping
significant miRNAs, pathways and GO analysis, revealing that the
top nine miRNAs were hsa-miR-497-5p, hsa-miR-29a-3p,
hsa-miR-124-3p, hsa-miR-4269, hsa-miR-145-5p, hsa-miR-204-5p,
hsa-miR-4510, hsa-miR-6840-3p and hsa-miR-6722-3p. In order to
validate that the nine candidate miRNAs identified were indeed
dysregulated in BCa tissues, another miRNA microarray was performed
by overlapping different expressed miRNAs and the top nine
candidate miRNAs suggesting that four miRNAs involving
hsa-miR-497-5p, hsa-miR-29a-3p, hsa-miR-145-5p and hsa-miR-204-5p
were significantly altered. Moreover, as illustrated in Figs. 4 and 5, the top 10 target genes were TPM3,
GXYLT1, SSH2, UBE2G1, CREB5, PARVA, TRAF3, IQSEC2, VANGL1 and
IGF1R. Above all, these data showed that the regulatory network
consisted of key miRNAs and genes may regulate biological process
such as cell cycle, apoptosis and proliferation of human bladder
cancer. Among the top ten target genes, tropomyosin 3 is a member
of the tropomyosin family of actin-binding protein, which has been
reported to relate to malignant transformation in BCa (36). VANGL planar cell polarity 1
(VANGL1), as an oncogene, is associated with many cancers. Park
et al (37) revealed that
miR-124 targeting VANGL could suppress colorectal cancer and Oh
et al (38) reported that
VANGL1 has correlation with tumor progression in human colorectal
cancer. Insulin-like growth factor 1 receptor (IGF1R), with
tyrosine kinase activity, binds insulin-like growth factor with a
high affinity, playing a critical role in tumorigenesis and
chemosensitivity (39,40). In the present study, IGF1R has high
correlation with has-miR-378 family. Some studies indicated that
miR-378 deficiency played a key role in the development of cardiac
hypertrophy by targeting IGF1R through negatively regulated Ras
signaling pathway (41). Parvin
alpha, a member of the parvin family of actin-binding proteins,
playing a role in cell adhesion, motility and survival, is
connected with different cancers, such as colorectal cancer and
lung cancer (42).
The four miRNAs screened out from the miRNA
microarray and verified by another miRNA microarray indeed had a
functional role in MIBC, which has been demonstrated by previous
studies. Zhang et al (43)
reported that miR-497 was downregulated in BCa tissues compared
with normal bladder tissues and may represent a novel prognostic
biomarker for the early detection of metastasis of bladder cancer.
Du et al (44) also
reported that miR-497 was decreased in plasma of bladder cancer
patients compared with healthy patients and could be a promising
novel circulating biomarkers in clinical detection of bladder
cancer. Chiyomaru et al (45) found that miR-145 was a tumor
suppressor and inhibited cell viability by targeting FSCN1 in BCa
cells. In addition, Avgeris et al (46) suggested that miR-145 could act as a
novel marker helpful for prediction of oncologic outcome for
bladder cancer patients. However, there are no related reports
about miR-29a-3p and miR-204-5p in BCa, therefore, the authors
would like to investigate and confirm the two miRNAs using human
BCa cell lines and mouse model in our next research article.
However, predicting the miRNAs targets merely by bioinformatics
analysis is not sufficient. Since the size of the MIBC samples used
in the present study is small, these results may have many
limitations. Thus, functional experiments should be performed
strictly to verify the miRNAs and its targets in the further
studies. As a result of that, the author group will select some of
the significantly dysregulated miRNAs and perform verification
experiments to confirm their targets and then figure out the
functional role of miRNAs and the underlying mechanisms in
MIBC.
Acknowledgements
The excellent technical assistance of Yuan Zhu,
Shanshan Zhang and Danni Shan is gratefully acknowledged. The
present study was supported in part by grants from the Zhongnan
Hospital of Wuhan University Science, Technology and Innovation
Seed Fund (grant no. cxpy20160031 and cxpy20160077) and the
National Natural Science Foundation of China (grant no. 81300578).
The funders had no role in study design, data collection and
analysis, decision to publish, or preparation of the
manuscript.
References
|
1
|
Griffiths TR; Action on Bladder Cancer, :
Current perspectives in bladder cancer management. Int J Clin
Pract. 67:435–448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pastorelli R, Saletta F, Carpi D, Campagna
R, del l'Osta C, Schiarea S, Vineis P, Airoldi L and Matullo G:
Proteome characterization of a human urothelial cell line resistant
to the bladder carcinogen 4-aminobiphenyl. Proteome Sci. 5:62007.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gu S, Jin L, Zhang F, Sarnow P and Kay MA:
Biological basis for restriction of microRNA targets to the 3′
untranslated region in mammalian mRNAs. Nat Struct Mol Biol.
16:144–150. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP and Chen CZ: Micromanagers of
gene expression: The potentially widespread influence of metazoan
microRNAs. Nat Rev Genet. 5:396–400. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs-the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Adlakha YK and Saini N: Brain microRNAs
and insights into biological functions and therapeutic potential of
brain enriched miRNA-128. Mol Cancer. 13:332014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He Y, Lin J, Kong D, Huang M, Xu C, Kim
TK, Etheridge A, Luo Y, Ding Y and Wang K: Current State of
circulating MicroRNAs as cancer biomarkers. Clin Chem.
61:1138–1155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chawla JP, Iyer N, Soodan KS, Sharma A,
Khurana SK and Priyadarshni P: Role of miRNA in cancer diagnosis,
prognosis, therapy and regulation of its expression by Epstein-Barr
virus and human papillomaviruses: With special reference to oral
cancer. Oral Oncol. 51:731–737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Andrew AS, Marsit CJ, Schned AR, Seigne
JD, Kelsey KT, Moore JH, Perreard L, Karagas MR and Sempere LF:
Expression of tumor suppressive microRNA-34a is associated with a
reduced risk of bladder cancer recurrence. Int J Cancer.
137:1158–1166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bandres E, Agirre X, Ramirez N, Zarate R
and Garcia-Foncillas J: MicroRNAs as cancer players: Potential
clinical and biological effects. DNA Cell Biol. 26:273–282. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Catto JW, Miah S, Owen HC, Bryant H, Myers
K, Dudziec E, Larré S, Milo M, Rehman I, Rosario DJ, et al:
Distinct microRNA alterations characterize high- and low-grade
bladder cancer. Cancer Res. 69:8472–8481. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pignot G, Cizeron-Clairac G, Vacher S,
Susini A, Tozlu S, Vieillefond A, Zerbib M, Lidereau R, Debre B,
Amsellem-Ouazana D and Bieche I: microRNA expression profile in a
large series of bladder tumors: Identification of a 3-miRNA
signature associated with aggressiveness of muscle-invasive bladder
cancer. Int J Cancer. 132:2479–2491. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gottardo F, Liu CG, Ferracin M, Calin GA,
Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, et
al: Micro-RNA profiling in kidney and bladder cancers. Urol Oncol.
25:387–392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hidaka H, Seki N, Yoshino H, Yamasaki T,
Yamada Y, Nohata N, Fuse M, Nakagawa M and Enokida H: Tumor
suppressive microRNA-1285 regulates novel molecular targets:
Aberrant expression and functional significance in renal cell
carcinoma. Oncotarget. 3:44–57. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoshino H, Seki N, Itesako T, Chiyomaru T,
Nakagawa M and Enokida H: Aberrant expression of microRNAs in
bladder cancer. Nat Rev Urol. 10:396–404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao R, Meng Z, Liu T, Wang G, Qian G, Cao
T, Guan X, Dan H, Xiao Y and Wang X: Decreased TRPM7 inhibits
activities and induces apoptosis of bladder cancer cells via ERK1/2
pathway. Oncotarget. 7:72941–72960. 2016.PubMed/NCBI
|
|
18
|
Chen L, Wang G, Luo Y, Wang Y, Xie C,
Jiang W, Xiao Y, Qian G and Wang X: Downregulation of LAPTM5
suppresses cell proliferation and viability inducing cell cycle
arrest at G0/G1 phase of bladder cancer cells. Int J Oncol.
50:263–271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang G, Cao R, Wang Y, Qian G, Dan HC,
Jiang W, Ju L, Wu M, Xiao Y and Wang X: Simvastatin induces cell
cycle arrest and inhibits proliferation of bladder cancer cells via
PPARgamma signalling pathway. Sci Rep. 6:357832016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tusher VG, Tibshirani R and Chu G:
Significance analysis of microarrays applied to the ionizing
radiation response. Proc Natl Acad Sci USA. 98:pp. 5116–5121. 2001;
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chu G, Seo M, Li J, Narasimhan B,
Tibshirani R and Tusher V: SAM: Significance Analysis of
Microarrays. Users Guide and technical document. http://statweb.stanford.edu/~tibs/SAM/sam.pdf
|
|
22
|
Storey JD: A direct approach to false
discovery rates. J Roy Stat Soc Ser. 64:479–498. 2002. View Article : Google Scholar
|
|
23
|
Li T, Pan H and Li R: The dual regulatory
role of miR-204 in cancer. Tumour Biol. 37:11667–11677. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang G, Xiong G, Cao Z, Zheng S, You L,
Zhang T and Zhao Y: miR-497 expression, function and clinical
application in cancer. Oncotarget. 7:55900–55911. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Y, Zhang X, Li H, Yu J and Ren X: The
role of miRNA-29 family in cancer. Eur J Cell Biol. 92:123–128.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang X, Wu Q, Xu B, Wang P, Fan W, Cai Y,
Gu X and Meng F: MiR-124 exerts tumor suppressive functions on the
cell proliferation, motility and angiogenesis of bladder cancer by
fine-tuning UHRF1. FEBS J. 282:4376–4388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shaham L, Binder V, Gefen N, Borkhardt A
and Izraeli S: miR-125 in normal and malignant hematopoiesis.
Leukemia. 26:2011–2018. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li S, Xu X, Xu X, Hu Z, Wu J, Zhu Y, Chen
H, Mao Y, Lin Y, Luo J, et al: MicroRNA-490-5p inhibits
proliferation of bladder cancer by targeting c-Fos. Biochem Biophys
Res Commun. 441:976–981. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yonemori M, Seki N, Yoshino H, Matsushita
R, Miyamoto K, Nakagawa M and Enokida H: Dual tumor-suppressors
miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in
bladder cancer. Cancer Sci. 107:1233–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin
C and Zhang W: microRNA-133 inhibits cell proliferation, migration
and invasion in prostate cancer cells by targeting the epidermal
growth factor receptor. Oncol Rep. 27:1967–1975. 2012.PubMed/NCBI
|
|
31
|
Kou B, Gao Y, Du C, Shi Q, Xu S, Wang CQ,
Wang X, He D and Guo P: miR-145 inhibits invasion of bladder cancer
cells by targeting PAK1. Urol Oncol. 32:846–854. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yoshino H, Chiyomaru T, Enokida H,
Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N and Nakagawa
M: The tumour-suppressive function of miR-1 and miR-133a targeting
TAGLN2 in bladder cancer. Br J Cancer. 104:808–818. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pierzynski JA, Hildebrandt MA, Kamat AM,
Lin J, Ye Y, Dinney CP and Wu X: Genetic Variants in the
Wnt/β-Catenin signaling pathway as indicators of bladder cancer
risk. J Urol. 194:1771–1776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee SJ, Lim JH, Choi YH, Kim WJ and Moon
SK: Interleukin-28A triggers wound healing migration of bladder
cancer cells via NF-κB-mediated MMP-9 expression inducing the MAPK
pathway. Cell Signal. 24:1734–1742. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pawlak G, McGarvey TW, Nguyen TB,
Tomaszewski JE, Puthiyaveettil R, Malkowicz SB and Helfman DM:
Alterations in tropomyosin isoform expression in human transitional
cell carcinoma of the urinary bladder. Int J Cancer. 110:368–373.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Park SY, Kim H, Yoon S, Bae JA, Choi SY,
Jung YD and Kim KK: KITENIN-targeting microRNA-124 suppresses
colorectal cancer cell motility and tumorigenesis. Mol Ther.
22:1653–1664. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Oh HH, Park KJ, Kim N, Park SY, Park YL,
Oak CY, Myung DS, Cho SB, Lee WS, Kim KK and Joo E: Impact of
KITENIN on tumor angiogenesis and lymphangiogenesis in colorectal
cancer. Oncol Rep. 35:253–260. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
XM, Zhu HY, Bai WD, Wang T, Wang L, Chen
Y, Yang AG and Jia LT: Epigenetic silencing of miR-375 induces
trastuzumab resistance in HER2-positive breast cancer by targeting
IGF1R. BMC Cancer. 14:1342014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Taliaferro-Smith L, Oberlick E, Liu T,
McGlothen T, Alcaide T, Tobin R, Donnelly S, Commander R, Kline E,
Nagaraju GP, et al: FAK activation is required for IGF1R-mediated
regulation of EMT, migration, and invasion in mesenchymal triple
negative breast cancer cells. Oncotarget. 6:4757–4772. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nagalingam RS, Sundaresan NR, Gupta MP,
Geenen DL, Solaro RJ and Gupta M: A cardiac-enriched microRNA,
miR-378, blocks cardiac hypertrophy by targeting Ras signaling. J
Biol Chem. 288:11216–11232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Huang AH, Pan SH, Chang WH, Hong QS, Chen
JJ and Yu SL: PARVA promotes metastasis by modulating ILK
signalling pathway in lung adenocarcinoma. PLoS One.
10:e01185302015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang Y, Zhang Z, Li Z, Gong D, Zhan B,
Man X and Kong C: MicroRNA-497 inhibits the proliferation,
migration and invasion of human bladder transitional cell carcinoma
cells by targeting E2F3. Oncol Rep. 36:1293–1300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Du M, Shi D, Yuan L, Li P, Chu H, Qin C,
Yin C, Zhang Z and Wang M: Circulating miR-497 and miR-663b in
plasma are potential novel biomarkers for bladder cancer. Sci Rep.
5:104372015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chiyomaru T, Enokida H, Tatarano S,
Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N
and Nakagawa M: miR-145 and miR-133a function as tumour suppressors
and directly regulate FSCN1 expression in bladder cancer. Br J
Cancer. 102:883–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Avgeris M, Mavridis K, Tokas T,
Stravodimos K, Fragoulis EG and Scorilas A: Uncovering the clinical
utility of miR-143, miR-145 and miR-224 for predicting the survival
of bladder cancer patients following treatment. Carcinogenesis.
36:528–537. 2015. View Article : Google Scholar : PubMed/NCBI
|