Introduction
Pancreatic cancer is the fourth leading cause of
cancer-related deaths in Western countries (1) and ranks sixth among cancer-related
diseases with the highest mortality rate in China (2). Approximately 39,590 and 227,000
deaths due to pancreatic cancer are recorded annually in the USA
and worldwide, respectively (3).
Pancreatic ductal adenocarcinoma, the most common type of
pancreatic cancer, accounts for more than 85% of all pancreatic
cancer cases (4). Currently,
surgery resection is the primary treatment for long-term survival
of patients with pancreatic cancer. However, only approximately
10–20% of pancreatic cancer cases are suitable for surgery at the
time of diagnosis because of difficulty in early diagnosis, local
aggression and rapid progression (5). Despite considerable efforts to
improve early detection and treatments for patients with pancreatic
cancer, the overall prognosis remains unsatisfactory with 5-year
survival rate of lower than 10% (6–8).
Moreover, the causes of pancreatic cancer remain unknown, although
our knowledge of pancreatic cancer biology has progressed (9). Therefore, the mechanisms underlying
pancreatic cancer formation and progression must be elucidated to
develop effective therapeutic strategies for patients with this
malignancy.
MicroRNAs (miRNAs) constitute a group of endogenous,
noncoding and approximately 20–22 nucleotide-long RNAs that
function as post-transcriptional regulators (10). MiRNAs modulate messenger RNA (mRNA)
expression by base pairing to complementary sites in the
3′-untranslated regions (3′-UTRs) of target mRNAs, thereby inducing
the degradation of the target mRNAs or inhibiting the translation
of the target mRNAs (11). MiRNAs
regulate over 30% coding genes (11). Moreover, miRNAs participate in
various physiological processes, including cell proliferation,
cycle, apoptosis, differentiation, metabolism and development of
cells and pathological conditions, such as cardiovascular diseases,
neurological diseases and tumours (12–14).
Studies have reported that miRNAs are aberrantly expressed in
various kinds of human cancers, such as pancreatic cancer (15), colorectal cancer (16), lung cancer (17) and bladder cancer (18). Depending on the characteristic of
their target mRNAs, miRNAs may function as either oncogenes or
tumour suppressors and thus play important roles in the occurrence
and development of cancer (19).
Therefore, the expression and roles of miRNAs in tumourigenesis and
tumour development must be determined to elucidate the formation
and progression of human cancers and improve the diagnosis and
therapy of cancers.
Deregulation of miR-720 deregulation has been
reported in several types of human cancers (20–22).
Nevertheless, no specific studies have been conducted to reveal the
expression and biological roles of miR-720 in pancreatic cancer.
The present study aims to investigate the expression and roles of
miR-720 in pancreatic cancer and determine the underlying
regulatory mechanism.
Material and methods
Tissue samples
The present study was approved by the Ethics
Committee of Yongcheng City People's Hospital (Ethics approval
number: 20140037). Written informed consent was obtained from all
pancreatic cancer patients enrolled in this research. Twenty-three
pairs of pancreatic cancer tissues and matched adjacent normal
pancreatic tissues were obtained from patients who received surgery
resection at Yongcheng City People's Hospital between June 2014 to
February 2016. All tissues were obtained from patients prior to
chemotherapy or radiation therapy. Tissues were immediately frozen
and stored at −80°C prior to further processed.
Cell lines and cell culture
Human pancreatic cancer cell lines, Panc-1, Bxpc-3,
Sw1990, and Aspc-1, were purchased from Cell Bank of Type Culture
Collection of Chinese Academy of Sciences, Shanghai Institute of
Cell Biology (Shanghai, China). The human normal pancreatic cell
line (HPDE6c7) were obtained from American Type Culture Collection
(Manassas, VA, USA). All cells were grown in Dulbecco's modified
Eagle's medium (DMEM; Gibco, Thermo Scientific, Shanghai, China)
supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo
Scientific, Shanghai, China), 100 U/ml penicillin and 100 U/ml
streptomycin, and maintained at 37°C in a humidified incubator
containing 5% CO2.
Transfection
The miR-720 mimic and miRNA mimic negative control
(NC) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai,
China). CCND1 expression vector (pcDNA 3.1-CCND1) and pcDNA3.1
empty vector were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). For transfection, cells were seeded into 6-well
plates and cultured until reaching 70–80% confluence. Cell
transfection was performed using Lipofectamine 2000 (Invitrogen,
Grand Island, NY, USA) according to the manufacturer's protocol.
After incubation at 37°C with 5% CO2 for 6 h, the medium
was replaced with DMEM containing 10% FBS.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissues or cells using
TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA)
according to the manufacturer's instructions. For miR-720
expression analysis, reverse transcription was performed using the
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems,
Foster City, CA, USA). MiR-720 expression was examined using the
TaqMan MicroRNA PCR kit (Applied Biosystems, Foster City, CA, USA)
on an ABI 7300 PCR Thermal Cycler (Thermo Fisher Scientific,
Waltham, MA, USA). U6 served as an internal reference for miR-720
expression level. To quantify CCND1 mRNA level, total RNA was
converted into cDNA using a PrimeScript RT Reagent kit (Takara
Biotechnology, Co., Ltd., Dalian, China). Quantitative PCR was then
performed to examine CCND1 mRNA expression using the Power SYBR
Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA).
GAPDH was used as an internal control for CCND1 mRNA. The sequences
of the primers used for PCR were as follows: miR-720 forward,
5′-GCGTGCTCTCGCTGGGG-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6
forward, 5′-CTCGCTTCGGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′; CCND1 forward,
5′-CGGAGGACAACAAACAGATC-3′ and reverse, 5′-GGGTGTGCAAGCCAGGTCCA-3′;
and GAPDH forward, 5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse,
5′-GCCATCACGCCACAGTTTC-3′. Each sample was performed in triplicate
and relative expression was analyzed using the 2−ΔΔCt
method (23).
Cell counting kit 8 (CCK 8) assay
CCK8 (Dojindo Laboratories, Kumamoto, Japan) assay
was used to determine cell proliferation according to the
manufacturers instructions. In brief, cells were plated into
96-well plates at a density of 3×103 cells per well.
Cells were then transfected with miRNA mimics or plasmid, and
incubated at 37°C with 5% CO2 for 0, 24, 48, or 72 h. At
every time point, 10 µl CCK8 regent was added to the medium and the
cells were incubated for a further 4 h. Following incubation, the
absorbance at a wavelength of 450 nm was detected using an
enzyme-linked immunosorbent assay reader (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). Three independent experiments were
performed with three replicates in each.
Matrigel invasion assay
Cell invasion ability was determined using a 24-well
Matrigel-coated Transwell chamber with an 8-µm pore-size
polycarbonate membrane (BD Biosciences, San Jose, CA, USA). After
incubation 48 h, 1×105 transfected cells in 200 µl
FBS-free DMEM were seeded into the upper chamber, after which 500
µl DMEM with 10% FBS was added into the lower chamber. Cells were
then incubated at 37°C with 5% CO2 for 20 h.
Subsequently, cells on the upper side of the membranes were removed
slightly by the dry cotton swabs. Invasive cells were fixed in
methanol, stained with 0.5% crystal violet, photographed and
counted in five representative microscopic fields under a light
microscope (Olympus IX53; Olympus, Tokyo, Japan).
Bioinformatic analysis
Target genes of miR-720 were searched using the
TargetScan (http://www.targetscan.org) and miRBase
(http://www.mirbase.org/).
Luciferase reporter assay
The targeting relationship between miR-720 and
3′-UTR of CCND1 was detected by using luciferase reporter assay.
Luciferase reporter vector containing the wild-type
(pMIR-CCND1-3′-UTR WT) or mutant (pMIR-CCND1-3′-UTR MUT) 3′-UTR of
CCND1 were synthesized by GenePharma Co., Ltd. Cells were seeded
into 24-well plates at a density of 60–70% confluence and then
co-transfected with the pMIR-CCND1-3′-UTR WT or pMIR-CCND1-3′-UTR
MUT and miR-720 mimics or NC by using Lipofectamine 2000. 48 h
after co-transfection, cells were harvested and luciferase
activities were detected using the Dual-Luciferase assay system
(Promega, Madison, WI, USA), according to the manufacturer's
protocol. Firefly luciferase activities were normalized to Renilla
luciferase activities. Each assay was performed as three
replicates.
Western blotting
Total protein were extracted from tissues or cells
using radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China) supplemented with a
protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA).
Protein concentration was examined using a BCA Protein Assay kit
(Beyotime Biotechnology, Haimen, China). Equal quantities of
protein were separated by 10% SDS-PAGE and electrotransferred onto
PVDF membranes (EMD Millipore, Billerica, MA, USA). Subsequently,
the membranes were blocked by 5% non-fat milk in Tris-based
saline-Tween 20 (TBST) for 1 h at room temperature and blotted with
primary antibodies: mouse anti-human CCND1 monoclonal antibody
(sc-450; 1:1,000 dilution; Santa Cruz Biotechnology, CA, USA) or
mouse anti-human GAPDH monoclonal antibody (sc-47724; 1:1,000
dilution; Santa Cruz Biotechnology, CA, USA). After washing thrice
in TBST for 5 min, the membranes were incubated with a
corresponding horseradish peroxidase-conjugated secondary antibody
(sc-2005; 1:5,000 dilution; Santa Cruz Biotechnology, CA, USA) at
room temperature for 1 h. Finally, proteins were detected by
enhanced chemiluminescence (ECL) using a Pierce™ ECL Western
Blotting detection system (Thermo Fisher Scientific, Inc.,
Rockford, IL, USA), and the protein intensities were quantified
using Quantity One software (Bio-Rad Laboratories, Inc., Hercules,
CA, USA).
Statistical analysis
All statistical analyses were performed using
student's t test or one way ANOVA test with SPSS software (version
16; SPSS, Inc., Chicago, IL, USA). Data are presented as mean ±
standard deviation. P value less than 0.05 were considered
significant.
Results
miR-720 expression is downregulated in
pancreatic cancer tissues and cell lines
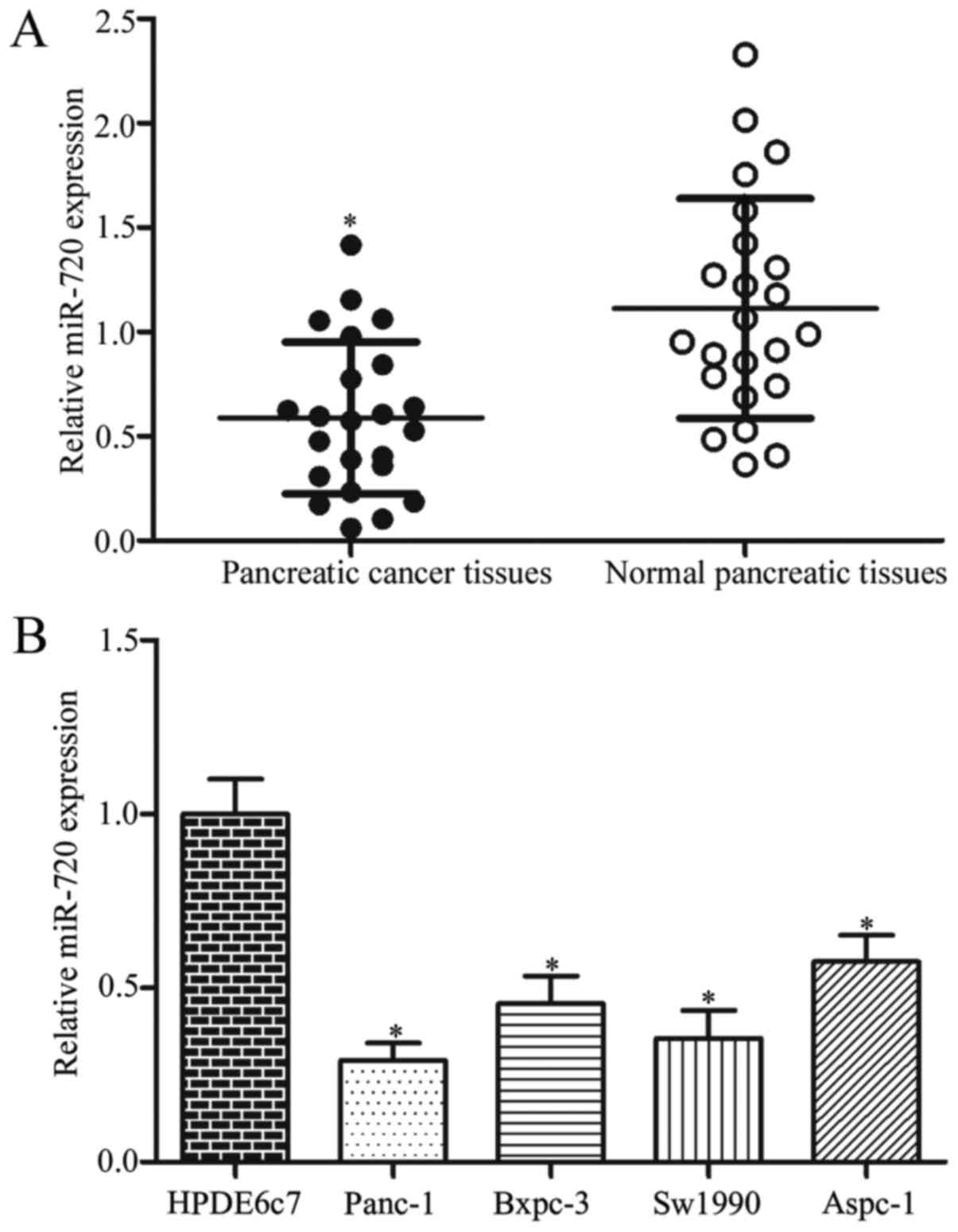
RT-qPCR analysis was performed in twenty-three pairs
of pancreatic cancer tissues and matched adjacent normal pancreatic
tissues to determine miR-720 expression. The results showed that
miR-720 was significantly downregulated in pancreatic cancer
tissues compared with that in their matched adjacent normal
pancreatic tissues (Fig. 1A;
P<0.05). Furthermore, miR-720 expression was detected in
pancreatic cancer cell lines (Panc-1, Bxpc-3, Sw1990, Aspc-1) and a
human normal pancreatic cell line (HPDE6c7). As shown in Fig. 1B, the expression levels of miR-720
decreased in pancreatic cancer cell lines than that in HPDE6c7
cells (P<0.05). The expression of miR-720 significantly
decreased in Panc-1 and Sw1990 cell lines, which were then selected
for subsequent experiments.
miR-720 inhibits the proliferation and
invasion of pancreatic cancer cells
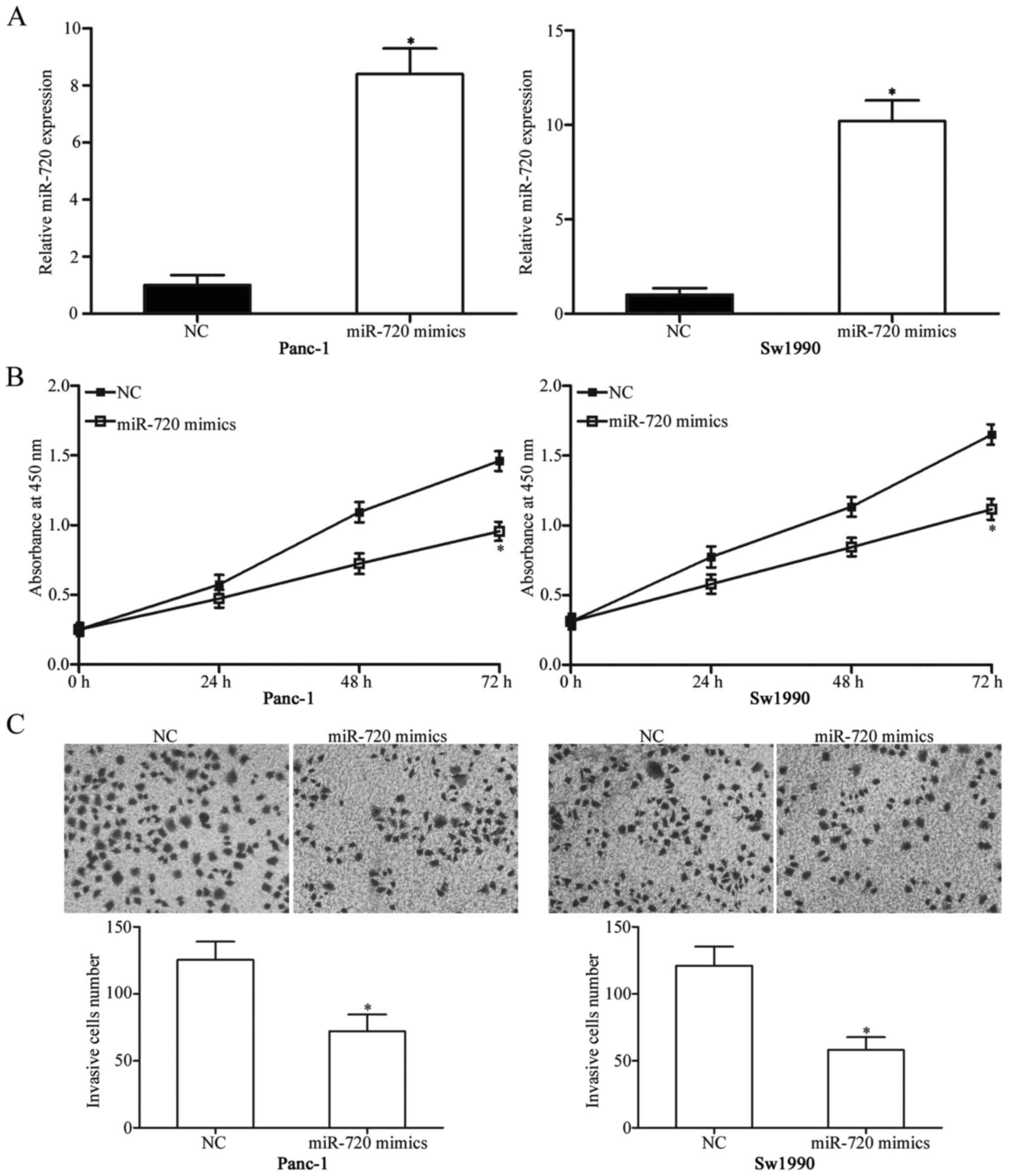
To delineate the potential roles of miR-720 in
pancreatic cancer, we firstly transfected miR-720 mimics into
Panc-1 and Sw1990 cells, which expressed relatively low miR-720
expression among four examined pancreatic cancer cell lines.
RT-qPCR analysis was conducted to determine transfection
efficiency. The results showed that miR-720 was markedly
upregulated in Panc-1 and Sw1990 cells after transfection with
miR-720 mimics (Fig. 2A;
P<0.05). CCK8 assay was used to examine the effect of miR-720
overexpression on pancreatic cancer cell proliferation. As shown in
Fig. 2B, upregulating miR-720
reduced the proliferation of Panc-1 and Sw1990 cells (P<0.05).
Moreover, we performed Matrigel invasion assay to assess the effect
of miR-720 on the invasion capacity of pancreatic cancer. Restoring
the miR-720 expression reduced the invasive capability in both
Panc-1 and Sw1990 cells (Fig. 2C;
P<0.05). Hence, miR-720 may function as a tumour suppressor in
pancreatic cancer.
miR-720 directly targets and mediates
CCND1 expression
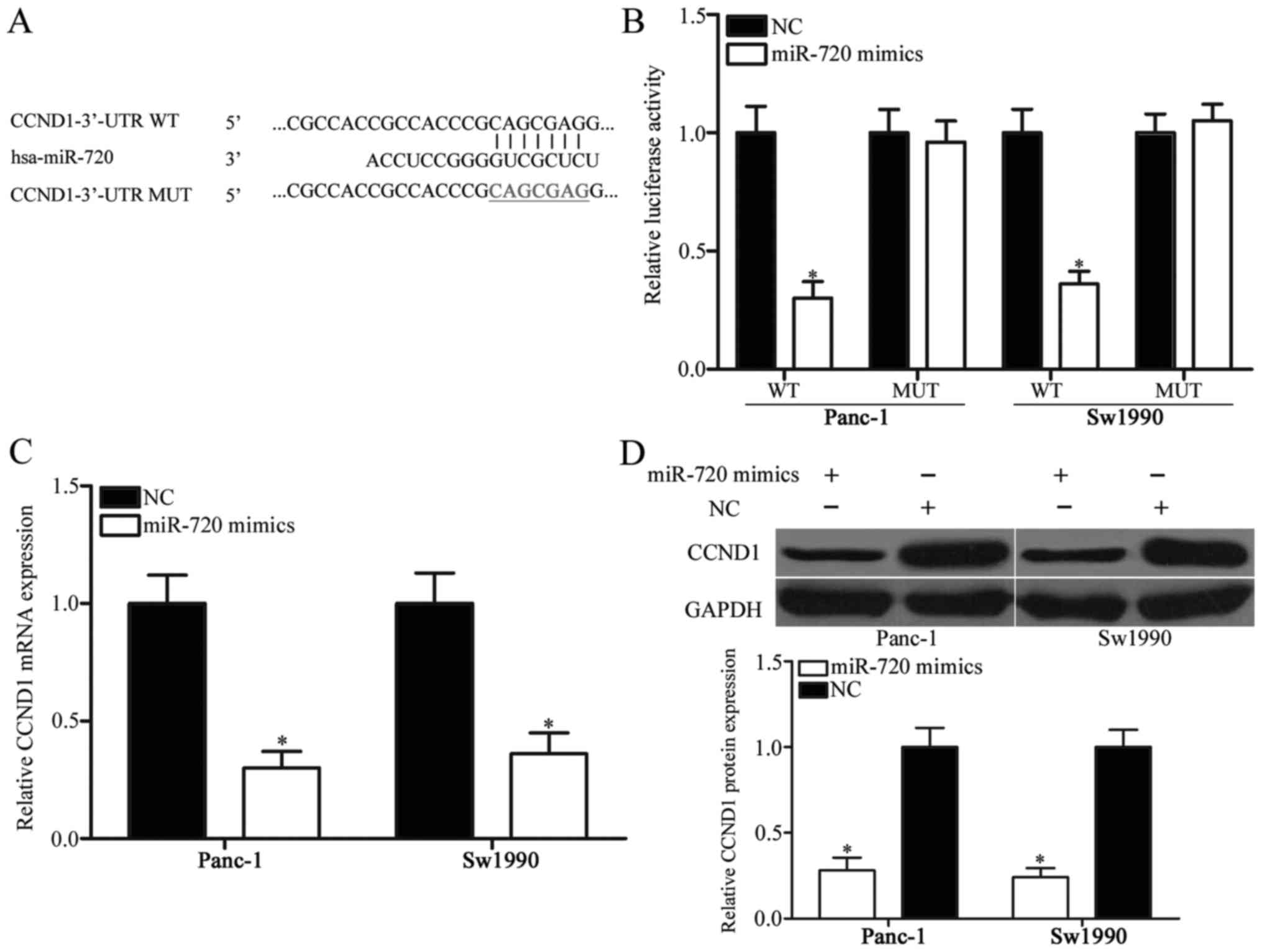
To investigate the molecular basis of miR-720 in
regulating pancreatic cancer, we screened the potential targets of
miR-720 by using bioinformatics analysis. We focused on CCND1 as
putative target (Fig. 3A) because
it plays important roles in pancreatic cancer initiation and
progression (24–26). Luciferase reporter assay was
performed in Panc-1 and Sw1990 cells transfected with miR-720
mimics or NC, along with pMIR-CCND1-3′-UTR WT or pMIR-CCND1-3′-UTR
MUT, to determine whether or not CCND1 is a direct target of
miR-720. As shown in Fig. 3B,
miR-720 introduction decreased the luciferase activity of the
wild-type CCND1 3′-UTR (P<0.05) but did not change of the
activity of the mutant CCND1 3′-UTR.
To determine whether miR-720 could regulate
endogenous CCND1 expression, we measured CCND1 mRNA and protein
expression levels in Panc-1 and Sw1990 cells transfected with
miR-720 mimics or NC. miR-720 overexpressing reduced the CCND1
expression in Panc-1 and Sw1990 cells at both mRNA (Fig. 3C; P<0.05) and protein (Fig. 3D; P<0.05) levels. Hence, miR-720
directly targets the 3′-UTR of CCND1 and downregulates CCND1 mRNA
and protein expression levels in pancreatic cancer cells.
CCND1 expression is upregulated and
negatively related to miR-720 expression in pancreatic cancer
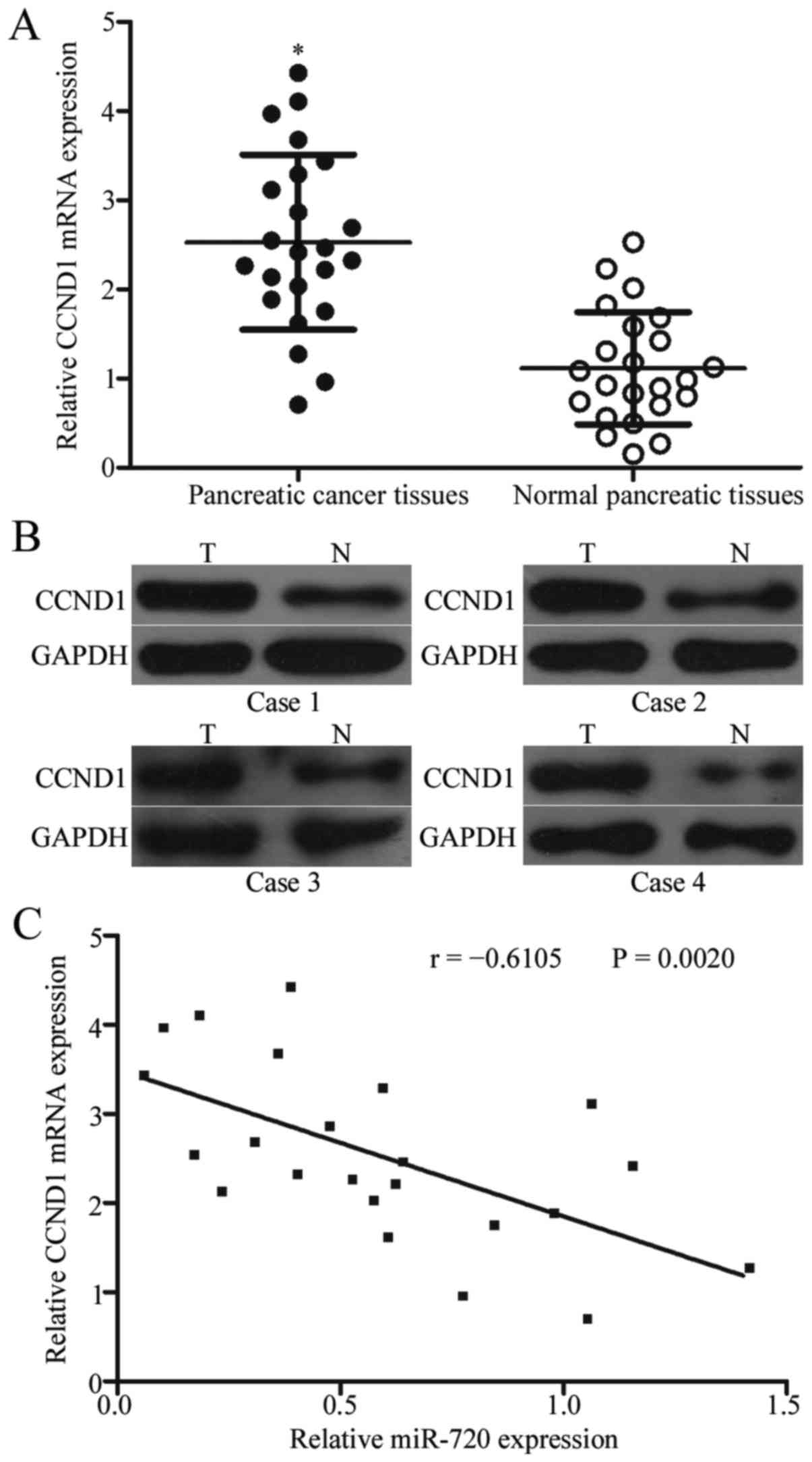
To further explore the functional significance of
miR-720 in pancreatic cancer, we evaluated its association with
CCND1 in pancreatic cancer tissues. RT-qPCR analysis was performed
to quantify CCND1 mRNA expression in 23 pairs of pancreatic cancer
tissues and matched adjacent normal pancreatic tissues. CCND1 mRNA
expression increased in pancreatic cancer tissues compared with
that in the adjacent normal pancreatic tissues (Fig. 4A; P<0.05). In addition, Western
blot analysis indicated that the protein expression level of CCND1
was significantly higher in pancreatic cancer tissues than that in
the adjacent normal pancreatic tissues (Fig. 4B; P<0.05). Furthermore,
Spearman's correlation analysis revealed that CCND1 mRNA was
inversely correlated with miR-720 expression in pancreatic cancer
tissues (Fig. 4C; r=-0.6105,
P=0.0020).
CCND1 reverses the tumour suppressive
roles of miR-720 in pancreatic cancer
To further identify whether the tumour-suppressing
roles of miR-720 in pancreatic cancer cell proliferation and
invasion were mediated by CCND1, we performed rescue experiments by
introducing pcDNA 3.1-CCND1 in the presence or absence of miR-720
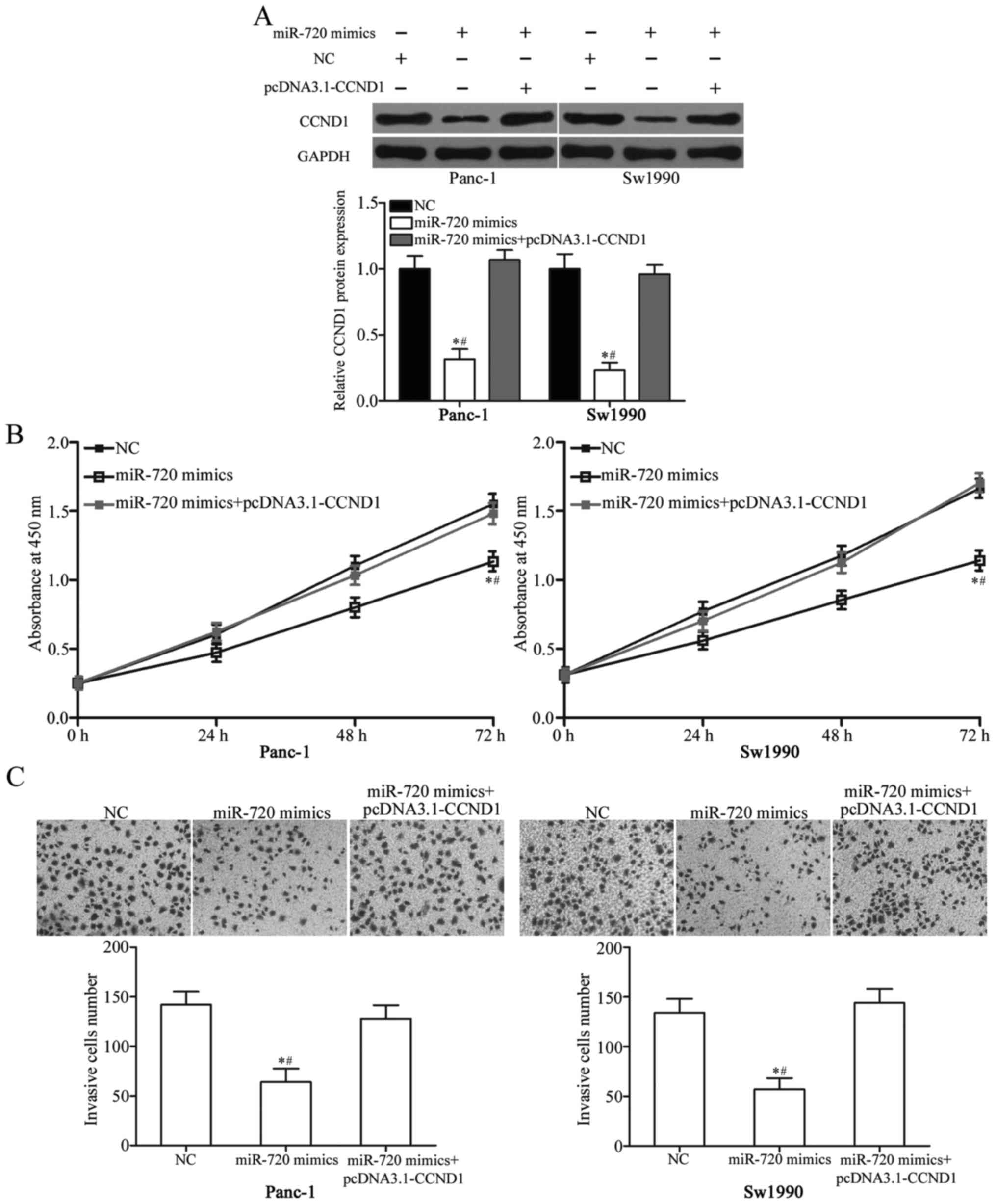
mimics in Panc-1 and Sw1990 cells. As shown in Fig. 5A, the ectopic expression of
miR-720-induced downregulation of CCND1 was rescued by transfection
of pcDNA 3.1-CCND1 (P<0.05). In addition, the increased
expression of CCND1 could partially abrogate the inhibitory effects
of miR-720 on the proliferation and invasion of pancreatic cancer
cells (Fig. 5B and C; P<0.05).
These findings suggested that miR-720 serves as a tumour suppressor
in pancreatic cancer, at least in part, by negatively regulating
CCND1.
Discussion
Deregulation of miRNA might contribute to the
occurrence and progression of many cancer types, including
pancreatic cancer (27–29). A large number of miRNAs play
crucial roles in the proliferation, apoptosis, cycle, metastasis
and angiogenesis of pancreatic cancer cells; hence, suggesting that
miRNA could be used in pancreatic cancer diagnosis and treatment
(30–32). In the present study, miR-720 was
downregulated in pancreatic cancer tissues and cell lines.
Resumption of miR-720 expression suppressed the proliferation and
invasion of pancreatic cancer cells in vitro. Furthermore,
CCND1 was validated as a direct and functional target of miR-720 in
pancreatic cancer. These results indicated that miR-720 acts as a
tumour suppressor in pancreatic cancer and thus can be used to
treat patients with pancreatic cancer.
Dysregulation of miR-720 has been reported in
several types of human cancer. For example, miR-720 expression
decreased in breast cancer tissues and was found to be
significantly correlated with lymph node metastasis (20). A previous study indicated that
miR-720 was upregulated in cervical cancer tissues compared with
that in normal cervical tissues (21). In colorectal cancer, the expression
levels of miR-720 was higher in tumour tissues than that in
corresponding normal-appearing tissues. High miR-720 expression is
correlated with tumour size, tumour-node-metastasis stage,
lymphatic metastasis and distant metastasis and could lead to poor
5-year overall survival rate in patients with colorectal cancer
(22). Additionally, miR-720
expression increased remarkably in the serum of patients with
colorectal cancer compared with that in healthy patients. The
expression level of miR-720 is also associated with male gender and
lymph node metastasis in patients with colorectal cancer. Thus,
miR-720 expression exhibits tissue specificity, and may be a
prognostic marker in these specific types of cancer.
Several studies have identified the functions of
miR-720 in cancer progression. For instance, miR-720 served as a
tumour suppressor in breast cancer by inhibiting tumour cell
metastasis both in vitro and in vivo (20). Nevertheless, miR-720 was validated
as an oncogene in tumourigenesis and development. Tang et al
reported that restoration expression of miR-720 decreased
E-cadherin expression, increased vimentin expression and promoted
cervical cancer cell migration (21). Wang et al found that miR-720
overexpression promoted cell proliferation, colony formation
ability and metastasis of colorectal cancer (22). These conflicting findings indicate
that miR-720 exhibits tissue specificity, which could be explained
by the non-ideal complementarity of the interactions between miRNA
and their direct target genes.
The targets of miR-720 include TWIST1 (20) in breast cancer, Rab35 (21) in cervical cancer, StarD13 in
colorectal cancer (22) and GATA3
in macrophages (33). The present
study demonstrated that CCND1 is a direct functional target of
miR-720 in pancreatic cancer, as supported through several lines of
evidence. Firstly, bioinformatics analysis predicted that CCND1 is
a potential target of miR-720. Secondly, miR-720 overexpression
decreased CCND1 3′-UTR luciferase activity, an effect abolished by
the mutation of the miR-720 seed region. Thirdly, miR-720
negatively regulated CCND1 expression in pancreatic cancer cells at
mRNA and protein levels. Furthermore, CCND1 was upregulated in
pancreatic cancer tissues and inversely correlated with miR-720
expression. The enforced expression of CCND1 could also partially
abrogate the inhibitory effects of miR-720 on pancreatic cancer
cells. These results demonstrate that CCND1 is a direct target gene
of miR-720 in pancreatic cancer.
The CCND1 gene, located on chromosome 11q13, is a
well-known oncogene that is frequently overexpressed in various
kinds of human cancers, such as breast cancer (34), lung cancer (35), gastric cancer (36) and bladder cancer (37). In pancreatic cancer, CCND1 was
overexpressed in tumour tissues; the expression of this gene is
significantly correlated with extent of differentiation and poor
prognosis (38,39). CCND1 also plays important roles in
pancreatic cancer formation and progression. Functional assays
demonstrated that CCND1 knockdown inhibited cell proliferation,
soft agar colony formation, metastasis, metabolism in vitro
and cell growth in vivo (24–26).
Moreover, CCND1 underexpression increased the chemosensitivity of
pancreatic cancer cells to 5-fluorouracil, 5-fluoro-2′-deoxyuridine
and mitoxantrone by downregulating multiple chemoresistance genes
(40). Hence, regulating
miR-720/CCND1 axis could be a novel and effective therapeutic
strategy for inhibiting the rapid growth and metastasis of
pancreatic cancer.
In conclusion, miR-720 acted as a tumour suppressor
in pancreatic cancer by directly targeting CCND1. The precise
mechanism of miR-720/CCND1 axis in pancreatic cancer must be
determined to elucidate the pathogenesis of pancreatic cancer and
develop new treatment approach for patients with pancreatic
cancer.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu
L and He J: Report of incidence and mortality in China cancer
registries, 2009. Chin J Cancer Res. 25:10–21. 2013.PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Modolell I, Guarner L and Malagelada JR:
Vagaries of clinical presentation of pancreatic and biliary tract
cancer. Ann Oncol. 10 Suppl 4:S82–S84. 1999. View Article : Google Scholar
|
|
5
|
Cartwright T, Richards DA and Boehm KA:
Cancer of the pancreas: Are we making progress? A review of studies
in the US Oncology Research Network. Cancer Control. 15:308–313.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paulson AS, Cao HS Tran, Tempero MA and
Lowy AM: Therapeutic advances in pancreatic cancer.
Gastroenterology. 144:1316–1326. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ansari D, Gustafsson A and Andersson R:
Update on the management of pancreatic cancer: Surgery is not
enough. World J Gastroenterol. 21:3157–3165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Puleo F, Maréchal R, Demetter P, Bali MA,
Calomme A, Closset J, Bachet JB, Deviere J and Van Laethem JL: New
challenges in perioperative management of pancreatic cancer. World
J Gastroenterol. 21:2281–2293. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greer JB, Lynch HT and Brand RE:
Hereditary pancreatic cancer: A clinical perspective. Best Pract
Res Clin Gastroenterol. 23:159–170. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Szafranska AE, Davison TS, John J, Cannon
T, Sipos B, Maghnouj A, Labourier E and Hahn SA: MicroRNA
expression alterations are linked to tumorigenesis and
non-neoplastic processes in pancreatic ductal adenocarcinoma.
Oncogene. 26:4442–4452. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Q, Wei L, Guan X, Wu Y, Zou Q and Ji
Z: Briefing in family characteristics of microRNAs and their
applications in cancer research. Biochim Biophys Acta.
1844:191–197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li M, Fu W, Wo L, Shu X, Liu F and Li C:
miR-128 and its target genes in tumorigenesis and metastasis. Exp
Cell Res. 319:3059–3064. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaplan BB, Kar AN, Gioio AE and Aschrafi
A: MicroRNAs in the axon and presynaptic nerve terminal. Front Cell
Neurosci. 7:1262013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chang W, Liu M, Xu J, Fu H, Zhou B, Yuan T
and Chen P: MiR-377 inhibits the proliferation of pancreatic cancer
by targeting Pim-3. Tumour Biol. 37:14813–14824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoo HI, Kim BK and Yoon SK:
MicroRNA-330-5p negatively regulates ITGA5 expression in human
colorectal cancer. Oncol Rep. 36:3023–3029. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W,
Liu Z and Huang JA: MicroRNA-205 targets SMAD4 in non-small cell
lung cancer and promotes lung cancer cell growth in vitro and in
vivo. Oncotarget. 8:30817–30829. 2017.PubMed/NCBI
|
|
18
|
Zhou Y, Wu D, Tao J, Qu P, Zhou Z and Hou
J: MicroRNA-133 inhibits cell proliferation, migration and invasion
by targeting epidermal growth factor receptor and its downstream
effector proteins in bladder cancer. Scand J Urol. 47:423–432.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bhardwaj A, Singh S and Singh AP:
MicroRNA-based Cancer Therapeutics: Big Hope from Small RNAs. Mol
Cell Pharmacol. 2:213–219. 2010.PubMed/NCBI
|
|
20
|
Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou
X, Zhang ZJ, Peng YH, Yang YZ and Yun JP: miR-720 inhibits tumor
invasion and migration in breast cancer by targeting TWIST1.
Carcinogenesis. 35:469–478. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tang Y, Lin Y, Li C, Hu X, Liu Y, He M,
Luo J, Sun G, Wang T, Li W and Guo M: MicroRNA-720 promotes in
vitro cell migration by targeting Rab35 expression in cervical
cancer cells. Cell Biosci. 5:562015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X, Kuang Y, Shen X, Zhou H, Chen Y,
Han Y, Yuan B, Zhou J, Zhao H, Zhi Q and Xue X: Evaluation of
miR-720 prognostic significance in patients with colorectal cancer.
Tumour Biol. 36:719–727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu Y, Liu T and Gao J: Effects of
antisense cyclin D1 expressing vector on the cell growth and
apoptosis of pancreatic carcinoma. Zhonghua Bing Li Xue Za Zhi.
27:348–351. 1998.(In Chinese). PubMed/NCBI
|
|
25
|
Yan L, Wang Y, Wang ZZ, Rong YT, Chen LL,
Li Q, Liu T, Chen YH, Li YD, Huang ZH and Peng J: Cell motility and
spreading promoted by CEACAM6 through cyclin D1/CDK4 in human
pancreatic carcinoma. Oncol Rep. 35:418–426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee Y, Ko D, Min HJ, Kim SB, Ahn HM, Lee Y
and Kim S: TMPRSS4 induces invasion and proliferation of prostate
cancer cells through induction of Slug and cyclin D1. Oncotarget.
7:50315–50332. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mizuguchi Y, Takizawa T, Yoshida H and
Uchida E: Dysregulated miRNA in progression of hepatocellular
carcinoma: A systematic review. Hepatol Res. 46:391–406. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu H, Wang H, Liu X and Yu T: miR-1271
inhibits migration, invasion and epithelial-mesenchymal transition
by targeting ZEB1 and TWIST1 in pancreatic cancer cells. Biochem
Biophys Res Commun. 472:346–352. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren ZG, Dong SX, Han P and Qi J: miR-203
promotes proliferation, migration and invasion by degrading SIK1 in
pancreatic cancer. Oncol Rep. 35:1365–1374. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Trehoux S, Lahdaoui F, Delpu Y, Renaud F,
Leteurtre E, Torrisani J, Jonckheere N and Van Seuningen I:
Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by
targeting the MUC1 mucin in pancreatic cancer cells. Biochim
Biophys Acta. 1853:2392–2403. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xia X, Zhang K, Cen G, Jiang T, Cao J,
Huang K, Huang C, Zhao Q and Qiu Z: MicroRNA-301a-3p promotes
pancreatic cancer progression via negative regulation of SMAD4.
Oncotarget. 6:21046–21063. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li L, Li B, Chen D, Liu L, Huang C, Lu Z,
Lun L and Wan X: miR-139 and miR-200c regulate pancreatic cancer
endothelial cell migration and angiogenesis. Oncol Rep. 34:51–58.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhong Y and Yi C: MicroRNA-720 suppresses
M2 macrophage polarization by targeting GATA3. Biosci Rep. 36:pii:
e003632016. View Article : Google Scholar
|
|
34
|
Li X, Huo X, Li W, Yang Q, Wang Y and Kang
X: Genetic association between cyclin D1 polymorphism and breast
cancer susceptibility. Tumour Biol. 35:11959–11965. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Betticher DC, Heighway J, Hasleton PS,
Altermatt HJ, Ryder WD, Cerny T and Thatcher N: Prognostic
significance of CCND1 (cyclin D1) overexpression in primary
resected non-small-cell lung cancer. Br J Cancer. 73:294–300. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kumari S, Puneet, Prasad SB, Yadav SS,
Kumar M, Khanna A, Dixit VK, Nath G, Singh S and Narayan G: Cyclin
D1 and cyclin E2 are differentially expressed in gastric cancer.
Med Oncol. 33:402016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kopparapu PK, Boorjian SA, Robinson BD,
Downes M, Gudas LJ, Mongan NP and Persson JL: Expression of cyclin
d1 and its association with disease characteristics in bladder
cancer. Anticancer Res. 33:5235–5242. 2013.PubMed/NCBI
|
|
38
|
Gansauge S, Gansauge F, Ramadani M, Stobbe
H, Rau B, Harada N and Beger HG: Overexpression of cyclin D1 in
human pancreatic carcinoma is associated with poor prognosis.
Cancer Res. 57:1634–1637. 1997.PubMed/NCBI
|
|
39
|
Li YJ and Ji XR: Relationship between the
expression of beta-cat, cyclin D1 and c-myc and the occurance and
biological behavior of pancreatic cancer. Zhonghua Bing Li Xue Za
Zhi. 32:238–241. 2003.(In Chinese). PubMed/NCBI
|
|
40
|
Kornmann M, Danenberg KD, Arber N, Beger
HG, Danenberg PV and Korc M: Inhibition of cyclin D1 expression in
human pancreatic cancer cells is associated with increased
chemosensitivity and decreased expression of multiple
chemoresistance genes. Cancer Res. 59:3505–3511. 1999.PubMed/NCBI
|