Introduction
Lactalbumin α has important nutritional and
physiological value: i) It contains numerous essential amino acids,
especially tryptophan, and benefits cognitive function and muscle
protein synthesis; ii) it interacts with
β-1,4-galactosyltransferase to promote lactose synthesis, and
thereby facilitates milk production and secretion (1); iii) it binds divalent cations like Ca
and Zn and facilitates mineral absorption (1); iv) it exerts strong gastroprotective
activity via stimulating mucus metabolism and increasing thickness
of the mucus gel layer; v) its digested peptides have
immunostimulatory and antibacterial properties, and thus possibly
protect against infection (1) and
iv) its folding variants can induce tumor apoptosis and remedy
cutaneous warts.
In contrast with its abundancy in human milk (25–35%
of total protein) (2), lactalbumin
α only accounts for 2–5% of the total protein in cow milk (1). In addition, endogenous cow
lactalbumin α is one of the major allergens for humans. In order to
increase the concentration of human lactalbumin α in cow milk,
genetically modified cows expressing human lactalbumin α have been
developed with the highest content up to 1.55 mg/ml in milk
(3). Subsequently, previous
studies have investigated the effect overexpression or transgenesis
of human lactalbumin α on the protein expression profile and milk
composition in cows, as well as the environmental risks.
Reportedly, overexpression of human lactalbumin α in cows does not
affect milk composition (4). It
has been determined that 166 proteins in the milk fat globule
membrane (MFGM) were specifically expressed only in cows
overexpressing human lactalbumin α and this was different from the
cloned non-transgenic cows and conventionally bred normal cows
(5). A previous study performed a
functional analysis that revealed that the up and downregulated
proteins in MFGM of cows overexpressing human lactalbumin α were
not associated with a specific biological pathway or function and
that human lactalbumin α overexpression had no deleterious effects
on the cattle mammary gland (5).
Additionally, the transgene of human lactalbumin α (LALBA)
cannot be detected in gut microbial DNA and thus has no influence
on microbial communities in cow gut or soil. However, the effect of
transgenic modification on the metabolomic profile in cow serum
remains to be elucidated. As somatic cell nuclear transfer was
performed during the genetic modification in a previous study
(3), LALBA was expressed in
all the somatic cells of the transgenic cows, including blood
cells. The presence of non-inherent LALBA may abnormally
regulate gene expression via endogenous microRNAs or transcription
factors and thus change the metabolomic profile of cow serum. This
is the hypothesis and rationale behind the present study.
Metabolomics has been successfully used to
investigate genetically modified organisms like transgenic rat
models of various diseases and genetically engineered goats that
produce human lysozyme (hLZ) in their milk (6). It is of note that when the serum
metabolomics of hLZ goats (lactating does) and controls were
compared, one metabolite was significantly different (6). The present study characterized and
compared serum metabolomic profiles in cows carrying and expressing
human LALBA (LALBA cows) and non-LALBA cows.
The findings of the present study may indirectly reflect the health
condition of the LALBA cows and provide complement evidence
for the future use of genetic modification in cows. The
humanization of bovine milk requires additional investigation of
multiple transgenesis in future studies, and the findings of the
present study may also contribute to the humanization of bovine
milk.
Materials and methods
Transgenic cloning of Chinese Holstein
cow
The protocol of the present study was approved by
the Institutional Animal Care and Use Committee of the Chinese
Academy of Inspection and Quarantine (Beijing, China). The
transgenic cloned Chinese Holstein cow (F0 generation, female) was
provided by the China Agricultural University (Beijing, China),
which expressed and characterized the bioactive recombinant human
lactalbumin α in cow milk (3). It
is of note that only human lactalbumin α was genetically
overexpressed in the cow referenced in Wang et al (3); however, human LALBA was
expressed in all the somatic cells of this cow. Both the
LALBA and non-LALBA cows used in the present study
were the offspring of the Wang et al transgenic cow.
LALBA and non-LALBA cows
The primary generation of transgenic cloned cows was
defined as the F0 generation (1 female). Following superovulation,
the ovum of F0 generation (female) was inseminated with the sperm
from one healthy male cattle. Embryos were transferred to ten
recipient cows, and a healthy F1 generation (male) was selected,
which expressed LALBA. The sperm from the F1 generation
(male) was used to artificially inseminate with the ova in 20
normal cows during their child-bearing periods, which were similar
in genetic background, age and feeding conditions. In this way,
LALBA and non-LALBA cows (F2 generation, female) were
generated, which were identified by using polymerase chain reaction
(PCR). The processes of superovulation, embryo transfer, and
artificial insemination were performed as previously described
(7).
To eliminate genetic background noise and reduce the
influence of embryo transfer and cloning process, only half-sibs
(F2 generation, female) with the same father (F1 generation, male)
were used in the present study. All cows in the present study were
similar in genetic background, sex (female) and age (~22 months),
and were housed under same conditions (temperature of 14°C and 60%
relative humidity) and had the same diet. Additionally, all the
cows used were not lactating and none of them were pregnant.
DNA extraction and PCR
PCR was performed to identify the LALBA and
non-LALBA cows. Total DNA was extracted from 5 ml cow blood
samples (namely, pellets following centrifugation at 3,500 × g for
30 min at 4°C) by using GenElute Blood Genomic DNA kit
(Sigma-Aldrich, Merck Millipore, Darmstadt, Germany). The 25 µl PCR
system consisted of 12.5 µl 2X Mix solution including buffer, dNTPs
and rTaq polymerase (Takara Biotechnology Co., Ltd., Dalian,
China), 1 µl forward primer (AGAGAGCACAGTGTTTGG; Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and 1 µl reverse primer
(GTCCAGCAGGTAAGTAAGG; Invitrogen; Thermo Fisher Scientific, Inc.),
1 µl DNA sample, and 9.5 µl ddH2O. The PCR conditions
were 95°C for 10 min then 30 cycles (95°C, 30 sec; 60°C, 30 sec;
72°C, 30 sec) and extension at 72°C for 30 min. Furthermore, PCR
products were electrophoresed on a 1.5% agarose gel (Takara
Biotechnology Co., Ltd.).
RNA extraction, reverse transcription
(RT)-PCR and sequencing
RT-PCR and sequencing were performed to validate the
transcription of LALBA in LALBA cows. Total RNA was
extracted from cow blood samples by using Blood RNA mini kit
(Qiagen, Hilden, Germany), and reverse transcription was performed
using a cDNA synthesis kit (Tiangen Biotech Co., Ltd., Beijing,
China). Then, PCR was conducted and cDNA was amplified using Taq
PCR Mater Mix kit (Qiagen), forward primer
(5′-GGTTCAGTGGTAGACAATCG-3′; Invitrogen; Thermo Fisher Scientific,
Inc.), and reverse primer (5′-GGTAAGGAGAAGGAGGATGG-3′; Invitrogen;
Thermo Fisher Scientific, Inc.). The conditions of PCR and
electrophoresis were as aforementioned and the corresponding
products were sequenced. Following alignment, the sequences of
these products were consistent with that of LALBA,
confirming the transcription of LALBA in LALBA
cows.
Serum samples
Blood samples (5 ml) were obtained from the jugular
vein of 6 LALBA cows (LALBA group) and 6
non-LALBA cows (non-LALBA group) at 10:00 am after
the animals were fasted for 10 h. After storing the blood at 4°C
for 12 h, blood samples were centrifuged at 3,500 × g for 30 min at
4°C. The supernatant was used as serum samples, which were
maintained at −80°C until processing.
Serum biochemistry indexes
In order to detect the effect of LALBA on the
blood chemistry of cows, serum biochemistry indexes were
quantified, including potassium, sodium, chlorine, calcium,
phosphorus, glucose, cholesterol, albumin, total bilirubin,
creatine phosphokinase, glutamyl transpeptidase, blood urea
nitrogen, alanine aminotransferase and creatinine. These indexes
were detected according to the protocols recommended by the
International Federation of Clinical Chemistry (IFCC) (8).
Metabolite detection
Sample preparation, metabolite extraction and
metabolite detection were performed by the SJTU-Metabolon Joint
Metabolomics Laboratory (Metabolon Inc., Durham, NC, USA; project
no. SJTUX-02-14VW) based on a global unbiased platform. The
metabolite detection, data processing, metabolite quantitation and
corresponding instrumentation have been were performed as
previously described (9). Briefly,
the samples were prepared by using Hamilton MicroLab STAR system
and the resulting extract was divided and prepared by using
TurboVap (Zymark, Palo Alto, CA, USA) and vacuum drying machine for
the following instrument: i) Ultra-Performance Liquid
Chromatography-Tandem Mass Spectrometry (UPLC-MS/MS) with positive
ion mode electrospray ionization (Waters Acquity UPLC and
Thermo-Finnigan LTQ mass spectrometer; scan range, 80–1000 m/z);
ii) UPLC-MS/MS with negative ion mode electrospray ionization; iii)
Liquid Chromatography (LC) polar platform; iv) Gas
Chromatography-Mass Spectroscopy (GC-MS, Thermo-Finnigan Trace DSQ
fast-scanning single-quadrupole mass spectrometer; scan range,
50–750 m/z). Data were extracted using Metabolon's hardware and
software (10) constructed via
Microsoft's NET technologies and compounds were identified based on
Metabolon library recording m/z, retention time/index, and
chromatographic data of more than 3,300 molecules.
Principal component analysis
(PCA)
As an unsupervised clustering approach, PCA reduces
the dimension of data based on orthogonal transformation,
generating principle components (11). In the present study, SIMCA-P
version 12.0 (Umetrics AB, Umea, Sweden) was used to perform the
PCA.
Hierarchical clustering analysis
As an unsupervised method for clustering data,
hierarchical clustering may reveal the large-scale differences
between groups based on Euclidean distance. Hierarchical clustering
analysis was conducted by using pheatmap package in R (version
0.7.7; R Core Team, Vienna, Austria) (12).
Altered metabolite screening
In order to generate metabolite levels, peaks were
quantified using area-under-the-curve. The metabolite levels were
log10 transformed, while missing values were imputed by
using the minimum observed value for each compound. The Welch's
two-sample t-test (13) was used
to identify significantly altered metabolites, namely, the
metabolites that significantly differed between LALBA and
non-LALBA groups. The threshold criteria used for this
analysis were P<0.05 and fold-change ≠ 1.
Random forest analysis
As a supervised classification approach based on
decision trees (14), random
forest analysis is a valuable statistical tool for compound
classification (15). Random
forest has several advantages: i) It makes no parametric
assumptions; ii) variable selection is not required; iii) it does
not overfit; iv) it is invariant to transformation and v) it is
fairly easy to implement in R. The present study used the
randomForest package (16) to
perform random forest analysis. To identify the metabolites making
a large contribution to classification, parameter ‘variable
importance’ was computed, and random forest analysis provides an
‘importance’ rank ordering of metabolite based on mean decrease
accuracy.
Pathway analysis
The significantly altered metabolites were mapped
onto general biological pathways using the Pathview software
package (version 1.8.0; bioconductor.org/packages/release/bioc/html/pathview.html)
(17) in R, generating the
potential pathways affected by LALBA transgenesis.
Results
LALBA and non-LALBA cows
The presence of LALBA and LALBA mRNA
in LALBA cows was confirmed by using PCR (Fig. 1A) and RT-PCR (Fig. 1B) and the quantity of LALBA
and LALBA mRNA in the 6 LALBA cows were similar
(Fig. 1).
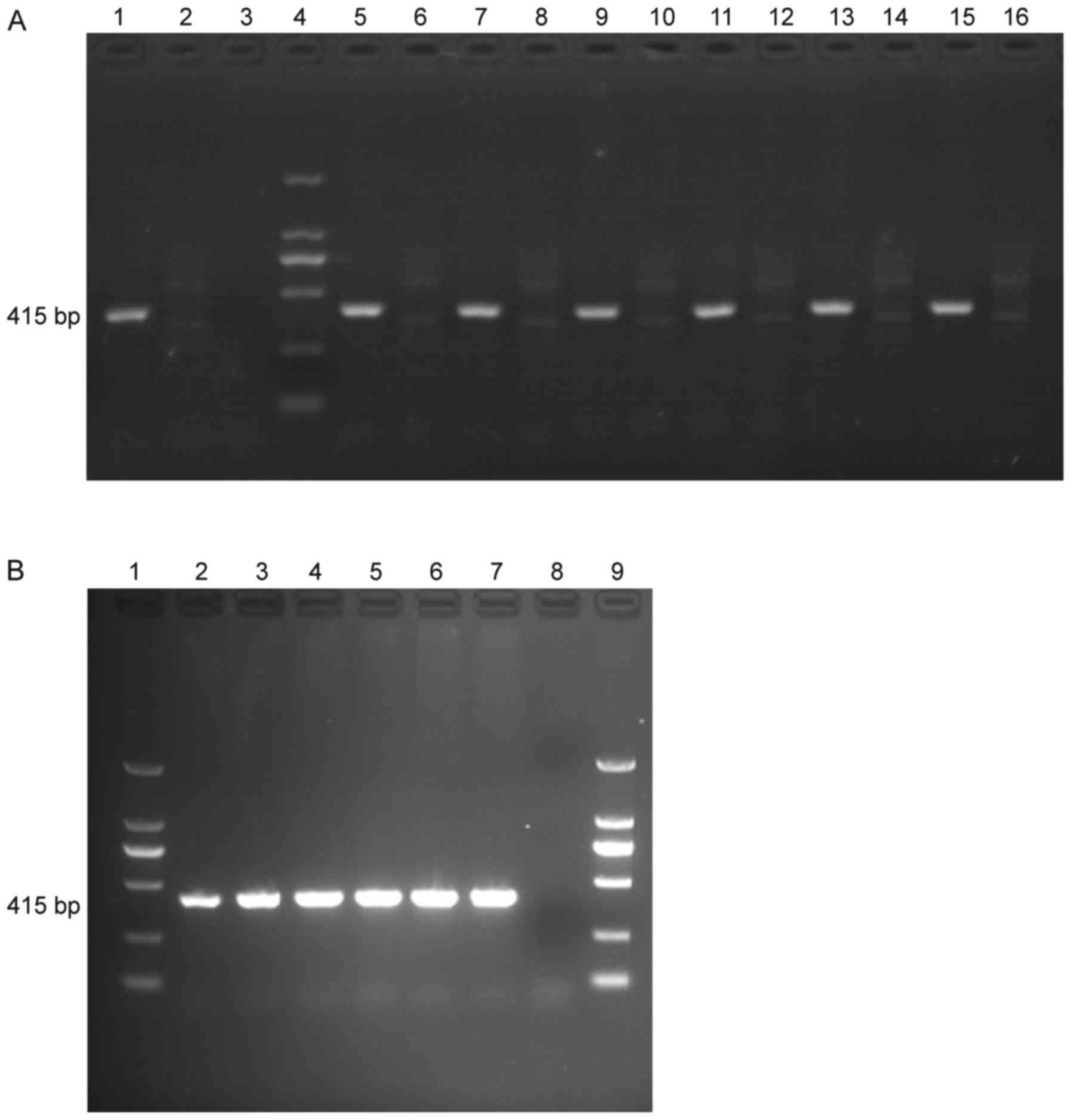 | Figure 1.PCR and RT-PCR products of
LALBA and non-LALBA cows. (A) PCR products. 1,
positive control; 2, negative control; 3, blank control; 4, DL2000
marker; 5, 7, 9, 11, 13, 15, samples from LALBA cows; 6, 8,
10, 12, 14, 16, samples from non-LALBA cows. (B) RT-PCR
products. 1, DL2000 marker; 2, 3, 4, 5, 6, 7, samples from
LALBA cows; 8, blank control; 9, DL2000 marker.
LALBA, human lactalbumin α; PCR, polymerase chain reaction;
RT, reverse transcription. |
Serum biochemistry indexes
Although differences of some serum biochemistry
indexes occurred between LALBA and non-LALBA cows,
all of the serum biochemistry indexes of LALBA and
non-LALBA cows were within the previously established normal
range (18) for healthy cows
(Table I).
 | Table I.Serum biochemistry indexes of
LALBA and non-LALBA cows. |
Table I.
Serum biochemistry indexes of
LALBA and non-LALBA cows.
| Index | Normal
rangea | LALBA |
non-LALBA |
|---|
| K, mmol/l | 3.9–5.8 |
4.07±0.47 |
4.84±0.53 |
| Na, mmol/l | 132–152 |
140.95±3.46 |
141.55±5.57 |
| Cl, mmol/l | 97–111 |
104.17±3.79 |
103.70±3.96 |
| Ca, mmol/l | 2.1–2.67 |
2.51±0.17 |
2.49±0.14 |
| P, mmol/l | 1.3–2.65 |
1.82±0.48 |
2.08±0.15 |
| GLU, mmol/l | 2.5–6.0 |
4.24±0.10 |
4.12±0.25 |
| CHO, mmol/l | 1.58–5.9 |
2.87±1.24 |
3.22±0.65 |
| ALB, g/l | 25–38 |
34.85±1.75 |
34.83±2.49 |
| TBIL, µmol/l | 0–8.6 |
5.58±1.05 |
7.08±1.33 |
| CK, U/l | 76–376 |
197.33±67.03 |
168.83±164.08 |
| GGT, U/l | 9–39 |
19.00±5.06 |
23.17±4.80 |
| BUN, mmol/l | 1.4–15.7 |
3.54±0.49 |
4.58±0.20 |
| ALT, U/l | 11–40 |
30.50±5.44 |
22.00±6.67 |
| CRE, µmol/l | 55–130 |
119.97±10.92 |
128.30±5.30 |
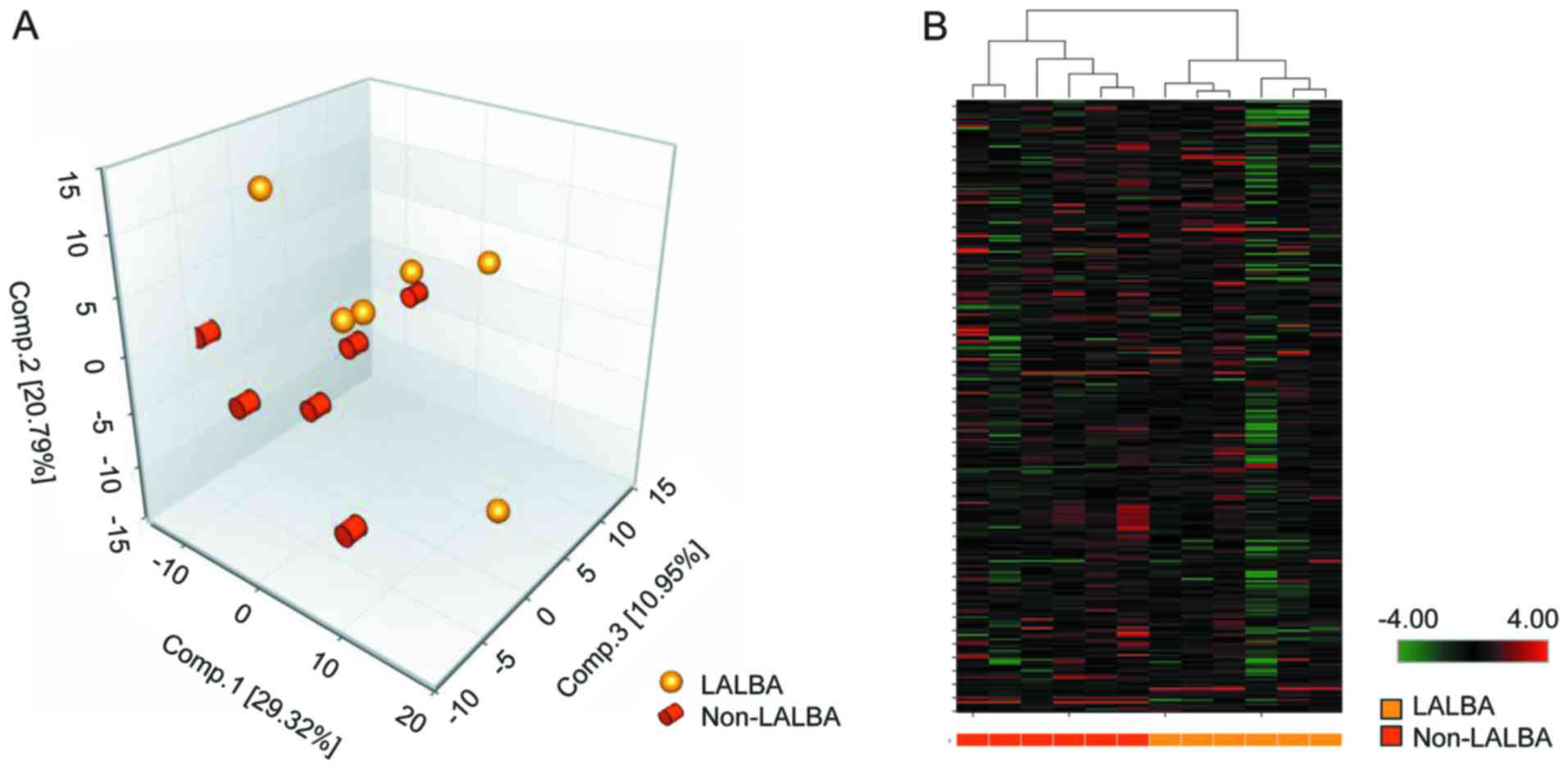
PCA and hierarchical clustering
analysis based on detected metabolites
Based on UPLC-MS/MS, LC and GC-MS platforms, a total
of 273 compounds with known identity (metabolites) were detected in
cow the extracted serum. PCA (Fig.
2A) and hierarchical clustering analysis (Fig. 2B) revealed a distinct separation
between LALBA and non-LALBA groups. In PCA, the 12
samples could be clearly classified into 2 groups (LALBA and
non-LALBA), and 5 samples from the two groups were closely
related (Fig. 2A), which may be
due to the small sample size of each group.
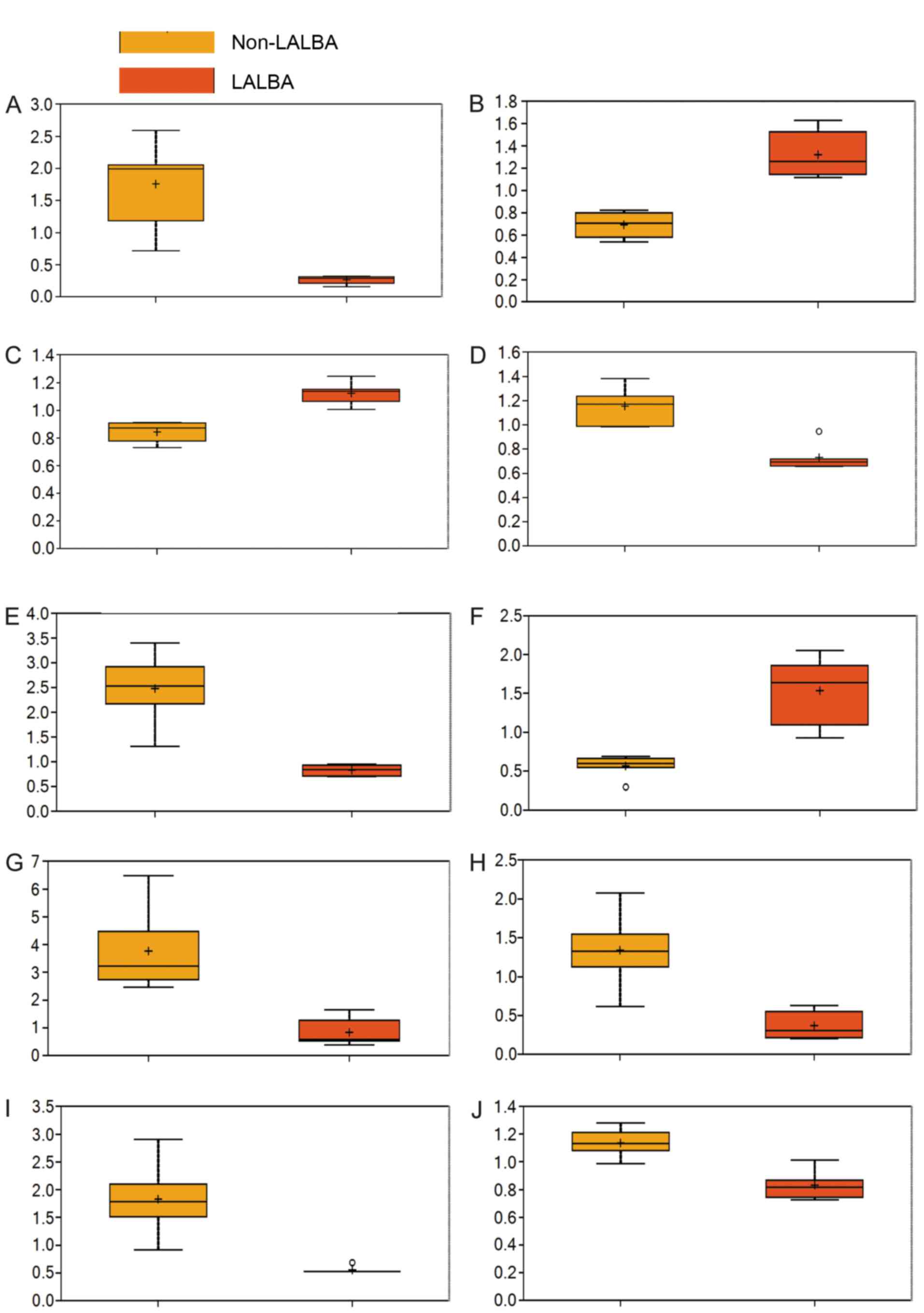
Significantly altered metabolites in
serum of LALBA cows
Among the 273 detected metabolites, 79 metabolites
were found to differ significantly between the LALBA and
non-LALBA groups based on Welch's two-sample t-test
(Table II; fold change ≠ 1 and
P<0.05), particularly erythritol, 4-hydroxyphenylacetyl glycine,
arginine, N-acetylglycine, α-hydroxyisocaproate, leucyltryptophan,
myristoleate (14:1n5), β-hydroxyisovalerate, phenyllactate and
glycerol (Fig. 3). Additionally,
46 metabolites were significantly increased, whereas 33 metabolites
were significantly reduced in the LALBA group, when compared
with the non-LALBA group.
 | Table II.Significantly altered metabolites
between LALBA and non-LALBA cows. |
Table II.
Significantly altered metabolites
between LALBA and non-LALBA cows.
| Metabolite
name |
LALBA/non-LALBA | P-value |
|---|
|
N-acetylglycine | 0.63 | 0.0002a |
|
3-methylhistidine | 0.75 | 0.0107 |
| Lysine | 1.76 | 0.0247 |
| Pipecolate | 1.52 | 0.0246 |
| Phenyllactate
(PLA) | 0.30 | 0.0005a |
|
4-hydroxyphenylacetate | 1.66 | 0.0090 |
| Tyrosine | 1.45 | 0.0159 |
|
4-hydroxyphenylacetyl glycine | 1.91 | 0.0001a |
| Tryptophan | 1.38 | 0.0184 |
|
Indolepropionate | 0.34 | 0.0073 |
| 3-indoxyl
sulfate | 1.71 | 0.0008a |
|
C-glycosyltryptophan | 0.76 | 0.0248 |
| Leucine | 1.32 | 0.0320 |
|
β-hydroxyisovalerate | 0.28 | 0.0004a |
|
α-hydroxyisovalerate | 0.50 | 0.0087 |
| Isoleucine | 1.27 | 0.0199 |
|
2-methylbutyrylcarnitine (C5) | 0.45 | 0.0110 |
|
2-methylbutyrylglycine | 0.59 | 0.0043a |
|
3-hydroxy-2-ethylpropionate | 0.39 | 0.0007a |
| Valine | 1.44 | 0.0040 |
|
Isobutyrylcarnitine | 0.68 | 0.0047a |
|
α-hydroxyisocaproate | 0.34 | 0.0002a |
|
2-aminobutyrate | 1.46 | 0.0468 |
|
S-methylcysteine | 1.60 | 0.0012a |
| Arginine | 1.33 | 0.0001a |
| Urea | 0.73 | 0.0482 |
| Ornithine | 2.48 | 0.0091a |
| N-methyl
proline | 0.31 | 0.0033a |
|
Guanidinoacetate | 1.68 | 0.0284 |
|
γ-glutamyltyrosine | 1.49 | 0.0009a |
|
γ-glutamylvaline | 1.40 | 0.0327 |
| Glycylleucine | 1.64 | 0.0384 |
| Leucylleucine | 1.67 | 0.0331 |
|
Leucyltryptophan | 2.70 | 0.0003a |
| 1,5-anhydroglucitol
(1,5-AG) | 0.49 | 0.0023 |
| Xylitol | 1.38 | 0.0112 |
| Lactose | 0.27 | 0.0473 |
| Erythronate | 0.76 | 0.0282 |
| Myristate
(14:0) | 0.69 | 0.0427 |
| Myristoleate
(14:1n5) | 0.22 | 0.0003a |
| Palmitoleate
(16:1n7) | 0.61 | 0.0165 |
| Eicosapentaenoate
(EPA; 20:5n3) | 1.68 | 0.0070a |
| Docosapentaenoate
(n3 DPA; 22:5n3) | 1.36 | 0.0387 |
| Mead acid
(20:3n9) | 1.60 | 0.0053 |
|
Hexadecanedioate | 1.66 | 0.0179 |
| Docosadioate | 1.48 | 0.0476 |
|
2-aminooctanoate | 0.45 | 0.0217 |
|
Butyrylcarnitine | 0.78 | 0.0174 |
|
Acetylcarnitine | 0.68 | 0.0046 |
| Deoxycarnitine | 1.38 | 0.0015a |
|
3-dehydrocarnitine | 0.57 | 0.0007a |
| Myo-inositol | 0.66 | 0.0416 |
| Choline | 1.17 | 0.0369 |
|
1-palmitoleoylglycero-phosphocholine
(16:1) | 2.20 | 0.0219 |
|
1-margaroylglycero-phosphocholine
(17:0) | 2.65 | 0.0236 |
|
2-stearoylglycero-phosphocholine | 2.33 | 0.0442 |
|
2-oleoylglycero-phosphocholine | 2.04 | 0.0387 |
|
1-palmitoylplasmenylethano-lamine | 2.16 | 0.0401 |
|
1-palmitoylglycero-phosphoethanolamine | 2.67 | 0.0048 |
|
1-stearoylglycero-phosphoethanolamine | 2.42 | 0.0014a |
|
1-oleoylglycero-phosphoethanolamine | 2.52 | 0.0149a |
|
1-arachidonoylglycero-phosphoethanolamine | 1.69 | 0.0115a |
| Glycerol | 0.73 | 0.0006a |
| Cholate | 2.05 | 0.0429 |
|
7-ketodeoxycholate | 2.79 | 0.0038a |
| Pseudouridine | 0.68 | 0.0013a |
|
2′-deoxycytidine | 1.48 | 0.0036 |
|
Methylphosphate | 1.33 | 0.0420 |
|
4-hydroxyhippurate | 1.25 | 0.0152 |
| 4-vinylphenol
sulfate | 2.00 | 0.0009a |
| 2,3-butanediol | 3.02 | 0.0387 |
| Gluconate | 0.53 | 0.0060 |
| Equol
glucuronide | 0.57 | 0.0068 |
| Equol sulfate | 0.23 | 0.0007a |
| Erythritol | 0.15 |
0.00003a |
|
Homostachydrine | 0.16 | 0.0007a |
|
N-(2-furoyl)glycine | 4.67 | 0.0062 |
| 4-acetylphenol
sulfate | 5.74 | 0.0046 |
|
N-methylpipecolate | 0.44 | 0.0038a |
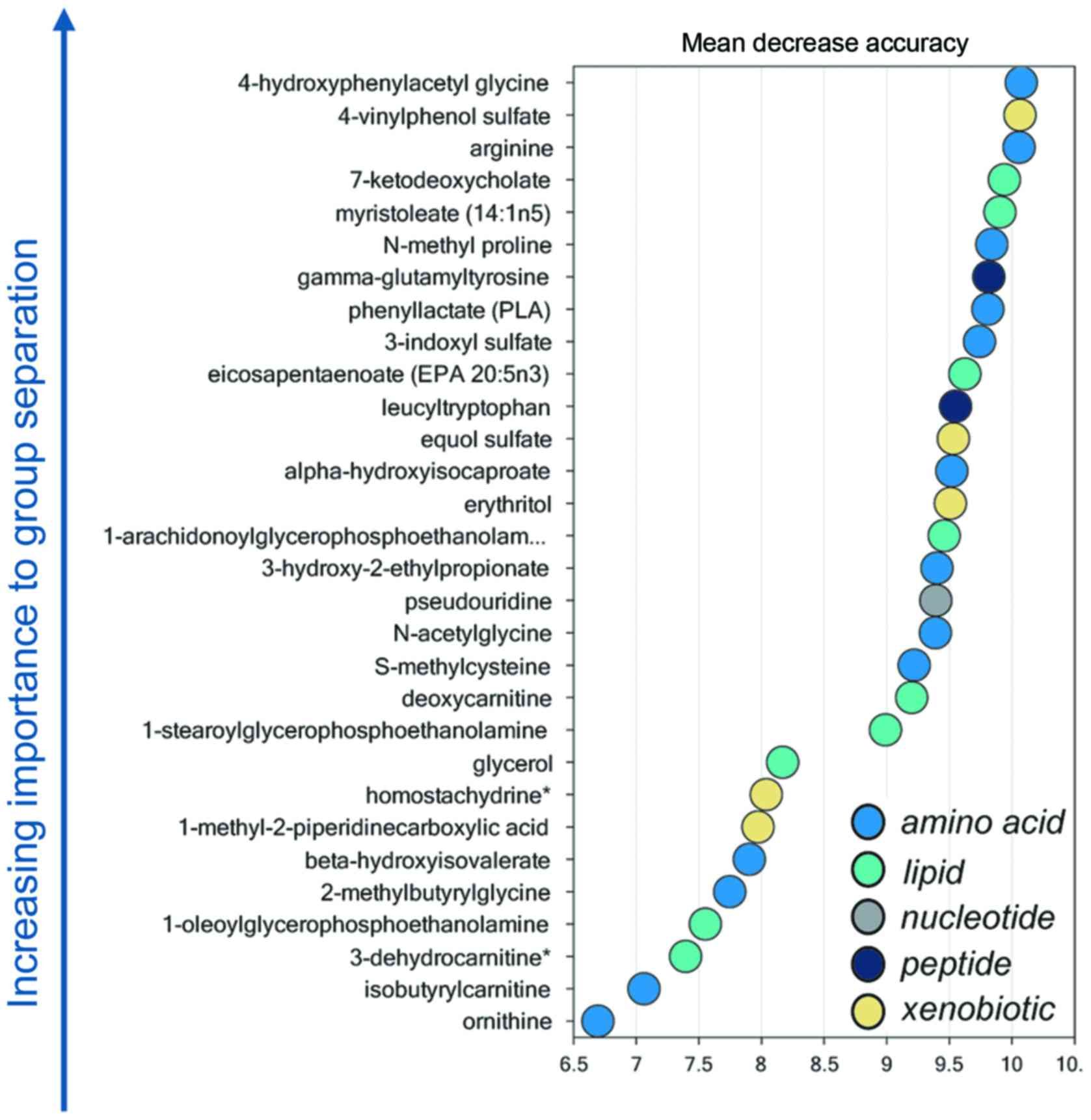
Random forest analysis
Random forest analysis identified a unique metabolic
signature between LALBA and non-LALBA groups with a
predictive accuracy of 100% (Fig.
4). Therefore, these 30 metabolites (14 elevated and 16
reduced) were defined as the potential key metabolites (Table II).
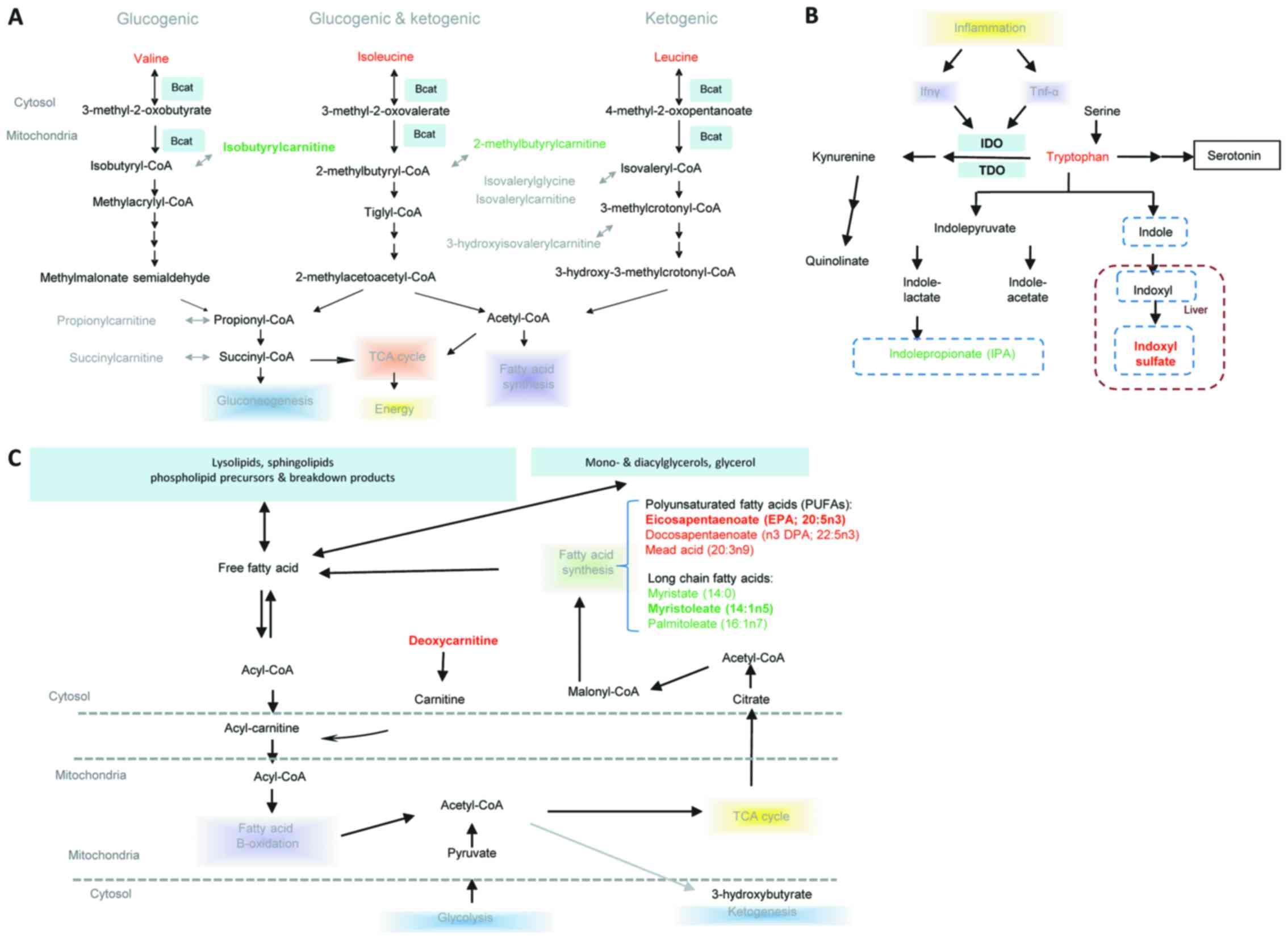
Pathway analysis
The significantly altered metabolites and the
potential key metabolites were determined to be primarily involved
in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways,
including leucine, isoleucine and valine, tryptophanand and lipid
metabolism (Fig. 5). Leucine,
isoleucine, and valine, also termed branched chain amino acids
(BCAA) were increased in LALBA serum compared with
non-LALBA controls, whereas their downstream catabolites
were reduced, including isobutyrylcarnitine and
2-methylbutyrylcarnitine (Fig.
5A). In the LALBA serum, tryptophan accumulation was
accompanied by reduced levels of indolepropionate and
C-glycosyltryptophan; however, elevated levels 3-indoxyl sulfate
was also identified (Fig. 5B). In
addition, long chain fatty acids including palmitoleate (16:1n7),
myristoleate (14:1n5) and myristate (14:0) were reduced in
LALBA serum when compared with non-LALBA controls.
Polyunsaturated fatty acids (PUFAs) including eicosapentaenoate
(EPA; 20:5n3), docosapentaenoate (n3 DPA; 22:5n3) and mead acid
(20:3n9) were elevated in LALBA serum (Fig. 5C).
Discussion
Quantifying the metabolomic profile in serum of cows
carrying and expressing LALBA may provide an insight into
the health condition of transgenic LALBA cows and provide
complement evidence for the usage of genetic modification in cows.
The present study was conducted to identify the serum metabolic
perturbations associated with the presence of human LALBA in
cows. A total of 79 metabolites were identified to differ
significantly between LALBA and non-LALBA groups, 30
of which were defined as potential key metabolites. These
metabolites were determined to primarily participate in the
metabolism of amino acids and lipids.
In the present study, valine, isoleucine, leucine
and tryptophan were found to be significantly increased in the
serum of LALBA cows. Reportedly, BCAA including valine,
isoleucine and leucine participate in muscle growth (19) and are associated with physical
functions and body composition (20). The supplementation of BCAA or
leucine enhances the resistance to fatigue and promotes muscle- and
liver-glycogen spares in trained rats (21). Therefore, the accumulation of BCAA
may benefit muscle growth and physical functions in LALBA
cows.
Additionally, tryptophan is the precursor of the
neurotransmitter serotonin. Serotonin receptors, including HTR1B,
HTR2A and HTR2B are expressed in small mammary blood vessels of
bovids and serotonin may regulate lactational homeostasis and
involution in mammary cells, along with milk yield and composition
in dairy cows (22). Tryptophan
supplementation may increase plasma tryptophan in heifers and dairy
cows, plasma melatonin in heifers and milk yield during morning
milking in dairy cows (23).
Subcutaneous melatonin treatment during the early dry-off period
may improve reproductive performance postpartum in dairy cows,
reducing repeat breeding syndrome, the number of days open and
pregnancy loss (24). Therefore,
an increase in tryptophan level may benefit milk yield and
reproductive performance in LALBA cows.
The present study determined that long chain fatty
acids were reduced in LALBA serum, including myristate
(14:0), myristoleate (14:1n5) and palmitoleate (16:1n7). The
upregulation of either fatty acid synthesis or lipolysis is a
critical feature of tumor metabolism. Saturated fatty acids, such
as palmitic acid have pro-inflammatory effects (25,26).
The addition of myristic acid, a 14-carbon saturated fatty acid,
may enhance the growth of hepatocellular carcinoma cells in
ACOT8-knockdown cell lines Huh7 and Hep3B with poor growth
(27). Therefore, the decrease of
long chain fatty acids, especially myristate, may benefit
LALBA cows via its anti-cancer effect.
PUFAs including EPA, n3 DPA and mead acid were
significantly elevated in LALBA serum, especially EPA, which
was a potential key metabolite based on the random forest analysis
performed in the present study. EPA may improve health via its
anti-cardiovascular diseases, anti-cancer, anti-depression,
anti-aging, anti-arthritis, anti-hypertensive, anti-inflammatory,
anti-oxidant and insulin-sensitizing effects. It is of note that
EPA inhibits the formation of pro-inflammatory prostaglandin
E2 from arachidonic acid via competitively binding with
cyclooxygenase-1 and cyclooxygenase-2, promoting the production of
less-inflammatory prostaglandin E3 (28). Furthermore, EPA induces breast
cancer cell apoptosis and inhibits the in vitro growth of
breast cancer cells (MDA-MB-231) by 20–25% at concentrations as low
as 25 µM and these effects are mediated by the activation of
neutral sphingomyelinase, enhancing ceramide formation, membrane
bleb formation and p21 expression. Therefore, an increase in EPA
may also benefit LALBA cows.
One of the limitations of the present study was that
based on previous studies on metabolite comparison between groups,
only P<0.05 and fold change ≠ 1 were selected as the criteria
for the screening of the significantly altered metabolites.
However, future studies should perform a correction of P-value to
false discovery rate (FDR) and FDR<0.05 may lead to more
accurate results.
In conclusion, the serum biochemistry indexes of
LALBA cows obtained in the present study were in the normal
range, indicating that serum biochemistry characteristics of the
cows were not significantly influenced by LALBA. This is
consistent with previous studies regarding cloned cattle and their
offspring (4,5). In addition, although the serum in
LALBA cows possessed a unique metabolomic signature compared
with their non-LALBA counterparts, the accumulation of PUFAs
and amino acids, and the decline of long chain saturated fatty
acids in LALBA serum may benefit LALBA cows.
Therefore, further investigations should focus on validating these
benefits and the corresponding mechanisms.
Acknowledgements
The present study was funded by the National Science
and Technology Major Project (grant no. 2014ZX0801203B). State Key
Laboratories for AgroBiotechnology China Agricultural University
aided in the sample collection for the present study.
References
|
1
|
Lönnerdal B and Lien EL: Nutritional and
physiologic significance of alpha-lactalbumin in infants. Nutr Rev.
61:295–305. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jackson JG, Janszen DB, Lonnerdal B, Lien
EL, Pramuk KP and Kuhlman CF: A multinational study of
alpha-lactalbumin concentrations in human milk. J Nutr Biochem.
15:517–521. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Yang P, Tang B, Sun X, Zhang R,
Guo C, Gong G, Liu Y, Li R, Zhang L, et al: Expression and
characterization of bioactive recombinant human alpha-lactalbumin
in the milk of transgenic cloned cows. J Dairy Sci. 91:4466–4476.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang R, Guo C, Sui S, Yu T, Wang J and Li
N: Comprehensive assessment of milk composition in transgenic
cloned cattle. PLoS One. 7:e496972012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sui S, Zhao J, Wang J, Zhang R, Guo C, Yu
T and Li N: Comparative proteomics of milk fat globule membrane
proteins from transgenic cloned cattle. PLoS One. 9:e1053782014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Clark M, Murray JD and Maga EA: Assessing
unintended effects of a mammary-specific transgene at the whole
animal level in host and non-target animals. Transgenic Res.
23:245–256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hayakawa H, Hirai T, Takimoto A, Ideta A
and Aoyagi Y: Superovulation and embryo transfer in Holstein cattle
using sexed sperm. Theriogenology. 71:68–73. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bergmeyer HU, Hørder M and Rej R:
International Federation of Clinical Chemistry (IFCC) Scientific
Committee, Analytical Section: Approved recommendation (1985) on
IFCC methods for the measurement of catalytic concentration of
enzymes. Part 2. IFCC method for aspartate aminotransferase
(L-aspartate: 2-oxoglutarate aminotransferase, EC 2.6.1.1). J Clin
Chem Clin Biochem. 24:497–510. 1986.PubMed/NCBI
|
|
9
|
Evans AM, DeHaven CD, Barrett T, Mitchell
M and Milgram E: Integrated, nontargeted ultrahigh performance
liquid chromatography/electrospray ionization tandem mass
spectrometry platform for the identification and relative
quantification of the small-molecule complement of biological
systems. Anal Chem. 81:6656–6667. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dehaven CD, Evans AM, Dai H and Lawton KA:
Organization of GC/MS and LC/MS metabolomics data into chemical
libraries. J Cheminform. 2:92010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu WW and Hu LP: Principal component
analysis and exploratory factor analysis using SAS software
package. Zhong Xi Yi Jie He Xue Bao. 8:589–593. 2010.(In Chinese).
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kolde R: Pheatmap: Pretty heatmaps. R
package version 0.7. 7.
|
|
13
|
Keselman HJ, Othman AR, Wilcox RR and
Fradette K: The new and improved two-sample T test. Psychol Sci.
15:47–51. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Breiman L: Random forests. Machine
Learning. 45:5–32. 2001. View Article : Google Scholar
|
|
15
|
Svetnik V, Liaw A, Tong C, Culberson JC,
Sheridan RP and Feuston BP: Random forest: A classification and
regression tool for compound classification and QSAR modeling. J
Chem Inf Comput Sci. 43:1947–1958. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liaw A and Wiener M: The randomforest
package. R News. 2:18–22. 2002.
|
|
17
|
Luo W and Brouwer C: Pathview: An
R/Bioconductor package for pathway-based data integration and
visualization. Bioinformatics. 29:1830–1831. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma T, Liu Y, Hao HS, Du WH, Dai YP, Zhao
XM, Wang D, Qin T, Zhu HB, Li FD and Wang ZL: Routine hematology
and serum biochemistry indexes of transgenic cloned cows carrying
human alpha-lactalbumin gene and the recipients. J Gansu
Agricultural Univ. 5:2012.
|
|
19
|
Layman DK: The role of leucine in weight
loss diets and glucose homeostasis. J Nutr. 133:261S–267S.
2003.PubMed/NCBI
|
|
20
|
Lustgarten M and Fielding R: Circulating
branched chain amino acids are associated with body composition and
physical function in older adults. FASEB J. 29(1 Suppl):
S1038–S1043. 2015.
|
|
21
|
Campos-Ferraz PL, Bozza T, Nicastro H and
Lancha AH: Distinct effects of leucine or a mixture of the
branched-chain amino acids (leucine, isoleucine, and valine)
supplementation on resistance to fatigue, and muscle and
liver-glycogen degradation, in trained rats. Nutrition.
29:1388–1394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hernandez LL, Limesand SW, Collier JL,
Horseman ND and Collier RJ: The bovine mammary gland expresses
multiple functional isoforms of serotonin receptors. J Endocrinol.
203:123–131. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kollmann MT, Locher M, Hirche F, Eder K,
Meyer HH and Bruckmaier RM: Effects of tryptophan supplementation
on plasma tryptophan and related hormone levels in heifers and
dairy cows. Domest Anim Endocrinol. 34:14–24. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garcia-Ispierto I, Abdelfatah A and
López-Gatius F: Melatonin treatment at dry-off improves
reproductive performance postpartum in high-producing dairy cows
under heat stress conditions. Reprod Domest Anim. 48:577–583. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nieman KM, Kenny HA, Penicka CV, Ladanyi
A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB,
Hotamisligil GS, et al: Adipocytes promote ovarian cancer
metastasis and provide energy for rapid tumor growth. Nat Med.
17:1498–1503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tirado-Vélez JM, Joumady I, Sáez-Benito A,
Cózar-Castellano and Perdomo G: Inhibition of fatty acid metabolism
reduces human myeloma cells proliferation. PloS One. 7:e464842012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hung YH, Chan YS, Chang YS, Lee KT, Hsu
HP, Yen MC, Chen WC, Wang CY and Lai MD: Fatty acid metabolic
enzyme acyl-CoA thioesterase 8 promotes the development of
hepatocellular carcinoma. Oncol Rep. 31:2797–2803. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Calder PC: Polyunsaturated fatty acids and
inflammatory processes: New twists in an old tale. Biochimie.
91:791–795. 2009. View Article : Google Scholar : PubMed/NCBI
|