Introduction
Persistent infection with hepatitis B virus (HBV) is
a leading cause of acute and chronic necroinflammatory liver
diseases, hepatic cirrhosis and hepatocellular carcinoma (HCC)
(1). HBV represents a major health
issue worldwide, >360 million chronically-infected patients at
risk of developing liver cirrhosis or HCC (1,2). HBV
is an hepadnavirus which replicates via reverse transcription of
pregenomic RNA (pgRNA), sharing a similar life cycle with human
immunodeficiency virus (HIV) (3).
Nucleos(t)ide analogs (NAs), extensively used to inhibit reverse
transcription in HBV and/or HIV, comprise lamivudine (LMV),
adefovirdipivoxil (ADV), entecavir (ETV), telbivudine (LdT),
tenofovir (TDF), zidovudine (AZT), stavudine (D4T) and
dideoxyinosine (DDI) (4–6).
In patients, HBV and/or HIV exists in the form of
quasispecies due to the error-prone reverse transcriptase (RT),
which is responsible for the variety of drug-resistant mutants
(7). In vivo and in
vitro, NAs mimic physiological nucleosides in terms of uptake
and metabolism and are incorporated into newly-synthesized DNA
chains, leading to synthesis inhibition via the termination of DNA
replication (8). The replication
of drug-resistant HBV/HIV variants during multidrug therapy is
considered to be a primary cause of treatment failure (9). Under antiviral pressure, variants may
contribute to the production and further selection of
replication-competent resistance mutants, which will spread to
other cells and may eventually replace the wild-type virus
(10). Long-term antiviral therapy
is therefore required, which frequently leads to the emergence and
selection of drug-resistant mutations in the HIV and HBV viral
polymerase (9,11).
NA-resistant mutations inhibit the anti-HBV effect
and induce virological advancement and hepatopathological
progression (6). A number of
mutations have been reported to account for drug resistance, as
reviewed below. Mutations of rtM204I/V in the YMDD motif of HBV-RT
lead to LMV resistance (LMVr) and LdT resistance (LdTr). ETV
resistance (ETVr) has additionally been observed in LMVr patients
(6). With rt M204I/V mutation, a
combination of mutations in the B, C or D domains of RT may lead to
ETVr, including rtI169T, rtL180M, rtS184G, rtA186T, rtS202I,
rtM204V, rtM250V (6). The
substitution mutations rtA181V/T, rtN236T, rtN238R, rtT240Y and
rtN248H are able to reduce the anti-HBV effects of ADV (12), while rtP177G and rtF249A have been
reported to induce TDF resistance; rtV214A, rtQ215S and rtA194T
required further confirmation (9).
Owing to the decreased replicative capacity of resistance mutants,
for survival, compensatory mutations of HBV which restore viral
replication are selected for, including rtL80V/I, rtI163V, rtI169T,
rtV173L, rtT184S/G, rtS202I, rtQ215S and rtQ267H (13).
As a commonly-used anti-HBV reagent, NAs were
originally used for the treatment of HIV as antiretroviral drugs
(9). Due to their similar
replication processes, involving the reverse transcription of
pgRNA, HIV is analogous to HBV, and resistance mutations occur
following long-term anti-HIV therapy with NAs (9). The majority of resistance mutations
are located within the palm and finger subdomains of HIV-1-RT. The
combination of M41L, L210W, and T215Y decreases TDF susceptibility
by ~4-fold (14). Mutation L210W
has been frequently detected in viruses sequenced from patients
undergoing AZT therapy (15).
In a previous study, given the homology between
HIV-RT and HBV polymerase, their primary amino acids sequences were
aligned, and corresponding positions were identified and mapped
(9). Based on this model, the
distances of a given amino acid (aa) residue in the HBV-RT to an NA
substrate were able to be estimated. Notably, positions 210–211 of
HIV-RT and 228–229 of HBV-RT are conserved double leucine residues.
As summarized above, L210W in HIV-RT is one of the six principal
mutations that confer in vivo resistance to certain NAs,
particularly AZT, which appears following T215Y/F (14,16).
Crystallographic studies suggested that the aromatic side chain of
Trp210 was able to stabilize the interaction between Phe/Tyr215 and
the dNTP-binding pocket (14). In
present study, the L228 and L229 mutations were investigated. A
series of HBV constructs harboring different mutations were
constructed. The replication capacity, antigen expression level and
resistance phenotype were analyzed in vitro. The results
demonstrated that rtL228 and/or L229 of HBV-RT did not affect
susceptibility to NAs, although replication and HBs/eAg secretion
were altered.
Materials and methods
Cells
Hepatoma cell lines (Huh7 and HepG2), which were
kindly provided by Professor Xinwen Chen (Wuhan Institute of
Virology, Chinese Academy of Sciences, Wuhan, China), were cultured
in Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 2 mM/l
glutamine, 100 IU/ml penicillin and 100 IU/ml streptomycin at 37°C
in a 5% CO2 atmosphere incubator (17).
NAs
A total of eight types of NA were used in the
present study: 3TC (GlaxoSmithKline, plc, Brentford, UK), ADV
(Gilead Sciences, Inc., Foster City, CA, USA), ETV (Bristol-Myers
Squibb Co., New York, NY, USA), LdT (Novartis International AG,
Basel, Switzerland), TDF (Gilead Sciences, Inc.), AZT (Teva Canada,
Ltd., Whitchurch-Stouffville, ON, Canada), D4T (Bristol-Myers
Squibb Co.) and DDI (Teva Canada, Ltd.). The NAs were dissolved in
the appropriate solution according to the manufacturer's protocol,
and were used in the assays at the indicated concentrations
(9,18).
Sequences and plasmid constructs
HIV-RT (GenBank accession no. HQ718313; https://www.ncbi.nlm.nih.gov/nuccore/HQ718313) and HBV
polymerases (GenBank accession no. CAA48354.1; https://www.ncbi.nlm.nih.gov/protein/CAA48354.1), were
used for their primary amino acids sequences alignment. For the
construction of the replication-competent plasmid with aa
substitutions, the pHBV1.3 plasmid (1.3-fold over-length HBV
genome; subtype, ayw) was used as a backbone (9,13).
Mutations were introduced into wild-type (WT) HBV via polymerase
chain reaction (PCR)-based mutagenesis using the primer pairs
P-RT-mt-F, P-RT-mt-R and primers with specific mutations listed in
Table I, as described previously
(9). Plasmids containing mutation
sites were identified by sequencing. Serial plasmids pHBV1.3-rtX
(X, mutations including rtL228W) were producedusing DH5α
Escherichia coli and extracted using a Plasmid Plus Midi kit
(Qiagen GmbH, Hilden, Germany) (13).
 | Table I.Primers for the construction of MT HBV
plasmids and RT-qPCR. |
Table I.
Primers for the construction of MT HBV
plasmids and RT-qPCR.
| Name | Sequence (5′-3′) |
|---|
| p-RT-mt-F | TCTTCTCGAGGATTGGGGACC |
| p-RT-mt-R | GCAGCCATGGAAACGATGTAT |
| p-rtL228C-F |
ATTTTTGTTTGTCTTTGGGTATACAT |
| p-rtL228C-R |
ATGTATACCCAAAGACAAACAAAAAT |
| p-rtL228W-F |
ATTTTTGGTTGTCTTTGGGTATACAT |
| p-rtL228W-R |
ATGTATACCCAAAGACAACCAAAAAT |
| p-rtL229W-F |
ATTTTCTTTGGTCTTTGGGTATACAT |
| p-rtL229W-R |
ATGTATACCCAAAGACCAAAGAAAAT |
|
p-rtL228W/L229W-F |
ATTTTTGGTGGTCTTTGGGTATACAT |
|
p-rtL228W/L229W-R |
ATGTATACCCAAAGACCACCAAAAAT |
| p-HBV-rc-F |
GTTGCCCGTTTGTCCTCTAATTC |
| p-HBV-rc-R |
GGAGGGATACATAGAGGTTCCTT |
ELISA analysis
HBsAg in the supernatant and the cell lysate, HBeAg
in the supernatant from indicated HBV-bearing plasmid-transfected
Huh7/HepG2 cells were analyzed using a diagnostic kit for HBsAg
(cat. no. 201606212) and HBeAg (cat. no. 201606122) detection
(Shanghai Kehua Bio-Engineering Co., Ltd., Shanghai, China)
according to the manufacturer's protocol (17).
Detection of virion-associated HBV DNA
by southern blotting and quantitative (q)PCR analysis
Huh7 or HepG2 cells (1–2×106) were
transfected with 4 µg replication-competent plasmid, with or
without the indicated concentration of the different NAs, using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.).
HBV-encapsid at edreplicative intermediates were collected and
purified from intracellular core particles, according to the method
described in a previous study (9).
Cells were washed using ice-cold PBS and lysed with 400 µl lysis
buffer at 4°C for 15 min. The nuclei were pelleted by
centrifugation, and the supernatant was adjusted to 10 mM
MgCl2 and treated with 100 mg/ml DNase I (Roche
Diagnostics GmbH, Mannheim, Germany) at 37°C for 30 min to digest
the input DNA (transfected plasmid), which was terminated with a
final concentration of 25 mM EDTA. A total of 0.5 mg/ml proteinase
K (Qiagen GmbH) and 1% SDS was used to digest the excess proteins
in the supernatant at 56°C for 2 h. HBV DNA was extracted using
phenol/chloroform followed by isopropanol precipitation,
resuspended in TE buffer, and subjected to agarose gel
electrophoresis, followed by denaturation and Southern blotting
with a 32P-labeled full length HBV DNA probe.
Hybridization signals were acquired and analyzed using Phosphor
Imager (Cyclon; Packard Instrument Company, Inc., Meriden, CT, USA)
(13).
HBV DNA was additionally quantified by qPCR analysis
using primers [relaxed circular (RC)-sense and RC-antisense;
Table I] specific to HBV RC
genomes in a SYBR-Green reaction mix (Roche Diagnostics GmbH).
Plasmid pBSK-HBV1.3-WT was quantified and 10-fold diluted as a
standard curve (13). All samples
were analyzed in triplicate, and every value is presented as the
mean of at least three independent experiments.
Statistical analysis
In the present study, the statistical analysis was
performed using GraphPad (GraphPad Software, Inc., La Jolla, CA,
USA). Each value is presented as the mean of three independent
experiments. Differences in pairwise comparisons were determined
using a Student's t-test for statistical significance. Differences
in multiple comparisons were determined using a one-way analysis of
variance followed by the Least Significant Difference post hoc
test. P<0.05 was considered to indicate a statistically
significant difference. Results are presented as the means ±
standard deviation.
Results
Sequence alignment of HBV and
HIV-RT
Given the homology between HIV-RT and HBV
polymerases, their primary amino acids sequences were aligned
(Fig. 1A), corresponding positions
were identified and mapped. As presented in Fig. 1A, in addition to YMDD which shared
by HBV and HIV, positions 210–211 of HIV-RT and 228–229 of HBV-RT
are conservative double leucines. HBV plasmids harboring different
mutations of L228 and/or L229 were constructed by fusion PCR, and
all mutations were located at the D domain of HBV-RT (Fig. 1B).
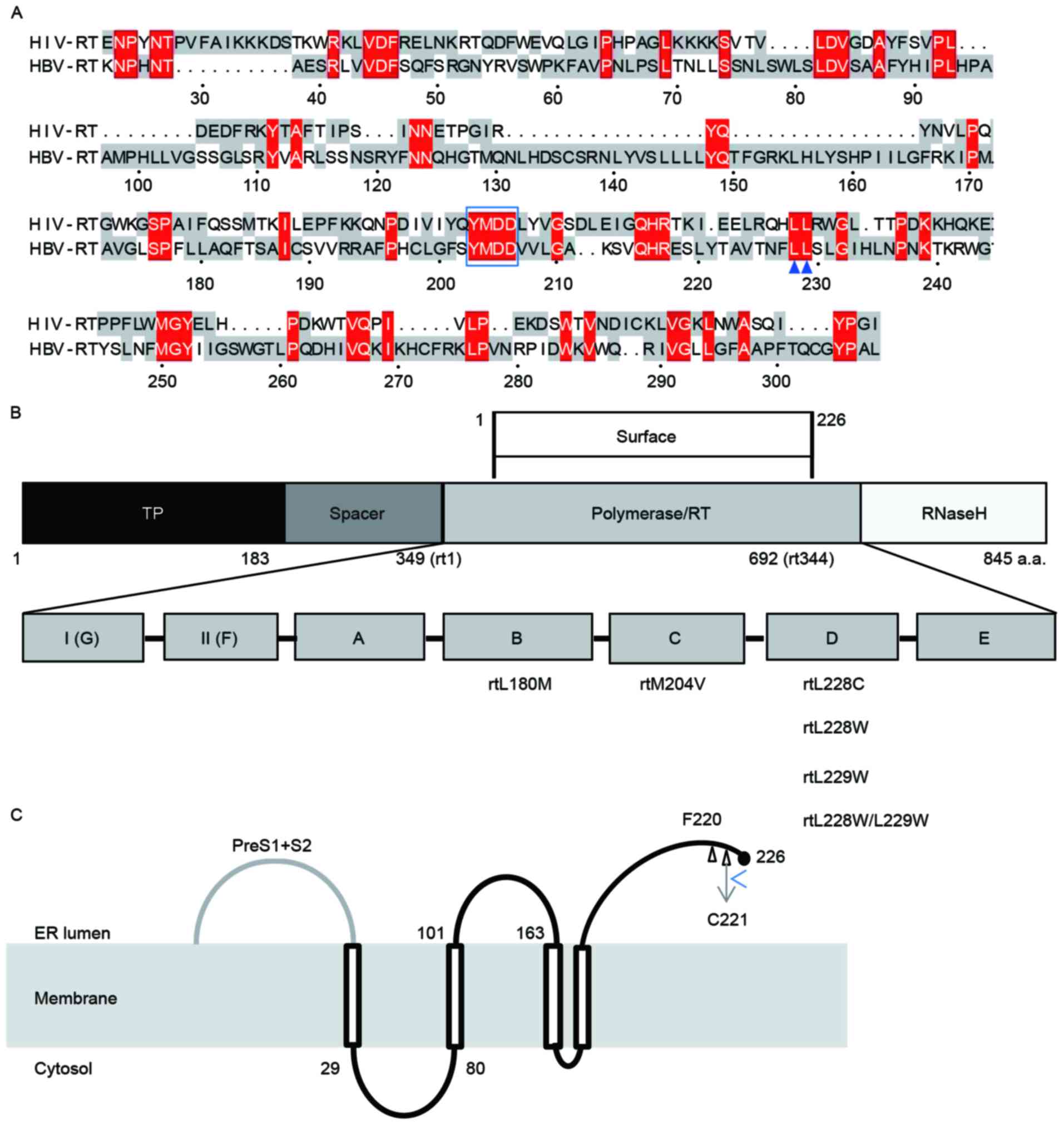 | Figure 1.Sequence alignment and structure of
HBV RT. (A) Amino acid sequence alignment of HBV-RT and HIV-RT.
Colored regions indicate areas of sequence conservation. Amino
acids in red were predicted to be potential regions which may
interact with NAs. Within these, L228 and L229, which are highly
conservative with the corresponding positions in HIV, are marked by
arrowheads. (B) Structure of HBV RT and location of target
mutations. HBV polymerase is divided into four regions: TP, spacer,
RT and RNaseH. RT, containing the whole surface protein, is
subdivided into seven regions, A-G. All the amino acid
substitutions examined in the present study are located in region
D. (C) Proposed topology of HBV small envelope protein. The small
envelope protein likely traverses the ER membrane four times:
TMD-I, between residues 4 and 28; TMD-II, between 100 and 164;
TMD-III, between173 and193; TMD-IV, between 202 and 222; and the C
terminal, between residues 223 and 226, where sF220 and sC221G are
located. HBV, hepatitis B virus; HIV, human immunodeficiency virus;
NA, nucleos(t)ide analogs; RT, reverse transcriptase; ER,
endoplasmic reticulum; TMD, transmembrane domain; TP, terminal
protein. |
Within the HBV genome, the S gene overlaps with the
P gene, meaning that one nucleotide substitution may alter the
surface protein and the polymerase (19). In the present study, rtL228C/W
resulted in sF220V/G, rtL229W gave rise to sC221G, while
rtL228W/L229W led to sF220G/C221G (Table II). The HBV envelope proteins
consist of small (S, 226 aa), middle (M, 281 aa), and large (L, 400
aa) proteins. S, M and L, closely-associated transmembrane
proteins, are in one single open reading frame (ORF) using three
different in-frame translation start codons and a common stop
codon.
 | Table II.List of co-mutations between surface
antigens and HBV polymerase. |
Table II.
List of co-mutations between surface
antigens and HBV polymerase.
| Mutations of
HBV-RT | HBsAg corresponding
mutations |
|---|
| rtL228C | sF220V |
| rtL228W | sF220G |
| rtL229W | sC221G |
| rtL228W/L229W | sF220G/sC221G |
The proposed topology of the HBV small envelope
protein demonstrated that the small envelope protein traversed the
endoplasmic reticulum (ER) membrane, probably four times (20,21),
between the N and C terminal (226 amino acids) (Fig. 1C), in order, there is the N
terminal (residues 1 to 3, exposed at the luminal side of the
membrane), transmembrane domain I (TMD-I; between residues 4 and
28), a cytosolic loop between residues 28 and 80, TMD-II (between
100 and 164, containing the antigenic loop and a glycosylation site
at Asn-146), TMD-III (between 173 and 193), TMD-IV (between 202 and
222) and the C terminal (between residues 223 and 226). As
indicated in Fig. 1C, sF220 and
sC221G were located at the C-end of the protein.
Replication of pHBV1.3-rtXin
transfected mammalian cells
Site substitutions in the P gene, including
pHBV1.3-rtL228C,-rtL228W, -rtL229W and -rtL228W/L229W, were
designed and constructed using fusion PCR to introduce into
pBSK-HBV-WT plasmids. In order to further analyze the influence of
these mutations on HBV replication, antigen production and
sensitivity to NAs, together with pBSK-HBV1.3-wt, -rtM204V and
-rtL180M/204V which were constructed in a previous study (13), WT and MT HBV replication-competent
plasmids were transfected into Huh7 cells and viral genome
replication competence was analyzed by Southern blotting. Southern
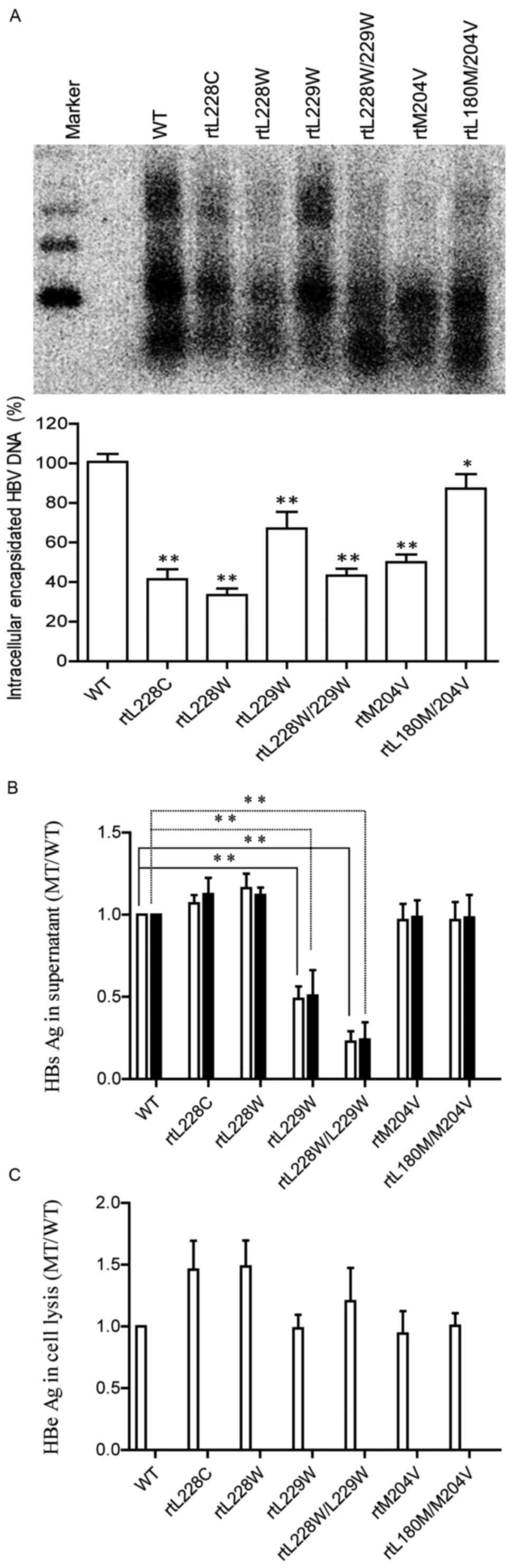
blotting and subsequent densitometric analysis (Fig. 2A) revealed that the substitutionsrt
L228C, -rtL228W, -rtL229W and -rtL228W/L229W decreasedthe HBV
replication capacity to ~46, 33, 74 and 52% of the WT,
respectively. The mutation rtM204V/I was demonstrated to weaken HBV
replication, while rtL180M was able to restore the impaired
replication capacity, in accordance with a previous report
(22).
HBsAg in the supernatant and cell lysate was
detected using a commercial ELISA kit (Fig. 2B), as was HBeAg in the supernatant
(Fig. 2C). The results indicated
that none of the mutations led to a decrease in the supernatant
HBeAg level compared with WT HBV. Compared with WT HBV, mutation
rtL228C/W (sF220V/G) enhanced the production and secretion of
HBsAg, while rtL229W and rtL228W/L229W significantly decreased
HBsAg expression in the supernatant and cell lysate. As the HBV
polymerase gene is overlapped by the surface protein gene,
mutations in polymerase may result in aa substitutions in HBsAg. In
current study, rtM204V led to sI195M, but pBSK-HBV1.3-rtM204V and
-rtL180M/204V exhibited an approximately consistent HBs/eAg level
with WT HBV (9,22). The results of the present study
demonstrated that different substitutions resulted in different
effects on HBV replication and HBsAg expression.
Inhibition of HBV replication with NAs
in vitro
In order to analyze the resistant phenotype of the
different HBV mutations to NAs, the NA concentration was optimized
usingp HBV1.3-WT in HepG2 cells. Following transfection with
pHBV1.3-WT for 6 h, different concentrations of the NAs were added
into the fresh medium; core-associated HBV DNA in HepG2 cells was
extracted and analyzed using the qPCR at 96 h (9). All the assayed drugs inhibited HBV
DNA replication and the half maximal effective concentration values
of 3TC, ADV, LdT, ETV, TDF, AZT, D4T and DDI were 1.08, 1.38,
11.60, 0.79, 0.19, 12.95, 9.85 and 34.52 µM, respectively (Fig. 3). The results of the present study
demonstrated that TDF was the most potent drug, followed by ETV,
and that LdT, AZT, D4T and DDI exerted mild effects on HBV. Apart
from pBSK-HBV1.3-rtM204V and -rtL180M/204V, which are classical
LMV/LdT resistance mutations, the other MT HBV with comparable
susceptibility were sensitive to the eight types of NA. In contrast
to HIV, substitution mutations on L228 and/or L229 did not
decreased the anti-HBV effect of the NAs. As reported previously,
the nucleoside analogues did not affect the HBsAg, HBeAg and HBcAg
expression (9).
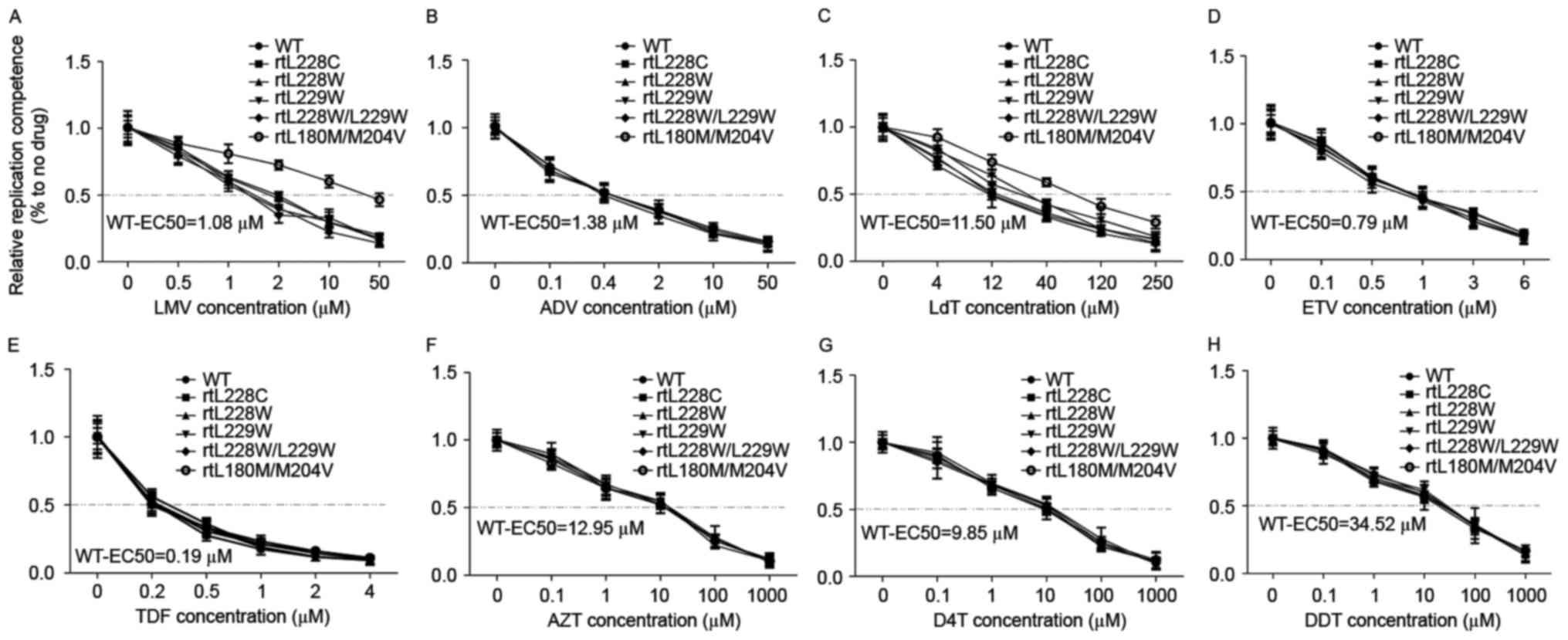 | Figure 3.RT-qPCR detection of the anti-HBV
effects of eight types of nucleos(t)ide analog. HepG2/Huh7 cells
were transfected with the plasmids pHBV1.3-WT, -rtL228C, -rtL228W,
-rtL229W, -rtL228W/L229W and -rtL180M/M204V, and subsequently
treated with anti-HBV drugs at the indicated concentrations.
Encapsulated HBV DNA was extracted and analyzed by RT-qPCR to
evaluate the anti-HBV effects of (A) LMV, (B) ADV, (C) LdT, (D)
ETV, (E) TDF, (F) AZT, (G) D4T and (H) DDI. RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; HBV,
hepatitis B virus; LMV, lamivudine; ADV, adefovirdipivoxil; LdT,
telbivudine; ETV, entecavir; TDF, tenofovir; AZT, zidovudine; D4T,
stavudine; DDI, dideoxyinosine; WT, wild-type; EC50,
half maximal effective concentration. |
Discussion
HBV replication is a complex dynamic process
involving the interaction of a number of factors, comprising
viruses, drugs and host factors (5). At present, thorough eradication of
HBV in patients is not achievable using the available agents,
including interferon and NAs (23). Certain NAs have been demonstrated
to be effective anti-HBV agents in vivo and in vitro,
although resistance mutations to NAs are selected during long-term
anti-HBV therapy (6). Due to
overlap between the polymerase gene and the surface protein gene,
mutations in HBV polymerase may result in aa substitutions in HBsAg
that potentially lead to immune evasion or modification of viral
fitness. In a previous study, it was found that some new resistance
mutations of HBV from the patients accepted anti-HBV therapy
(6).
Previously, when a virological breakthrough occurred
in a patient with chronic hepatitis B infection receiving one type
of NA therapy, the HBV sequence was analyzed by sequencing and
resistance mutations, if present, were detected. Not all resistance
mutations were identified; certain mutations may be undetectable
due to low frequency, and not all the detectable mutations exerted
a marked effect due to error-prone reverse transcription and the
phenomenon of quasispecies (6,9).
Additionally, this retrospective method is time-consuming and
disadvantageous for clinical therapy. Therefore, prospective
studies are required. Based on 3D modeling and the conservative
sequence alignment of HBV and HIV, certain resistance mutations may
be predicted, as described in a previous study (9). Both rtL210 and rtL211 in HIV possess
a double conservative leucine, according to the sequence alignment
of HBV and HIV-RT, and the corresponding positions of HBV (rtL228
and rtL229) also possess a double conservative leucine.
Furthermore, substitution rtL210W in HIV is an important resistance
mutation (24). In the present
study, in order to determine alterations in L228 and/or L229 of
HBV, rtL228C, -rtL228W, -rtL229W and -rtL228W/L229W were designed
and introduced into a WT HBV plasmid, termed pBSK-HBV1.3-WT
(GenBank accession no. U95551), and LMV resistance mutations
containing rtM204V and rtL180M/204V were used as the control.
Biomarkers of the viral life cycle and susceptibility to commonly
used NAs were analyzed systematically. Due to the overlap between
the polymerase gene and the surface protein gene of HBV, mutations
sF220V/G, sC221G and sF220G/C221G were driven by rtL228C/W, rtL229W
and rtL228W/L229W, respectively. Amino acid sF220 and sC221 were
located at TMD-IV, the carboxyl-terminal (between residues 164 and
226) of HBsAg, which has been reported to be highly hydrophobic and
complex, and important for virion packaging (20). Cysteine is a natural amino acid
containing sulfhydryl, which is able to form intra chain and inter
chain disulfide bonds; these are important for the folding and
stability of proteins, including in HBs (25). This may be the reason why rtL229W
(sC221G) rtL228W/L229W (sF220G/C221G) significantly decrease HBsAg
secretion. An altered sequence and structure of HBV-RT and HBs
protein led by substitutions may directly influence viral
replication capacity via alterations to the polymerase, or by
indirect influences driven by HBV packaging through the interaction
between surface and core protein of HBV, or a combination. In the
present study, single or amphimutationsinrt L228 and rtL229
significantly impaired HBV replication. The detailed mechanism
underlying this observation requires further study.
The HepG2 cell line used in the present study is
thought to be derived from hepatoblastoma rather than
hepatocarcinoma (26). HepG2 and
Huh7 cell lines were used in present study. With regard to the
replication capacity, the HBs/e antigen level and the
susceptibility to NAs of the WT and MT HBV, similar conclusions
were drawn from HepG2 and Huh7; therefore, only the results of
HepG2 are presented, and those of Huh7 are not shown. In a previous
study, HBs/eAg levels in the supernatant from Huh7 or HepG2
transfected with HBV plasmids were not consistent with the level of
HBV replication. Notably, in transfected systems, NAs selectively
inhibit HBV DNA and not HBs/eAg (9). However, specific small interfering
RNA may inhibit HBV DNA and HBs/eAg in the same system (27). In the present study, rtL229W and
rtL228W/L229W inhibited HBsAg production due to point mutations in
the surface protein. Different from HBsAg, for HBeAg in
supernatant, due to the intact polypeptide sequence of the HBe
antigen, there was no significant difference between WT and MT HBV.
These results were consistent previous research (9). However, the results of the resistance
phenotype analysis were not in line with previous findings.
Mutations in L228 and/or L229 did not decrease the anti-HBV effect
of NAs, while in the same experimental system, rtM204V and
rtL180M/204V were resistant to LMV and LdT compared with WT HBV.
Despite the similar RT structure shared by HBV and HIV, one aa
alteration may exert different influences on the RT of HBV and HIV.
The results of the present study demonstrated that 2D sequence
alignment alone is insufficient for the prediction of resistance
mutations, and requires the addition of 3D spatial structure
analysis.
Acknowledgements
The present study was supported by the Medical
Scientific Research Foundation of Zhejiang Province, China (grant
no. 2013KYA208), and the Shaoxing Science and Technology Bureau,
Zhejiang Province of China (grant no. 2013B70063).
References
|
1
|
Li W and Urban S: Entry of hepatitis B and
hepatitis D virus into hepatocytes: Basic insights and clinical
implications. J Hepatol. 64(1 Suppl): S32–S40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Q, Liao Y, Chen J, Cai B, Su Z, Ying
B, Lu X, Tao C and Wang L: Epidemiology study of HBV genotypes and
antiviral drug resistance in multi-ethnic regions from Western
China. Sci Rep. 5:174132015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartholomeusz A, Tehan BG and Chalmers DK:
Comparisons of the HBV and HIV polymerase and antiviral resistance
mutations. Antivir Ther. 9:149–160. 2004.PubMed/NCBI
|
|
4
|
Dandri M, Lutgehetmann M and Petersen J:
Experimental models and therapeutic approaches for HBV. Semin
Immunopathol. 35:7–21. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zoulim F and Durantel D: Antiviral
therapies and prospects for a cure of chronic hepatitis B. Cold
Spring Harb Perspect Med. 5:a0215012015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zoulim F and Locarnini S: Hepatitis B
virus resistance to nucleos(t)ide analogues. Gastroenterology.
137:1593–1608.e1-2. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cabuang LM, Shaw T, Littlejohn M, Colledge
D, Sozzi V, Soppe S, Warner N, Thompson A, Preiss S, Lam N, et al:
In vitro replication phenotype of a novel (−1G) hepatitis B virus
variant associated with HIV co-infection. J Med Virol.
84:1166–1176. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Petersen J, Thompson AJ and Levrero M:
Aiming for cure in HBV and HDV infection. J Hepatol. 65:835–848.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qin B, Budeus B, Cao L, Wu C, Wang Y,
Zhang X, Rayner S, Hoffmann D, Lu M and Chen X: The amino acid
substitutions rtP177G and rtF249A in the reverse transcriptase
domain of hepatitis B virus polymerase reduce the susceptibility to
tenofovir. Antiviral Res. 97:93–100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aragri M, Alteri C, Battisti A, Di Carlo
D, Minichini C, Sagnelli C, Bellocchi MC, Pisaturo MA, Starace M,
Armenia D, et al: Multiple Hepatitis B virus (HBV) quasispecies and
immune-escape mutations are present in HBV surface antigen and
reverse transcriptase of patients with acute hepatitis B. J Infect
Dis. 213:1897–1905. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Caviglia GP, Abate ML, Noviello D, et al:
Hepatitis B core-related antigen kinetics in chronic HBV-genotype-D
infected patients treated with nucleos(t)ide analogues or
pegylated-interferon-alpha. Hepatol Res. 2016.
|
|
12
|
Kim HS, Yim HJ, Jang MK, Park JW, Suh SJ,
Seo YS, Kim JH, Kim BH, Park SJ, Lee SH, et al: Management of
entecavir-resistant chronic hepatitis B with adefovir-based
combination therapies. World J Gastroenterol. 21:10874–10882. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qin B, Zhang B, Zhang X, He T, Xu W, Fu L
and Tu C: Substitution rtq267h of hepatitis B virus increases the
weight of replication and lamivudine resistance. Hepat Mon.
13:e121602013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yahi N, Tamalet C, Tourres C, Tivoli N and
Fantini J: Mutation L210W of HIV-1 reverse transcriptase in
patients receiving combination therapy. Incidence, association with
other mutations and effects on the structure of mutated reverse
transcriptase. J Biomed Sci. 7:507–513. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Clavel F and Hance AJ: HIV drug
resistance. N Engl J Med. 350:1023–1035. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Delviks-Frankenberry KA, Lengruber RB,
Santos AF, Silveira JM, Soares MA, Kearney MF, Maldarelli F and
Pathak VK: Connection subdomain mutations in HIV-1 subtype-C
treatment-experienced patients enhance NRTI and NNRTI drug
resistance. Virology. 435:433–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qin B, He T, Chen Z, Xu W, Pan G and Tu C:
A novel method for the analysis of drug-resistant phenotypes of
hepatitis B virus. Int J Mol Med. 31:975–981. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pokrovsky AG, Pronayeva TR, Fedyuk NV,
Shirokova EA, Khandazhinskaya AL, Tarusova NB, Karpenko IL and
Krayevsky AA: Anti-HIV activity of novel phosphonate derivatives of
AZT, d4T and ddA. Nucleosides, Nucleotides Nucleic Acids.
20:767–769. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Su M, Xiang K, Li Y, Li Y, Deng J, Xu X,
Yan L, Zhuang H and Li T: Higher detection rates of amino acid
substitutions in HBV reverse transcriptase/surface protein
overlapping sequence is correlated with lower serum HBV DNA and
HBsAg levels in HBeAg-positive chronic hepatitis B patients with
subgenotype B2. Infect Genet Evol. 40:275–281. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Khan N, Guarnieri M, Ahn SH, Li J, Zhou Y,
Bang G, Kim KH, Wands JR and Tong S: Modulation of hepatitis B
virus secretion by naturally occurring mutations in the S gene. J
Virol. 78:3262–3270. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abou-Jaoudé G, Molina S, Maurel P and
Sureau C: Myristoylation signal transfer from the large to the
middle or the small HBV envelope protein leads to a loss of HDV
particles infectivity. Virology. 365:204–209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li MW, Hou W, Wo JE and Liu KZ: Character
of HBV (hepatitis B virus) polymerase gene rtM204V/I and rtL180M
mutation in patients with lamivudine resistance. J Zhejiang Univ
Sci B. 6:664–667. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lucifora J, Xia Y, Reisinger F, Zhang K,
Stadler D, Cheng X, Sprinzl MF, Koppensteiner H, Makowska Z, Volz
T, et al: Specific and nonhepatotoxic degradation of nuclear
hepatitis B virus cccDNA. Science. 343:1221–1228. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fourati S, Malet I, Guenzel CA, Soulie C,
Maidou-Peindara P, Morand-Joubert L, Wirden M, Sayon S, Peytavin G,
Simon A, et al: E17A mutation in HIV-1 Vpr confers resistance to
didanosine in association with thymidine analog mutations.
Antiviral Res. 93:167–174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Michel M, Lone YC, Centlivre M, Roux P,
Wain-Hobson S and Sala M: Optimisation of secretion of recombinant
HBsAg virus-like particles: Impact on the development of HIV-1/HBV
bivalent vaccines. Vaccine. 25:1901–1911. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lopez-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pei R, Qin B, Zhang X, Zhu W, Kemper T, Ma
Z, Trippler M, Schlaak J, Chen X and Lu M: Interferon-induced
proteins with tetratricopeptide repeats 1 and 2 are cellular
factors that limit hepatitis B virus replication. J Innate Immun.
6:182–191. 2014. View Article : Google Scholar : PubMed/NCBI
|