Introduction
Ovarian cancer (OC) is the fifth leading cause of
cancer-related deaths among women in developed countries and the
primary cause of gynecological cancer death (1). Epithelial OC (EOC), the most common
type of OC, accounts for approximately 90% of all OCs (2). According to histological origins, EOC
can be divided into many different subgroups, including serous,
mucinous, endometrioid, undifferentiated, clear cell and Brenner
carcinomas (3,4). EOC is generally called a ‘silent
killer’ because it does not create symptoms in patients with EOC;
consequently, numerous patients are diagnosed in advanced stage
with metastasis (5). Although
advancements in the diagnosis of EOC patients in early stages have
been achieved and efficient treatments have been developed, the
prognosis of patients with EOC remains poor, and their 5-year
survival rate is less than 50% (6). The exact mechanisms underlying EOC
formation and progression are also unknown (7). Thus, further investigations on the
molecular mechanism of EOC occurrence and development may be
advantageous to identify novel biomarkers and therapeutic
strategies to improve the survival of patients with this
disease.
MicroRNAs (miRNAs) are endogenous, single-stranded,
noncoding and short RNA molecules of approximately 22 nucleotides
long (8). miRNAs serve as critical
gene regulators by binding to the 3′-untranslated regions (3′-UTRs)
of their target gene mRNAs in a base pairing sequence-specific
manner; as a result, translational repression or mRNA degradation
occurs (9). More than 60% of human
protein-coding genes have been regulated by miRNAs (10). Half of the human miRNAs are located
in cancer-related genomic regions, such as regions of homozygous
deletion, loss of heterozygosity, oncogene and around tumor
suppressor gene, breakpoint area and amplified region, and are
implicated in tumorigenesis and tumor development (11). miRNAs are also involved in
modulating different cancer-related biological processes, such as
cell proliferation, cycle, apoptosis, angiogenesis, invasion,
migration and metastasis (12–14).
Moreover, numerous miRNAs are aberrantly expressed in various kinds
of human cancers, such as EOC (15), cervical cancer (16), gastric cancer (17), lung cancer (18) and bladder cancer (19). miRNAs may act as either oncogenes
or tumor suppressors in tumor initiation and progression on the
basis of the biological roles of their target genes (20). Therefore, miRNAs can be
investigated as diagnostic markers or potential therapeutic targets
for various cancers.
miR-455 is abnormally expressed in several types of
human cancer (21–23). This finding suggests that miR-455
may play important roles in these types of cancer. However, the
detailed expression level, biological roles and underlying
mechanism of miR-455 in EOC remain unknown. In this study, the
miR-455 expression in EOC was detected, and its association with
clinical characteristics was analysed. The functional roles and
underlying mechanisms of miR-455 in EOC were also investigated.
Materials and methods
Clinical tissue samples
This study was approved by the Ethical Committee of
the Sixth People's Hospital of Jinan, and written informed consent
was obtained from all of the patients prior to the study. A total
of 45 paired EOC tissues and their adjacent normal ovarian
epithelial tissues were obtained from patients who received
surgical resection at the Sixth People's Hospital of Jinan between
January 2014 and February 2016. None of these EOC patients were
treated with radiotherapy, chemotherapy or immunotherapy prior to
operation. After surgery was completed, all of the tissue samples
were immediately snap-frozen in liquid nitrogen and then stored at
−80°C until use.
Cell lines, culture conditions and
transfection
Four EOC cell lines, namely, OVCAR3, SKOV3, ES-2 and
CAOV-3, were purchased from the Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences (Shanghai, China) and
cultured in Dulbecco's modified Eagles medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS), 100 U/ml penicillin and 100 mg/ml streptomycin
(all from Gibco; Thermo Fisher Scientific, Inc.). Human normal
ovarian epithelial NOEC cell line was obtained from the American
Type Culture Collection (Manassas, VA, USA) and grown in Ham's F-12
with 20% FBS, 120 mg/ml streptomycin and 120 U/ml penicillin
(Gibco; Thermo Fisher Scientific, Inc.). All of the cells were
grown at 37°C in a humidified environment with 5%
CO2.
miR-455 mimics and negative control miRNA mimics
(miR-NC) were synthesised by Shanghai GenePharma Co., Ltd.
(Shanghai, China). Notch1-expressing vector (pcDNA3.1-Notch1) and
empty vector (pcDNA3.1) were obtained from RiboBio (Guangzhou,
China). The cells were seeded in a 6-well plate (Corning, Inc.,
Corning, NY, USA) at a density of 60–70% confluence, incubated
overnight at 37°C and transfected with Lipofectamine 2000
(Invitrogen Life Technologies, Carlsbad, CA, USA) according to the
manufacturer's instructions.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The total RNA of tissue samples and cell lines was
extracted according to the instruction manual of TRIzol reagent
(Invitrogen Life Technologies). To quantify miR-455, total RNA was
reverse-transcribed using a TaqMan MicroRNA Reverse Transcription
kit (Applied Biosystems, Foster City, CA, USA) to synthesize cDNA,
and qPCR was performed with TaqMan MicroRNA PCR kit (Applied
Biosystems). For Notch1 mRNA expression, cDNA was synthesized using
a PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd.,
Dalian, China), and quantification of Nocth1 mRNA was performed
using a SYBR Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd.).
U6 and GAPDH were used as the internal control for miR-455 and
Notch1 mRNA, respectively. The primers used for miR-455 were
5′-GAACTGCAGTCCATGGGGATA-3′ (forward) and 5′-GCAGGGTCCGAGGTATTC-3′
(reverse). The primers for U6 5′-CTCGCTTCGGCAGCACA-3′ (forward) and
5′-AACGCTTCACGAATTTGCGT-3′ (reverse). The primers for Notch1 were
5′-GTGGATGACCTAGGCAAGTCG-3′ (forward) and
5′-GTCTCCTCCTTGTTGTTCTGC-3′ (reverse). The primers for GAPDH were
5′-GCACCGTCAAGGCTGAGAAC-3′ (forward) and 5′-TGGTGAAGACGCCAGTGGA-3′
(reverse). 2−ΔΔCT method was used to calculate the
relative expression (24).
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan).was used for analyzing the cell proliferation
following the manufacture's instructions. Briefly, transfected
cells were collected at 24 h post-transfection, and reseeded into
96-well plates at a density of 3×103 cells/well. Cells
were incubated at 37°C and cell proliferation was examined every 24
h according to the manufacture's instructions (0, 24, 48, 72 h). 10
µl of CCK-8 solution was added into each group of cells and
incubated at 37°C for 4 h. The absorbance of each sample was
determined at a wavelength of 450 nm using an automatic multi-well
spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
All samples were analyzed in triplicate and repeated three
times.
Cell invasion assay
Cell invasion assays were conducted using Transwell
chamber inserts (Costar; Corning Life Sciences, Cambridge, MA, USA)
with Matrigel (BD Biosciences, San Jose, CA, USA) according to the
manufacturer's protocol. In briefly, transfected cells were
collected at 48 h post-transfection, and seeded into the upper
chamber of the insert at a density 5×104 in 200 µl
FBS-free medium. The bottom of the insert was filled with DMEM
containing 20% FBS to serve as chemoattractant. After 20 h of
incubation, the cells remaining on the upper membrane were
carefully removed using cotton swabs. The invasive cells were fixed
in 100% methanol, stained with 0.5% crystal violet, washed with
PBS, and dried in air. Photographs of five manually selected fields
of the invasive cells were taken at ×200 magnification and counted
under an inverted light microscope (Olympus Corporation, Tokyo,
Japan).
miR-455 target prediction
miRNA target prediction algorithms: TargetScan
(release 7.0, http://www.targetscan.org/) and PicTar (http://pictar.mdc-berlin.de/) were used to forecast
the putative targets of miR-455.
Luciferase reporter assay
Luciferase reporter vector, pGL3-Notch1-3′UTR
wild-type (Wt) and pGL3-Notch1-3′UTR mutant (Mut), were synthesized
by Shanghai GenePharma Co., Ltd. For luciferase assays, cells were
plated in 24-well plates at a density of 1.5×105
cells/well, and then were co-transfected with pGL3-Notch1-3′UTR Wt
or pGL3-Notch1-3′UTR Mut, and miR-455 mimic or miR-NC, using
Lipofectamine 2000 following to the manufacturer's instructions.
After 48 h of incubation, luciferase activity was detected using
the Dual-Luciferase reporter assay system (Promega, Mannheim,
Germany) according to the manufacturer's protocol. Renilla
luciferase activity was used to normalize to firefly luciferase
activity.
Western blot analysis
Protein was extracted from tissues or cells with
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) containing proteinase inhibitor
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) on ice for 30 min.
After centrifugation at 4°C for 15 min at 13,000 rpm, the protein
concentration in the supernatant was examined using the BCA protein
assay (Pierce Biotechnology, Inc., Rockford, IL, USA). Equal
amounts of protein were resolved on a 10% SDS denaturing
polyacrylamide gel and transferred onto polyvinylidene difluoride
membranes (EMD Millipore, Billerica, MA, USA). Subsequently, the
membranes were blocked with 5% nonfat milk in Tris-buffered saline
with 0.05% Tween-20 (TBST) for 1 h at room temperature, and next
incubated with primary antibodies overnight at 4°C: Mouse
anti-human monoclonal Notch1 antibody (dilution, 1:1,000; cat no.
sc-373891) and mouse anti-human monoclonal GAPDH antibody
(dilution, 1:1,000; cat no. sc-47724) (both from Santa Cruz
Biotechnology, Inc., Dallas, TX, USA). The membranes were then
washed with TBST and incubated with horseradish peroxidase
(HRP)-conjugated goat anti-mouse IgG second antibodies (dilution,
1:5,000; cat no. sc-2005; Santa Cruz Biotechnology, Inc.) at room
temperature for 2 h. Finally, bands were visualized using an
enhanced luminol-based chemiluminescence detection kit (Pierce
Biotechnology, Inc.). GAPDH was used as a loading control.
Statistical analysis
Data are presented as the mean ± standard deviation
and statistical analysis was performed with Student's t-tests or
one-way analysis of variance plus multiple comparisons using SPSS
14.0 (SPSS Inc., Chicago, IL, USA). Spearman's correlation analysis
was used to analyze the inverse relationship of miR-455 and Notch1
mRNA expression level. P<0.05 was regarded as statistically
significant.
Results
miR-455 is downregulated in EOC tissue
samples and cell lines
miR-455 expression was detected in 45 paired EOC
tissues and their adjacent normal ovarian epithelial tissues
through quantitative reverse transcription polymerase chain
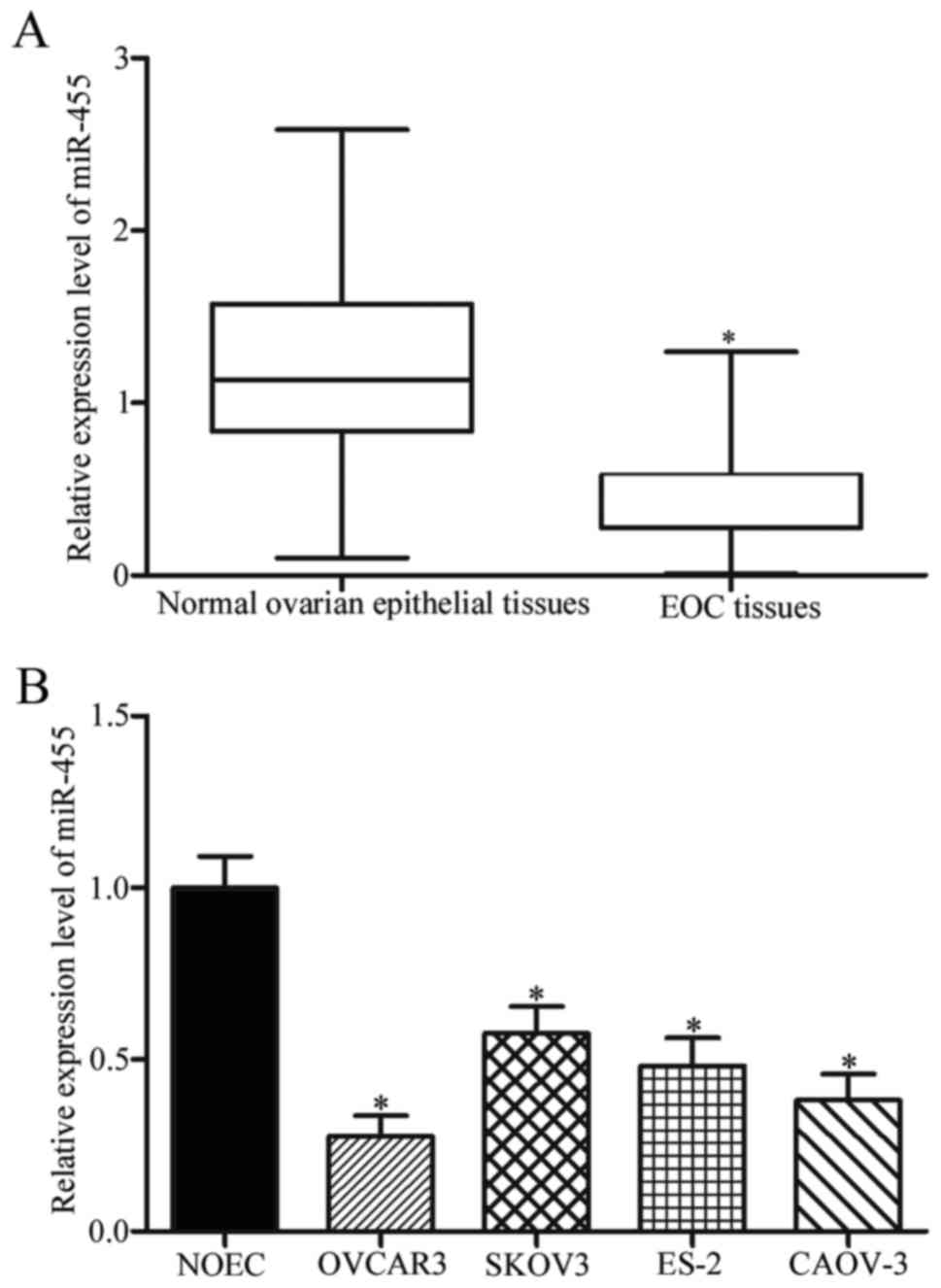
reaction (RT-qPCR). Compared with the levels in adjacent normal
ovarian epithelial tissues, miR-455 was significantly downregulated
in EOC tissues (Fig. 1A,
P<0.05). Consistent with this observation, our results confirmed
that miR-455 was also downregulated in OVCAR3, SKOV3, ES-2 and
CAOV-3 in comparison to the normal ovarian epithelial NOEC cell
line (Fig. 1B, P<0.05). These
results suggested that miR-455 expression was impaired in EOC and
might participate in EOC progression.
Association between miR-455 and
clinicopathological features in EOC patients
To evaluate the associations between miR-455
downregulation and various clinicopathological features of EOC
patients, we chose the median values of miR-455 in EOC tissues as
cutoff values and divided the EOC patients into low miR-455 group
(n=23) and high miR-455 group (n=22). In Table I, the low expression levels of
miR-455 were significantly correlated with the tumor size
(P=0.023), FIGO stage (P=0.004) and lymph node metastasis (P=0.020)
of patients with EOC. Conversely, no significant associations were
identified between miR-455 expression levels and other
clinicopathological variables, including age (P=0.300),
differentiation (P=0.449) and histological subtype (P=0.242).
 | Table I.Correlation between the expression of
microRNA-455 and clinicopathological variables of epithelial
ovarian cancer patients. |
Table I.
Correlation between the expression of
microRNA-455 and clinicopathological variables of epithelial
ovarian cancer patients.
|
|
| microRNA-455
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
variables | Case no. | Low | High | P-value |
|---|
| Age (years) |
|
|
| 0.300 |
|
<50 | 21 | 9 | 12 |
|
|
≥50 | 24 | 14 | 10 |
|
| Tumor size
(cm) |
|
|
| 0.023 |
|
<5 | 20 | 6 | 14 |
|
| ≥5 | 25 | 17 | 8 |
|
| FIGO stage |
|
|
| 0.004 |
|
I–II | 25 | 9 | 16 |
|
|
III–IV | 20 | 14 | 6 |
|
|
Differentiation |
|
|
| 0.449 |
|
I–II | 21 | 12 | 9 |
|
|
III | 24 | 11 | 13 |
|
| Histological
subtype |
|
|
| 0.242 |
|
Serous | 38 | 18 | 20 |
|
|
Non-serous | 7 | 5 | 2 |
|
| Lymph node
metastasis |
|
|
| 0.020 |
| No | 22 | 8 | 14 |
|
|
Yes | 23 | 15 | 8 |
|
miR-455 overexpression inhibits cell
proliferation and invasion of EOC
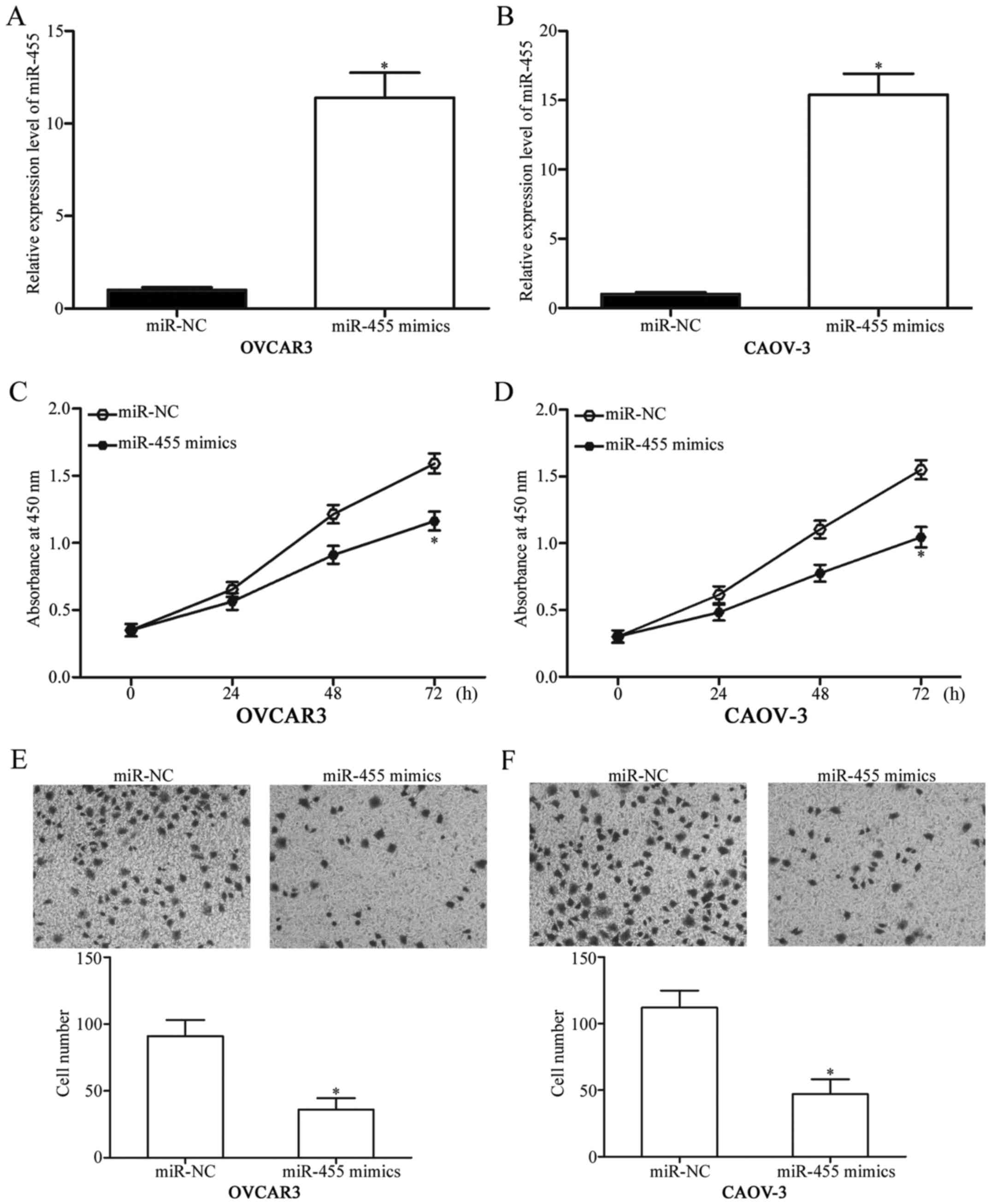
The cells were transfected with miR-455 mimics to
increase the endogenous miR-455 expression and to investigate the
potential biological roles of miR-455 in EOC, OVCAR3 and CAOV-3.
Using RT-qPCR, we found that miR-455 was markedly upregulated in
OVCAR3 and CAOV-3 cells transfected with the miR-455 mimics
compared with the cells transfected with miR-NC (Fig. 2A and B, P<0.05). The effects of
miR-455 overexpression on the proliferation of EOC cells were
examined through CCK-8 assay. In Fig.
2C and D, the miR-455 upregulation inhibited the proliferation
of OVCAR3 and CAOV-3 cells. A cell invasion assay was conducted to
evaluate the effects of miR-455 on the invasion capacity of EOC
cells. In Fig. 2E and F, the
restored expression of miR-455 decreased the invasion capacities of
OVCAR3 and CAOV-3 cells (P<0.05). These observations revealed
that miR-455 functioned as a tumor suppressor in EOC.
Notch1 is a direct target of miR-455
in EOC
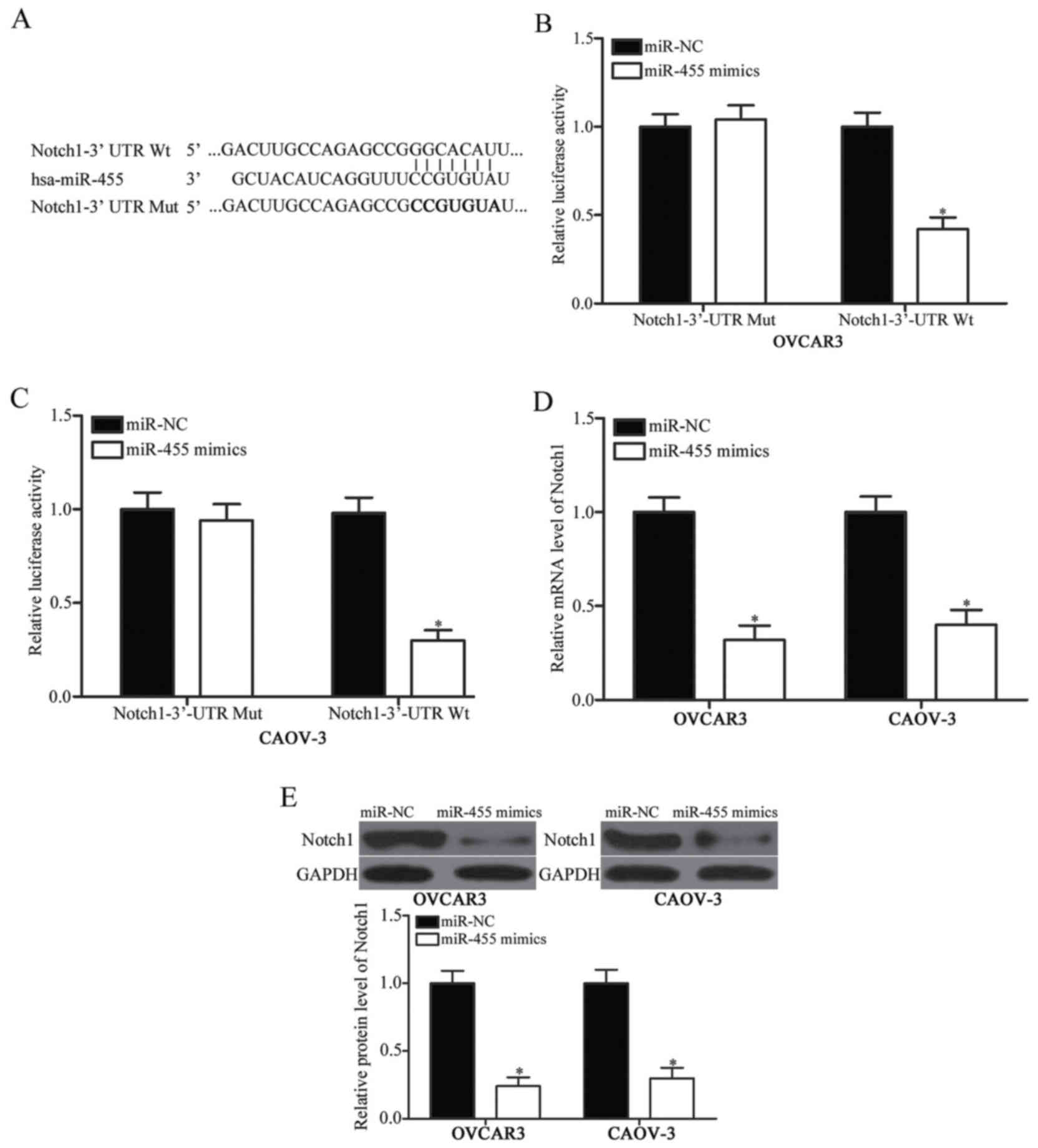
Bioinformatics analysis was conducted to predict the
putative targets of miR-455 and to investigate the molecular
mechanism by which miR-455 repressed EOC cell proliferation and
invasion. Numerous genes were identified as potential targets of
miR-455, and Notch1 (Fig. 3A) was
selected for further confirmation because Notch1 was involved in
EOC initiation and progression. A luciferase reporter assay was
carried out to investigate whether Notch1 was a target for miR-455.
OVCAR3 and CAOV-3 cells were co-transfected with pGL3-Notch1-3′-UTR
Wt or pGL3-Notch1-3′-UTR Mut and miR-455 mimics or miR-NC. We found
that the upregulation of miR-455 significantly reduced the
luciferase activity of pGL3-Notch1-3′-UTR Wt (Fig. 3B and C, P<0.05). By contrast,
the cells transfected with pGL3-Notch1-3′-UTR Mut were unaffected.
This finding suggested that miR-455 could directly target the
3′-UTR of Notch1. To confirm the endogenous regulatory role of
miR-455 in relation to Notch1 in EOC cells, we determined the
Notch1 expression in OVCAR3 and CAOV-3 cells transfected with
miR-455 mimic or miR-NC. In Fig. 3D
and 3E, the mRNA and protein levels of Notch1 were
downregulated in OVCAR3 and CAOV-3 cells transfected with miR-455
mimic (P<0.05). These results suggested that Notch1 might be a
direct target of miR-455 in EOC.
Inverse correlation between Notch1 and
miR-455 expression in EOC tissues
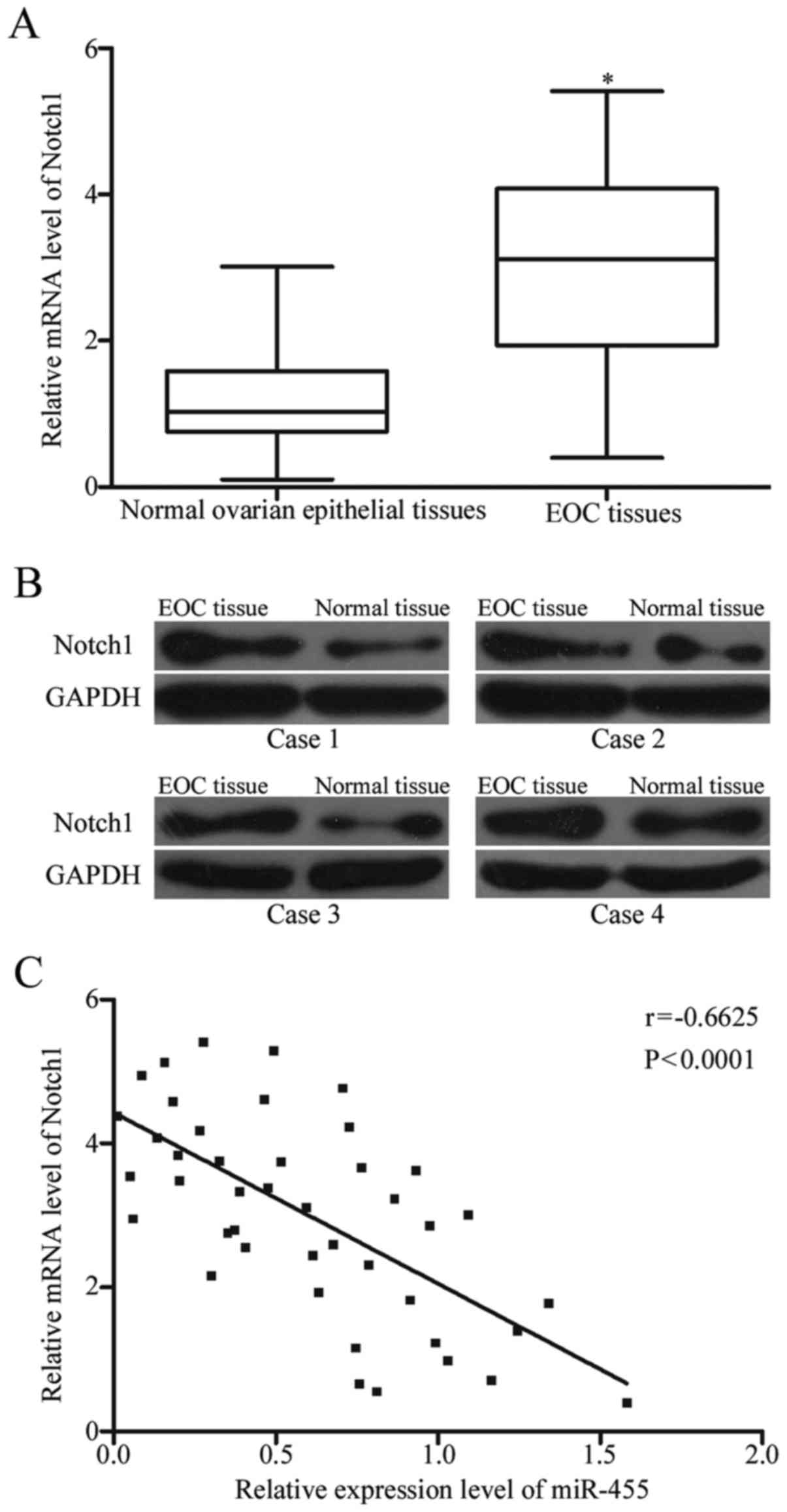
To explore the association between Notch1 and
miR-455, we examined the Notch1 expression in 45 paired EOC tissues
and their adjacent normal ovarian epithelial tissues through
RT-qPCR and Western blot analysis. The results showed that the
expression of Notch1 was higher in EOC tissues at mRNA and protein
levels than in normal ovarian epithelial tissues (Fig. 4A and B, P<0.05). Spearman's
correlation analysis was conducted to validate the relationship
between Notch1 mRNA and miR-455 expression in these clinical
tissues. We found that miR-455 expression was negatively correlated
with the mRNA level of Notch1 (Fig.
4C; r=-0.6625, P<0.0001). This observation supported that
Notch1 was a target gene of miR-455 in EOC.
Restored expression of Notch1 can
rescue the inhibitory effects on EOC cells induced by miR-455
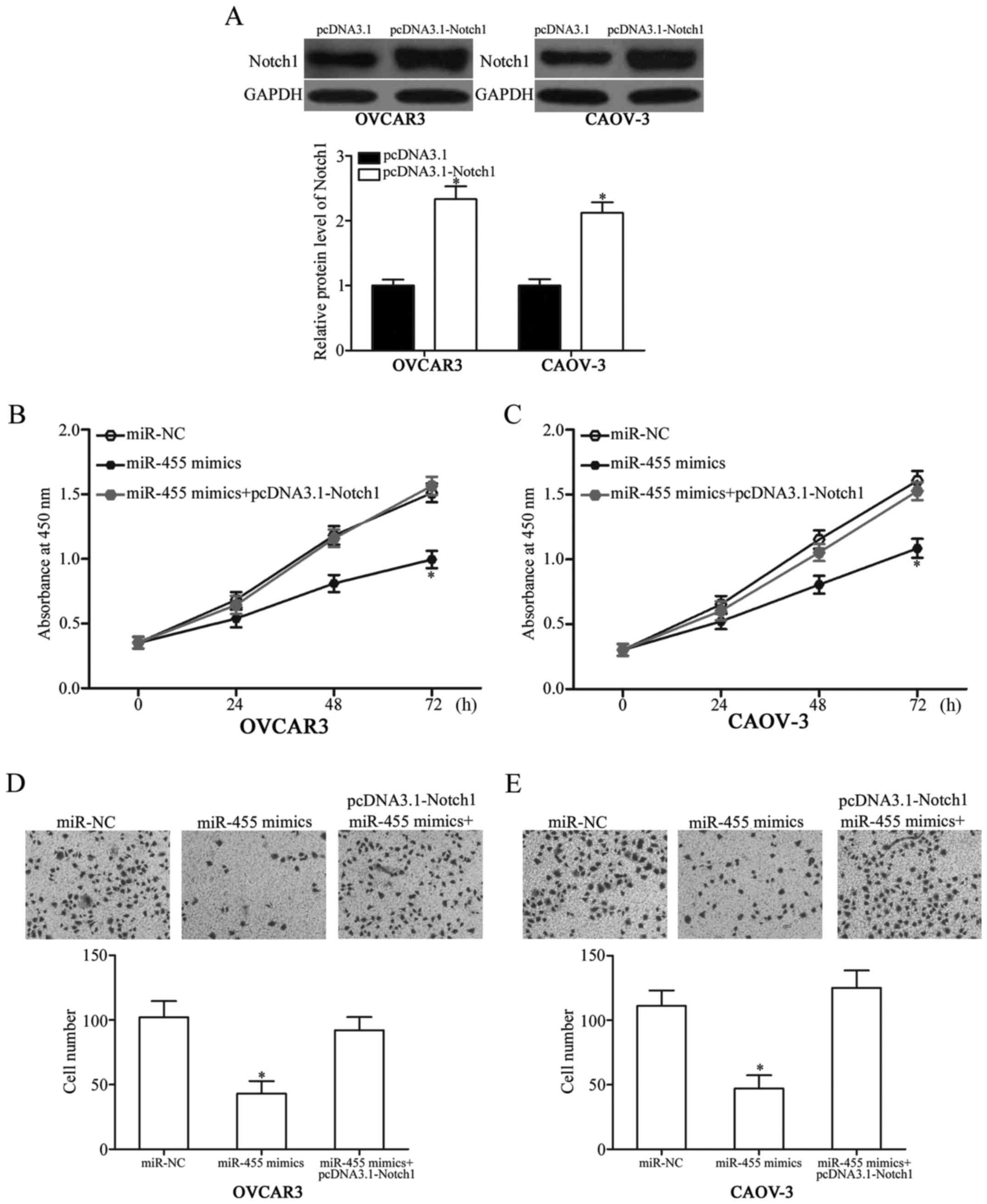
pcDNA3.1-Notch1 was transfected into OVCAR3 and
CAOV-3 cells and rescue experiments were performed to validate
whether Notch1 can mediate the tumor-suppressive effects of miR-455
on EOC cells. After transfection was completed, western blot
analysis confirmed that Notch1 was significantly upregulated in
OVCAR3 and CAOV-3 cells transfected with pcDNA3.1-Notch1 (Fig. 5A, P<0.05). We also found that
the upregulation of Notch1 could reverse the inhibitory effects of
miR-455 overexpression on cell proliferation (Fig. 5B and C, P<0.05) and invasion
(Fig. 5D and E, P<0.05) in
OVCAR3 and CAOV-3 cells. Overall, these results implied that
miR-455 inhibited the cell proliferation and invasion of EOC
partially by downregulating Notch1.
Discussion
miRNAs play important roles in cancer by acting as
tumor suppressors or oncogenes (25–27).
Many aberrantly expressed miRNAs have been reported in EOC and
involved in EOC cell proliferation, invasion, migration, metastasis
and apoptosis (13,28,29).
Therefore, miRNAs may be investigated as diagnostic and prognostic
molecular biomarkers and therapeutic targets for patients with EOC.
In this study, the miR-455 expression was downregulated in EOC
tissues and cell lines. Low miR-455 expression was correlated with
the tumor size, FIGO stage and lymph node metastasis of EOC
patients. In addition, the ectopic expression of miR-455 inhibited
the cell proliferation and invasion of EOC. Notch1 was also
identified as a direct target of miR-455 in EOC. These data
suggested that miR-455 might be implicated in the carcinogenesis
and progression of EOC.
Aberrant miR-455 expression has been observed in
various human cancers. For instance, miR-455 is weakly expressed in
hepatocellular carcinoma. miR-455 expression is correlated with
multiple tumor nodes, high Edmondson-Steiner grading, advanced
tumor node metastasis stage and venous infiltration of
hepatocellular carcinoma patients. miR-455 is also validated as a
novel prognostic indicator to predict the 5-year overall and
disease-free survival of hepatocellular carcinoma patients
(21). In gastric cancer, the
miR-455 expression level is downregulated in tumor tissues and
related to advanced clinical stage for patients with gastric cancer
(22). miR-455 downregulation is
also observed in nonsmall cell lung cancer (23) and colorectal cancer (30). However, miR-455 expression is
increased in oral squamous cancer tissues and cell lines (31). These findings suggested that
miR-455 expression exhibits tissue specificity and may be a
diagnostic and prognostic marker for cancers.
miR-455 is implicated in the development of many
tumors. Qin et al (21)
found that miR-455 upregulation suppresses cell migration and
invasion in hepatocellular carcinoma. Liu et al (22) demonstrated that miR-455
overexpression inhibits gastric cancer cell proliferation and
invasion in vitro. Li et al (23) revealed that enforced miR-455
expression attenuates the cell growth and metastasis of nonsmall
cell lung cancer. Chai et al (30) reported that the restored miR-455
expression represses the cell proliferation and invasion of
colorectal cancer. On the contrary, miR-455 performs oncogenic
roles in oral squamous cancer by promoting tumor cell proliferation
and invasion (31). This
contradiction may be explained by the ‘imperfect complementarity’
of the interactions between miRNAs and their target genes (32). These findings also suggested that
miR-455 is involved in the tumorigenesis and progression of these
cancer types and is a promising therapeutic target for the
treatment of cancer.
miRNAs perform its biological roles by negatively
regulating their target genes (9).
Therefore, the identification and characterisation of the targets
of altered miRNAs may help elucidate the molecular mechanisms
involved in carcinogenesis and progression. Previous studies
identified several miR-455 targets, including Runx2 (21) in hepatocellular carcinoma, RAB18
(22) in gastric cancer, ZEB1
(23) in lung cancer, RAF1
(30) in colorectal cancer and
UBE2B (31) in oral squamous
cancer. In this study, Notch1 was validated as a novel direct
target of miR-455 in EOC. TargetScan and Pictar predicted that
Notch1 was a potential miR-455 target. Secondly, luciferase
reporter assay revealed that miR-455 could directly target the
3′-UTR of Notch1. Thirdly, RT-qPCR and Western blot analysis
revealed that miR-455 reduced Notch1 expression at the mRNA and
protein levels in EOC cells. Fourthly, Notch1 was upregulated in
EOC tissues and inversely correlated with miR-455 expression level.
Finally, rescue experiments demonstrated that the Notch1
upregulation could reverse the inhibitory effects of miR-455
overexpression in EOC cells. These findings indicated that miR-455
was involved in EOC carcinogenesis and progression by directly
targeting Notch1.
Notch signalling pathway consists of three
components: Notch ligands, Notch receptors 1–4 and downstream
target genes (33). Notch1, a
member of Notch receptors, is a highly conserved I type
transmembrane glycoprotein (34).
Notch1 is aberrantly and highly expressed in multiple human
cancers, such as breast cancer (35), colorectal cancer (36), bladder cancer (37), gastric cancer (38), renal cell carcinoma (39) and oesophageal squamous cell cancer
(40). Moreover, Notch1
upregulation contributes to cancer cell proliferation, apoptosis,
angiogenesis, migration, invasion and metastasis (41–43).
In EOC, Notch1 expression is higher in tumor tissues and cell lines
than in matched normal tissues and normal tissues, respectively
(44,45). Notch1 expression is associated with
EOC tumor stage, differentiation status and FIGO stage (44,45).
High EOC expression is correlated strongly with poor prognosis
(45). Functional assays revealed
that Notch1 participates in regulating EOC cell proliferation,
apoptosis, invasion and angiogenesis (46–49).
Indeed, our work confirmed that miR-455 upregulation inhibited EOC
cell proliferation and invasion through the negative regulation of
Notch1. These findings suggested that the miR-455/Notch1 signalling
axis may provide efficient therapeutic targets for the treatment of
EOC patients.
In conclusion, miR-455 was downregulated in EOC
tissues and cell lines. Low Notch1 expression was correlated with
tumor size, FIGO stage and lymph node metastasis. miR-455 inhibited
EOC cell proliferation and invasion by directly targeting Notch1.
.However, the weakness of this study is that we do not explore the
effects of miR-450 on EOC cell growth and metastasis in
vivo. Besides, future studies needed to investigate whether the
miR-455/Notch1 pathway might be exploited in a therapeutic approach
for the treatment of patients with EOC.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mutch DG and Prat J: 2014 FIGO staging for
ovarian, fallopian tube and peritoneal cancer. Gynecol Oncol.
133:401–404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nodin B, Zendehrokh N, Sundström M and
Jirström K: Clinicopathological correlates and prognostic
significance of KRAS mutation status in a pooled prospective cohort
of epithelial ovarian cancer. Diagn Pathol. 8:1062013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei
Y, Li Y, Yang Q, Liu J, Wei JJ, et al: miR-145 inhibits tumor
growth and metastasis by targeting metadherin in high-grade serous
ovarian carcinoma. Oncotarget. 5:10816–10829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pereira A, Magrina JF, Magtibay PM,
Pérez-Medina T, Fernández A and Peregrin I: The impact of
peritoneal metastases in epithelial ovarian cancer with positive
nodes. Int J Gynecol Cancer. 21:1375–1379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Candido-dos-Reis FJ, Song H, Goode EL,
Cunningham JM, Fridley BL, Larson MC, Alsop K, Dicks E, Harrington
P, Ramus SJ, et al: Germline mutation in BRCA1 or BRCA2 and
ten-year survival for women diagnosed with epithelial ovarian
cancer. Clin Cancer Res. 21:652–657. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bi L, Yang Q, Yuan J, Miao Q, Duan L, Li F
and Wang S: MicroRNA-127-3p acts as a tumor suppressor in
epithelial ovarian cancer by regulating the BAG5 gene. Oncol Rep.
36:2563–2570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moss EG: MicroRNAs: Hidden in the genome.
Curr Biol. 12:R138–R140. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zaman MS, Maher DM, Khan S, Jaggi M and
Chauhan SC: Current status and implications of microRNAs in ovarian
cancer diagnosis and therapy. J Ovarian Res. 5:442012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen L, Zhang F, Sheng XG, Zhang SQ, Chen
YT and Liu BW: MicroRNA-106a regulates phosphatase and tensin
homologue expression and promotes the proliferation and invasion of
ovarian cancer cells. Oncol Rep. 36:2135–2141. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang T, Li L, Cheng Y, Ren C and Zhang G:
MicroRNA-194 promotes the growth, migration, and invasion of
ovarian carcinoma cells by targeting protein tyrosine phosphatase
nonreceptor type 12. Onco Targets Ther. 9:4307–4315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin ZY, Chen G, Zhang YQ, He HC, Liang YX,
Ye JH, Liang YK, Mo RJ, Lu JM, Zhuo YJ, et al: MicroRNA-30d
promotes angiogenesis and tumor growth via MYPT1/c-JUN/VEGFA
pathway and predicts aggressive outcome in prostate cancer. Mol
Cancer. 16:482017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cong J, Liu R, Wang X, Wang J, Wang H and
Hou J: Low miR-498 expression levels are associated with poor
prognosis in ovarian cancer. Eur Rev Med Pharmacol Sci.
19:4762–4765. 2015.PubMed/NCBI
|
|
16
|
Zhou X, Yue Y, Wang R, Gong B and Duan Z:
MicroRNA-145 inhibits tumorigenesis and invasion of cervical cancer
stem cells. Int J Oncol. 50:853–862. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li C, Lu S and Shi Y: MicroRNA-187
promotes growth and metastasis of gastric cancer by inhibiting
FOXA2. Oncol Rep. 37:1747–1755. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W,
Liu Z and Huang JA: MicroRNA-205 targets SMAD4 in non-small cell
lung cancer and promotes lung cancer cell growth in vitro and in
vivo. Oncotarget. 8:30817–30829. 2017.PubMed/NCBI
|
|
19
|
Guo J, Cao R, Yu X, Xiao Z and Chen Z:
MicroRNA-223-3p inhibits human bladder cancer cell migration and
invasion. Tumour Biol. 39:10104283176916782017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qin L, Zhang Y, Lin J, Shentu Y and Xie X:
MicroRNA-455 regulates migration and invasion of human
hepatocellular carcinoma by targeting Runx2. Oncol Rep.
36:3325–3332. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu J, Zhang J, Li Y, Wang L, Sui B and
Dai D: miR-455-5p acts as a novel tumor suppressor in gastric
cancer by down-regulating RAB18. Gene. 592:308–315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li YJ, Ping C, Tang J and Zhang W:
MicroRNA-455 suppresses non-small cell lung cancer through
targeting ZEB1. Cell Biol Int. 40:621–628. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Y, Dong Q and Wang E: MicroRNA-320
inhibits invasion and induces apoptosis by targeting CRKL and
inhibiting ERK and AKT signaling in gastric cancer cells. Onco
Targets Ther. 10:1049–1058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen Y, Ye YF, Ruan LW, Bao L, Wu MW and
Zhou Y: Inhibition of miR-660-5p expression suppresses tumor
development and metastasis in human breast cancer. Genet Mol Res.
16:2017. View Article : Google Scholar
|
|
27
|
Zhuo HC, Song YF, Ye J, Lai GX and Liu DL:
MicroRNA-154 functions as a tumor suppressor and directly targets
HMGA2 in human non-small cell lung cancer. Genet Mol Res. 15:2016.
View Article : Google Scholar
|
|
28
|
Xiaohong Z, Lichun F, Na X, Kejian Z,
Xiaolan X and Shaosheng W: MiR-203 promotes the growth and
migration of ovarian cancer cells by enhancing glycolytic pathway.
Tumour Biol. 37:14989–14997. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao X, Zhou Y, Chen YU and Yu F: miR-494
inhibits ovarian cancer cell proliferation and promotes apoptosis
by targeting FGFR2. Oncol Lett. 11:4245–4251. 2016.PubMed/NCBI
|
|
30
|
Chai J, Wang S, Han D, Dong W, Xie C and
Guo H: MicroRNA-455 inhibits proliferation and invasion of
colorectal cancer by targeting RAF proto-oncogene
serine/threonine-protein kinase. Tumour Biol. 36:1313–1321. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng CM, Shiah SG, Huang CC, Hsiao JR and
Chang JY: Up-regulation of miR-455-5p by the TGF-β-SMAD signalling
axis promotes the proliferation of oral squamous cancer cells by
targeting UBE2B. J Pathol. 240:38–49. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yu Z, Ni L, Chen D, Zhang Q, Su Z, Wang Y,
Yu W, Wu X, Ye J, Yang S, et al: Identification of miR-7 as an
oncogene in renal cell carcinoma. J Mol Histol. 44:669–677. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiao L and Wong BC: Role of Notch
signaling in colorectal cancer. Carcinogenesis. 30:1979–1986. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Purow BW, Haque RM, Noel MW, Su Q, Burdick
MJ, Lee J, Sundaresan T, Pastorino S, Park JK, Mikolaenko I, et al:
Expression of Notch-1 and its ligands, Delta-like-1 and Jagged-1,
is critical for glioma cell survival and proliferation. Cancer Res.
65:2353–2363. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhong Y, Shen S, Zhou Y, Mao F, Lin Y,
Guan J, Xu Y, Zhang S, Liu X and Sun Q: NOTCH1 is a poor prognostic
factor for breast cancer and is associated with breast cancer stem
cells. Onco Targets Ther. 9:6865–6871. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chu D, Zhou Y, Zhang Z, Li Y, Li J, Zheng
J, Zhang H, Zhao Q, Wang W, Wang R and Ji G: Notch1 expression,
which is related to p65 Status, is an independent predictor of
prognosis in colorectal cancer. Clin Cancer Res. 17:5686–5694.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sima J, Zhu MY, Ai Q, Zhang C, Yao ZY,
Huang QB and Zhang X: Expression analysis of NOTCH1/HES1/PTEN
signaling pathway in invasive bladder transitional cell carcinoma.
Zhonghua Yi Xue Za Zhi. 92:964–967. 2012.(In Chinese). PubMed/NCBI
|
|
38
|
Zhang H, Wang X, Xu J and Sun Y: Notch1
activation is a poor prognostic factor in patients with gastric
cancer. Br J Cancer. 110:2283–2290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ai Q, Ma X, Huang Q, Liu S, Shi T, Zhang
C, Zhu M, Zhang Y, Wang B, Ni D, et al: High-level expression of
Notch1 increased the risk of metastasis in T1 stage clear cell
renal cell carcinoma. PLoS One. 7:e350222012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ogawa R, Ishiguro H, Kimura M, Funahashi
H, Wakasugi T, Ando T, Shiozaki M and Takeyama H: NOTCH1 expression
predicts patient prognosis in esophageal squamous cell cancer. Eur
Surg Res. 51:101–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Won HY, Lee JY, Shin DH, Park JH, Nam JS,
Kim HC and Kong G: Loss of Mel-18 enhances breast cancer stem cell
activity and tumorigenicity through activating Notch signaling
mediated by the Wnt/TCF pathway. FASEB J. 26:5002–5013. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sharma A, Paranjape AN, Rangarajan A and
Dighe RR: A monoclonal antibody against human Notch1 ligand-binding
domain depletes subpopulation of putative breast cancer stem-like
cells. Mol Cancer Ther. 11:77–86. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Al-Hussaini H, Subramanyam D, Reedijk M
and Sridhar SS: Notch signaling pathway as a therapeutic target in
breast cancer. Mol Cancer Ther. 10:9–15. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang M, Wang J, Wang L, Wu L and Xin X:
Notch1 expression correlates with tumor differentiation status in
ovarian carcinoma. Med Oncol. 27:1329–1335. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Alniaimi AN, Demorest-Hayes K, Alexander
VM, Seo S, Yang D and Rose S: Increased Notch1 expression is
associated with poor overall survival in patients with ovarian
cancer. Int J Gynecol Cancer. 25:208–213. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rose SL, Kunnimalaiyaan M, Drenzek J and
Seiler N: Notch 1 signaling is active in ovarian cancer. Gynecol
Oncol. 117:130–133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Liang T, Guo Q, Li L, Cheng Y, Ren C and
Zhang G: MicroRNA-433 inhibits migration and invasion of ovarian
cancer cells via targeting Notch1. Neoplasma. 63:696–704. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gao Y, Rankin GO, Tu Y and Chen YC:
Theaflavin-3, 3′-digallate decreases human ovarian carcinoma
OVCAR-3 cell-induced angiogenesis via Akt and Notch-1 pathways, not
via MAPK pathways. Int J Oncol. 48:281–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zou W, Ma X, Hua W, Chen B and Cai G:
Caveolin-1 mediates chemoresistance in cisplatin-resistant ovarian
cancer cells by targeting apoptosis through the Notch-1/Akt/NF-κB
pathway. Oncol Rep. 34:3256–3263. 2015. View Article : Google Scholar : PubMed/NCBI
|