Introduction
Tumor necrosis factor (TNF) ligand superfamily
member 10 (TRAIL), a member of the TNF superfamily, is considered
to be an optimal candidate for cancer therapy due to its tumor cell
specificity and negligible cytotoxicity to normal cells in
vitro and in vivo (1).
TRAIL has antitumor activity in a variety of tumor cell lines
(2,3). However, a large number of cancer
cells, particularly in highly malignant tumors, are resistant to
apoptosis induction by TRAIL, and cancer cells that were originally
sensitive to TRAIL-induced apoptosis may become resistant following
repeated exposure (4). Therefore,
it is of importance to develop a more efficient form of TRAIL for
cancer therapy.
Crossing biological barriers represents a principal
limitation for the clinical application of biomolecules including
nucleic acids, peptides or proteins (5). Previous studies have used
cell-penetrating peptides (CPPs) as an efficient method for
delivering therapeutic targets into cells (6,7).
CPPs, additionally termed protein transduction domains, comprise
short and usually basic amino acid-rich peptides originating from
proteins which are able to cross biological barriers. Typically,
these peptides are cationic, rich in lysine or arginine residues,
or amphipathic in nature (8,9).
CPPs have emerged as a novel class of non-viral vectors allowing
for the delivery of various biomolecules across biological barriers
from low molecular weight drugs to nano-sized particles (10).
In the present study, a number of amino acids of the
N-terminal in soluble fragments (114-281aa) of the TRAIL protein
were selectively altered to form penetrating peptide-like amino
acid sequences. A total of >10 TRAIL penetrating peptide like
mutants were synthesized. Following further detection and
screening, one TRAIL penetrating peptide like mutant with eight
consecutive Arg sequences, which approximately maintained the
conformation of the TRAIL protein and had a penetrating peptide
like structure, was identified. This TRAIL mutant membrane
penetrating peptide alike (TMPPA) was termed TRAIL-Mu3. The present
study examined the design, synthesis, expression, identification
and purification of TRAIL-Mu3. In addition, the antitumor effects
of TRAIL-Mu3 in colorectal cancer was detected in vitro and
in vivo.
Materials and methods
Materials
The high-efficiency prokaryotic expression vector
pET32a plasmid and the BL21 (DE3) competent Escherichia coli
cells were purchased from Invitrogen (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). The pMD19-T vector was purchased from
Takara Bio, Inc. (Otsu, Japan). The pMD19/TRAIL plasmid and
wild-type TRAIL protein were obtained from Chengdu Huachuang
Biotechnology Co., Ltd. (Chengdu, China). The TRAIL polyclonal
antibody was purchased from Santa Cruz Biotechnology, Inc. (Dallas,
TX, USA). The HCT-15, COLO 205, SW620, HT-29 and HCT 116 colon
cancer cell lines were purchased from Wuhan Institute of Virology,
Chinese Academy of Sciences (Wuhan, China). Female nude mice (6
weeks old) were purchased from the Animal Experimental Center of
the Chinese Academy of Sciences (Shanghai, China).
Sequence and primer design of
TRAIL-Mu3
The 114–121 amino acid coding sequence VRERGPQR of
the wild-type TRAIL protein was selected and altered to become
RRRRRRRR. The upstream primer (Mu3-TR-NdeI) was 5′-ggt cat
atg cgt cgt cgt cgt cgt cgt cgt cgt gtg gct gct cac atc a-3′ and
the downstream primer (TR-EcoR) is 5′-gtt gaa ttc tta tta
acc aac aag gaa agc acc gaa gaa ag-3′.
Amplification of the TRAIL-Mu3
gene
The genomic DNA of the pMD19/TRAIL plasmid was used
as template DNA to perform polymerase chain reaction (PCR) analysis
for the amplification of the TRAIL-Mu3 gene. Amplified products
were purified and separated via electrophoresis (3% agarose gel) to
confirm whether the amplified products had the desired size
(11).
Ligation of pMD19-T vector and
TRAIL-Mu3 gene, and identification
The recovered TRAIL-Mu3 gene was ligated to the
Takara pMD19-T vector and digested using EcoRI and
HindIII enzymes. The gene was identified by electrophoresis.
The plasmids of the corrected transformed bacteria containing the
inserted gene segment were used for sequencing. The correctly
sequenced bacteria were stored for further study (12).
Ligation of pET32a plasmid and
TRAIL-Mu3 gene, and identification
The TRAIL-Mu3 DNA was ligated to the pET32a plasmid
and digested by XbaI and EcoRI enzymes. Subsequently,
the gene was identified by electrophoresis. The plasmids of the
corrected transformed bacteria containing the inserted gene segment
were used for sequencing. The correctly sequenced bacteria were
stored for further study (12).
pET32a/TRAIL-Mu3 expression
The plasmid pET32a/TRAIL-Mu3 was transformed into
BL21 (DE3) competent E. coli cells, which were cultured in
Luria-Bertani solid medium containing Ampicillin at 4°C overnight.
A single colony was isolated and cultured. The cultured liquid was
centrifuged (13,800 × g, 4°C, 10 min) and the precipitate was
resuspended to generate the prior-to-induction electrophoresis
sample. The remaining liquid was cultured and induced with
isopropyl β-D-thiogalactopyranoside to generate the post-induction
electrophoresis sample.
The culture liquid was centrifuged (13,800 × g, 4°C,
10 min) and the supernatant was discarded. The precipitate was
resuspended, broken down using ultrasound and further centrifuged
(13,800 × g, 4°C, 10 min). The precipitate was resuspended. The
20-µl supernatant and resuspended precipitate was used to make
separate electrophoresis samples.
All the above electrophoresis samples were disposed
in boiling water bath for 10 min. Subsequently, the samples were
centrifuged (13,800 × g, 4°C, 10 min) and analyzed by
electrophoresis (15% gel, 200 V, 35 min).
Purification of TRAIL-Mu3 protein
The TRAIL-Mu3 expression bacteria were broken down
by ultrasound and centrifuged. The supernatant was removed and
filtered with a 0.45-µm membrane. Subsequently, the filtered
supernatant was purified via cation exchange purification,
hydroxyapatite purification and anion exchange purification
methods, successively (13).
Western blot analysis of TRAIL-Mu3
protein
The TRAIL-Mu3 protein exhibited only 5 amino acids
which were altered compared with the N terminal of the wild-type
TRAIL protein, and the epitopes of the wild-type TRAIL protein were
conserved. Therefore, a TRAIL polyclonal antibody was used for the
detection and identification of TRAIL-Mu3 protein by western
blotting (14,15). The primary antibody used rabbit
anti-human TRAIL polyclonal antibody (cat no. bs-1214R) (1:500) at
4°C overnight. The secondary antibody used was a goat anti-rabbit
IgG-HRP (cat no. D2313; 1:5,000) at room temperature for 2 h.
Finally, the results were detected using an enhanced
chemiluminescence detection reagent (Beyotime Institute of
Biotechnology, Haimen, China).
Immunofluorescence analysis
The subcellular localization of TRAIL and TRAIL-Mu3
was detected in SW620 colon cancer cells by immunofluorescence
(16).
TRAIL-Mu3-mediated toxicity in
colorectal cancer cell lines
The antitumor effects of TRAIL-Mu3 and TRAIL were
detected in 32 different tumor cell lines (including many TRAIL
resistant tumor cell lines) through a Cell Counting Kit-8 (CCK-8).
The cell growth inhibition rates of TRAIL-Mu3 and TRAIL in
colorectal cancer cell lines (including HCT-15, COLO 205, SW620,
HT-29 and HCT 116) measured by CCK-8 assay were discussed in this
manuscript (17). The
concentrations of reagents that induced a 50% reduction in cell
viability [half-maximal inhibitory concentration,
(IC50)] were determined from the curves of reagent
concentration compared with the cell growth inhibition rate at 48 h
of incubation for the cell line analyzed. The sensitivity of cells
to a drug is evaluated by calculating the IC50 value;
IC50 <10 µg/ml indicates that cells are sensitive to
a drug, while IC50 ≥10 µg/ml suggests that cells are
relatively resistant to a drug (18–19).
Antitumor effects of HT-29 xenograft
in nude mice
All animal procedures were approved by the Animal
Care and Scientific Committee of Sichuan University (Chengdu,
China). A total of 48 HT-29-bearing nude mice (SPF, aged 6–8 weeks,
18–22 g) were divided into 6 groups (8 mice/group): Group 1,
treated with vehicle (saline); group 2, treated with TRAIL-Mu3 at a
concentration of 5 mg/kg; group 3, treated with TRAIL-Mu3 at a
concentration of 15 mg/kg; group 4, treated with TRAIL-Mu3 at a
concentration of 45 mg/kg; and group 5, treated with TRAIL at a
concentration of 45 mg/kg. The mice were bred at 23±2°C, with a
humidity of 40–70% and a 12/12 light/dark cycle with free access to
food and water. The vehicle, TRAIL and TRAIL-Mu3 were injected
through the tail vein five times in 5 days. The length, width, and
weight of the tumor was measured using a slide caliper every 3 or 4
days. Tumor volume (TV) was estimated using the formula: TV
(mm3) = (width2 × length)/2. The growth
inhibition rate of the tumor was calculated using the formula:
Inhibition rate of tumor (%) = (1 - average weight in treated
group/average weight in control) ×100 (11,12).
Side effects
During the experimental period, side effects,
including weight loss, mental state, appetite, behavior change and
reactions, were observed.
Statistical analysis
The experiments were repeated at least three times.
All data are expressed as the mean ± standard error of the mean
unless otherwise stated. Comparisons were analyzed using one-way
analysis of variance and Least Significant Difference post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
PCR amplification of TRAIL-Mu3
gene
The amplified products of the TRAIL-Mu3 gene were
detected by 3% agarose gel electrophoresis, and the results
demonstrated that a specific segment of ~500 bp (Fig. 1A) was obtained.
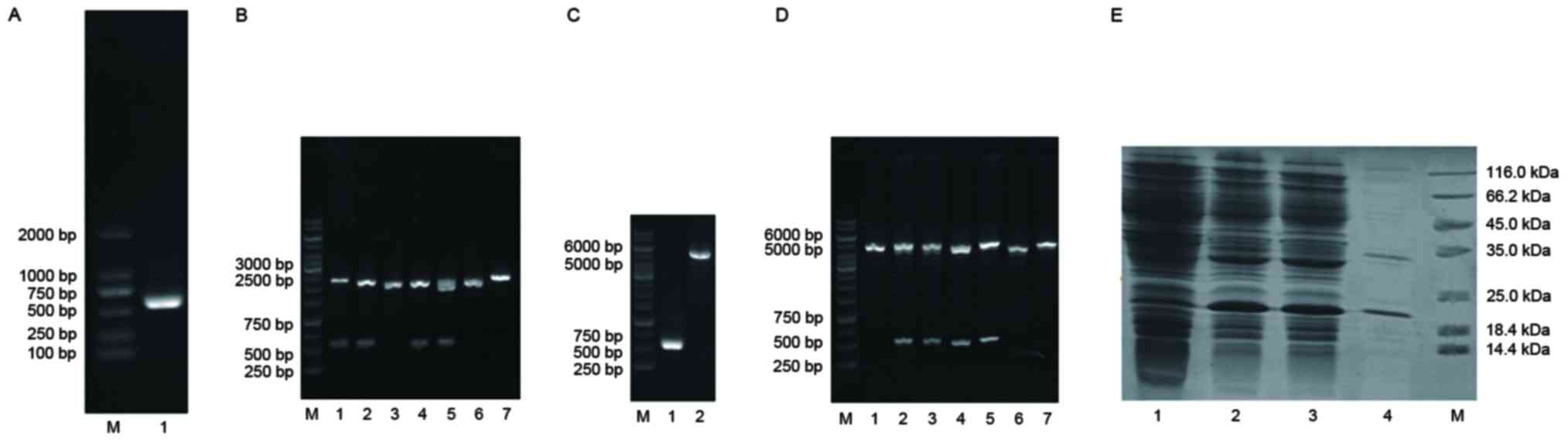 | Figure 1.Results of electrophoresis
identification during TRAIL-Mu3 synthesis. (A) Polymerase chain
reaction products of the TRAIL-Mu3 gene. M, DNA marker; lane 1,
TRAIL-Mu3. (B) Digestion of pMD19/TRAIL-Mu3 and electrophoresis
products. M, DNA marker; lanes 1, 2, 4 and 5, four colonies
transformed with pMD19/TRAIL-Mu3; lanes 3, 6 and 7, three colonies
transformed with pMD19-T vector. (C) Digestion of the pET32a
plasmid and TRAIL-Mu3 DNA. M, marker; lane 1, TRAIL-Mu3; lane 2,
pET32a plasmid. (D) Digestion of pET32a/TRAIL-Mu3 and
electrophoresis products. Lane M, marker; lanes 2, 3, 4 and 5, four
colonies transformed with pET32a/TRAIL-Mu3; lanes 1, 6 and 7, three
colonies transformed with the pET32a plasmid. (E) pET32a/TRAIL-Mu3
expression electrophoresis. Line M, marker; lane 1, induction
electrophoresis sample; lane 2, post-electrophoresis sample; lane
3, supernatant; lane 4, precipitate. TRAIL, tumor necrosis factor
ligand superfamily member 10. |
Identification of the enzyme-digested
products of pMD19/ TRAIL-Mu3
Following ligation of the pMD19-T vector and
TRAIL-Mu3 genes, they were transformed into bacteria and cultured.
A total of seven single colonies were isolated and the plasmids
were extracted. Subsequently, the plasmids were digested and
detected by agarose gel electrophoresis. The results demonstrated
that there were two segments of ~500 bp and ~2.7 kb in lines 1, 2,
4 and 5 (Fig. 1B), which were
equal to the size of the TRAIL-Mu3 gene and pMD19-T vector plasmid,
respectively. Therefore, these four colonies were transformed with
pMD19/TRAIL-Mu3, and were further assayed by sequencing.
Identification of enzyme-digested
products of pET32a/TRAIL-Mu3
The pET32a plasmid and TRAIL-Mu3 DNA were digested
by NdeI and EcoRI enzymes and detected by
electrophoresis. The results demonstrated that there were two
segments of ~500 bp and ~5.4 kb (Fig.
1C), which were equal to the size of the TRAIL-Mu3 gene and
pET32a plasmid, respectively. Subsequently, the plasmids were
ligated, transformed into bacteria and cultured. A total of seven
single colonies were isolated and the plasmids were extracted. The
plasmids were digested and detected by agarose gel electrophoresis.
The results demonstrated that there were two segments of ~500 bp
and ~5.4 kb in lines 2, 3, 4 and 5 (Fig. 1D), which were equal to the size of
the TRAIL-Mu3 gene and pET32a plasmid, respectively. Therefore,
these four colonies were transformed with pET32a/TRAIL-Mu3, and
they were further analyzed by sequencing.
pET32a/TRAIL-Mu3 expression
As presented in Fig.
1E, there was strong expression in the prior-to-induction
electrophoresis sample, post-electrophoresis sample, supernatant
and precipitate, and that this was most apparent in the
supernatant.
Purification of TRAIL-Mu3 protein
The results of this analysis are presented in
Fig. 2. Following cation exchange
purification (Fig. 2A), 15 ml
eluent with a concentration of 2.273 mg/ml was obtained. The purity
of TRAIL-Mu3 was thus demonstrated to be high. Via hydroxyapatite
purification (Fig. 2B), 12 ml
hydroxyapatite eluent with a concentration of 2.080 mg/ml was
obtained. The purpose of this step was to attempt to remove
contaminating proteins and pyrogen. Following anion exchange
purification (Fig. 2C) 20 ml flow
through fluid with a concentration of 0.846 mg/ml was obtained. In
this step, the pyrogen was further removed. Following repeated
purification, sufficient protein was obtained to evaluate its
bioactivity in vitro and in vivo.
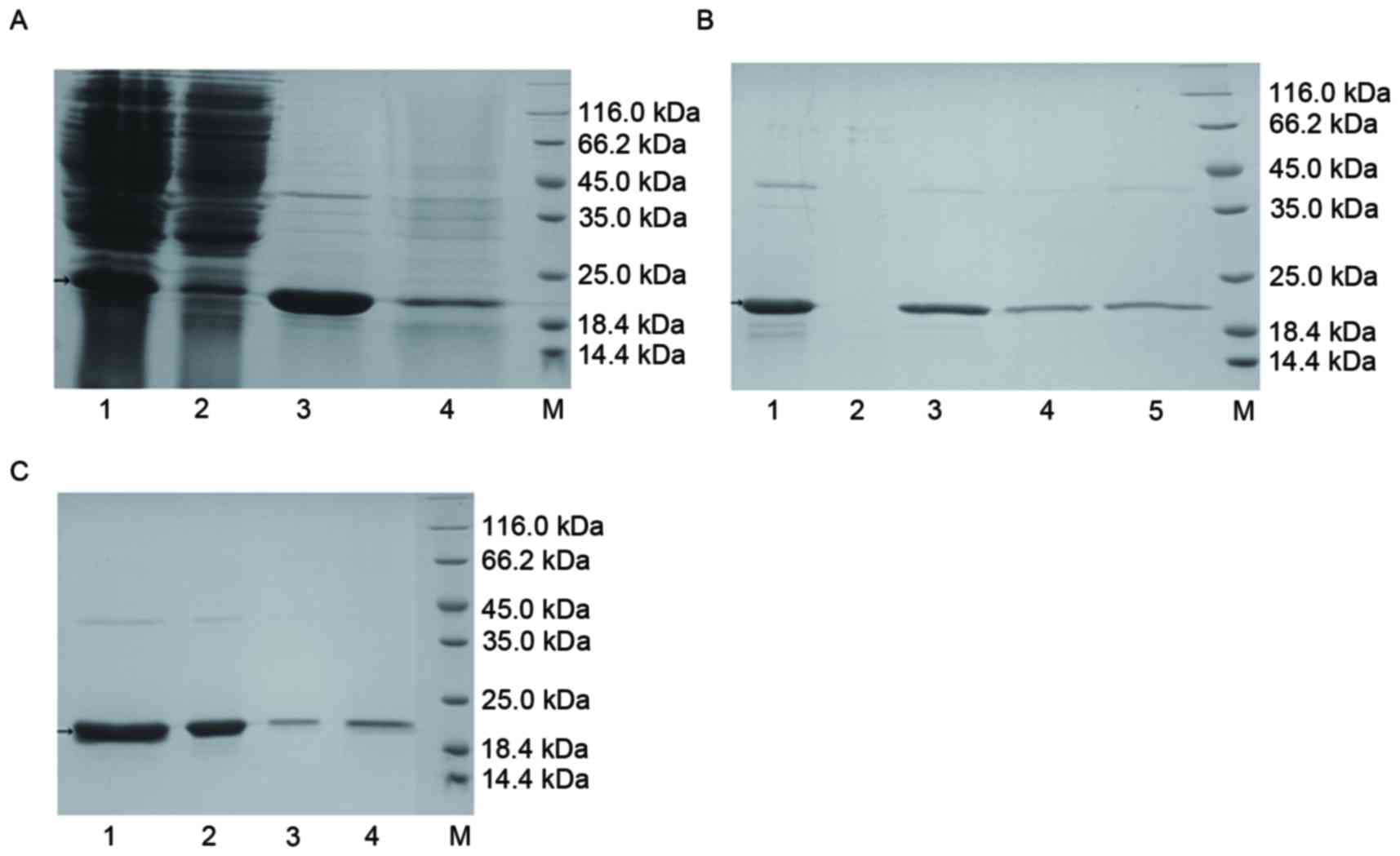 | Figure 2.Purification of TRAIL-Mu3 protein. (A)
Cation exchange purification. Lane 1, cation exchange liquid; lane
2, cation exchange flow-through liquid; lane 3, cation exchange
eluate; lane 4, cation exchange sodium hydroxide eluent; M,
unstained protein molecular weight marker. (B) Hydroxyapatite
purification. Lane 1, hydroxyapatite sample solution; lane 2,
hydroxyapatite flow-through liquid; lane 3, hydroxyapatite sodium
chloride eluent; lane 4, hydroxyapatite phosphate eluent; M,
unstained protein molecular weight marker. (C) Anion exchange
purification. Lane 1, anion exchange liquid; lane 2, anion exchange
flow-through liquid; lane 3, sodium chloride eluent; lane 4, sodium
hydroxide eluent; M, unstained protein molecular weight marker.
TRAIL, tumor necrosis factor ligand superfamily member 10. |
Western blot analysis of TRAIL-Mu3
protein
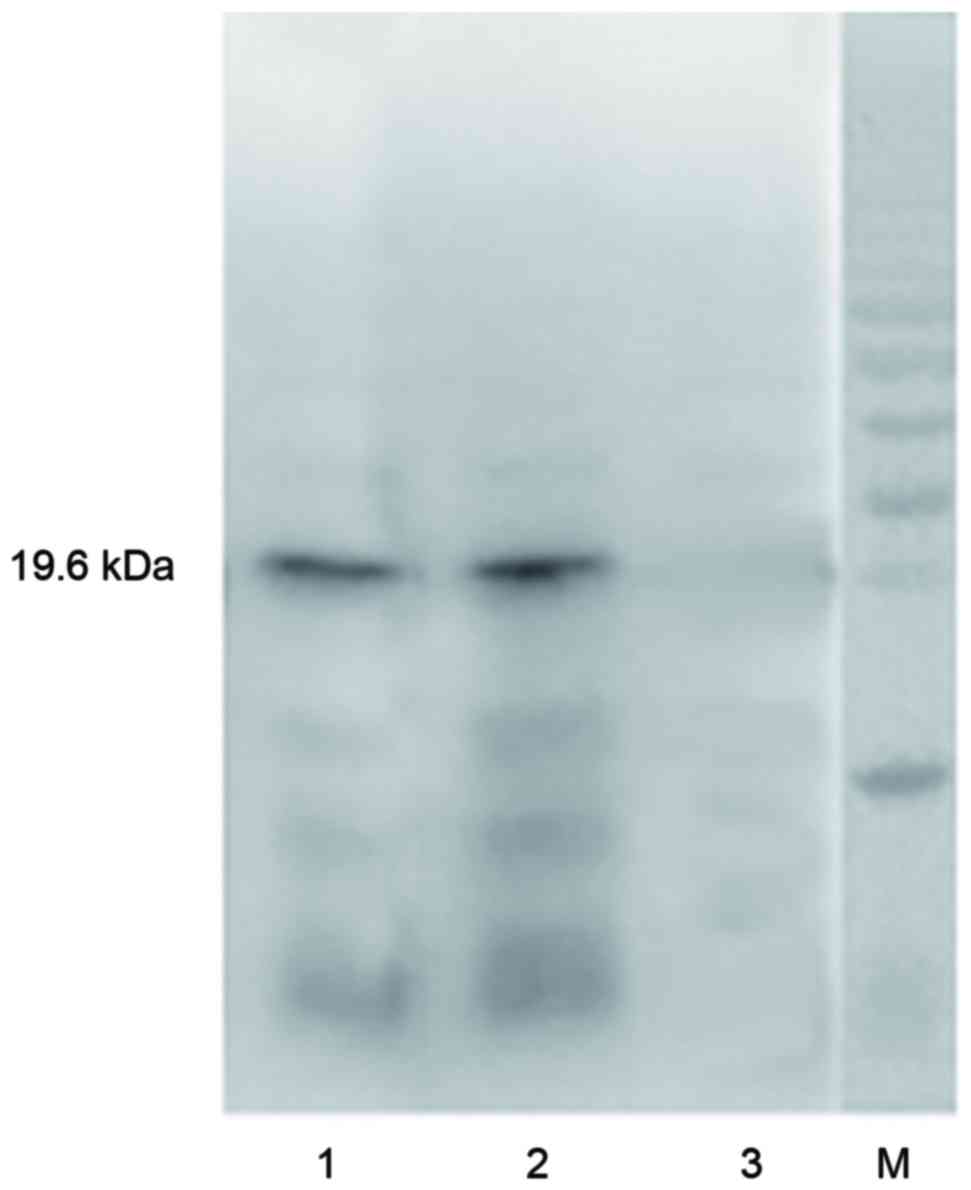
The results of the western blotting (Fig. 3) demonstrated that there was a
positive band of ~19.6 kDa for TRAIL-Mu3 and the TRAIL protein.
However, the same band was not present for the supernatant of lysed
BL21 (DE3) E. coli.
Immunofluorescence analysis
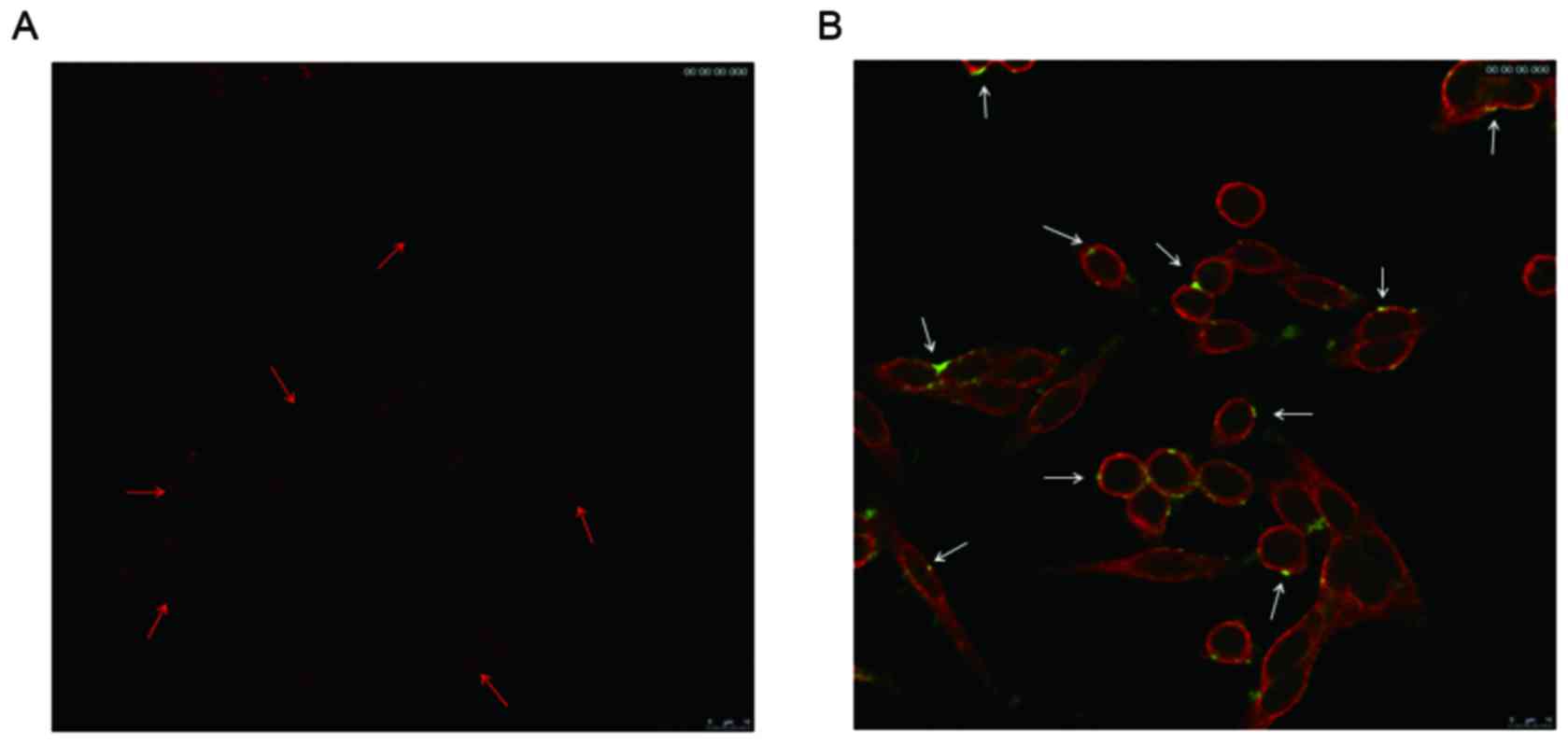
The results demonstrated that there was little TRAIL
aggregated on the cell membrane of SW620 cells (Fig. 4A); however, extensive TRAIL-Mu3 was
observed around the cell membrane of the cells (Fig. 4B).
TRAIL-Mu3 mediated toxicity on
colorectal cancer cell lines
The IC50values of TRAIL and TRAIL-Mu3 in
various colorectal cancer cell lines are presented in Table I. The results demonstrated that
TRAIL-Mu3 was able to enhance the antitumor effects of TRAIL in all
five colorectal cancer cell lines. In addition, TRAIL-Mu3 was able
to reverse the resistance of TRAIL-resistant HT-29 colorectal
cancer cells.
 | Table I.The IC50 values of TRAIL
and TRAIL-Mu3 in various colorectal cancer cell lines. |
Table I.
The IC50 values of TRAIL
and TRAIL-Mu3 in various colorectal cancer cell lines.
|
| IC50,
µg/ml |
|---|
|
|
|
|---|
| Cell line | TRAIL | TRAIL-Mu3 |
|---|
| HCT-15 | 0.008 | 0.001 |
| COLO 205 | 0.008 | 0.002 |
| SW620 | 0.009 | 0.002 |
| HT-29 | >100 | 0.030 |
| HCT 116 | 0.015 | 0.002 |
Antitumor effects in HT-29 xenograft
nude mice
The treatment commenced on day 10 following the
injection of tumor cells. The tumor growth curves and inhibitive
rates are presented in Fig. 5. The
results demonstrated that the tumor growth inhibition rate
increased with the increase in TRAIL-Mu3 concentration. In
addition, the tumor growth inhibition rate of TRAIL-Mu3 was
significantly increased compared with TRAIL at the same
concentration of 45 mg/kg.
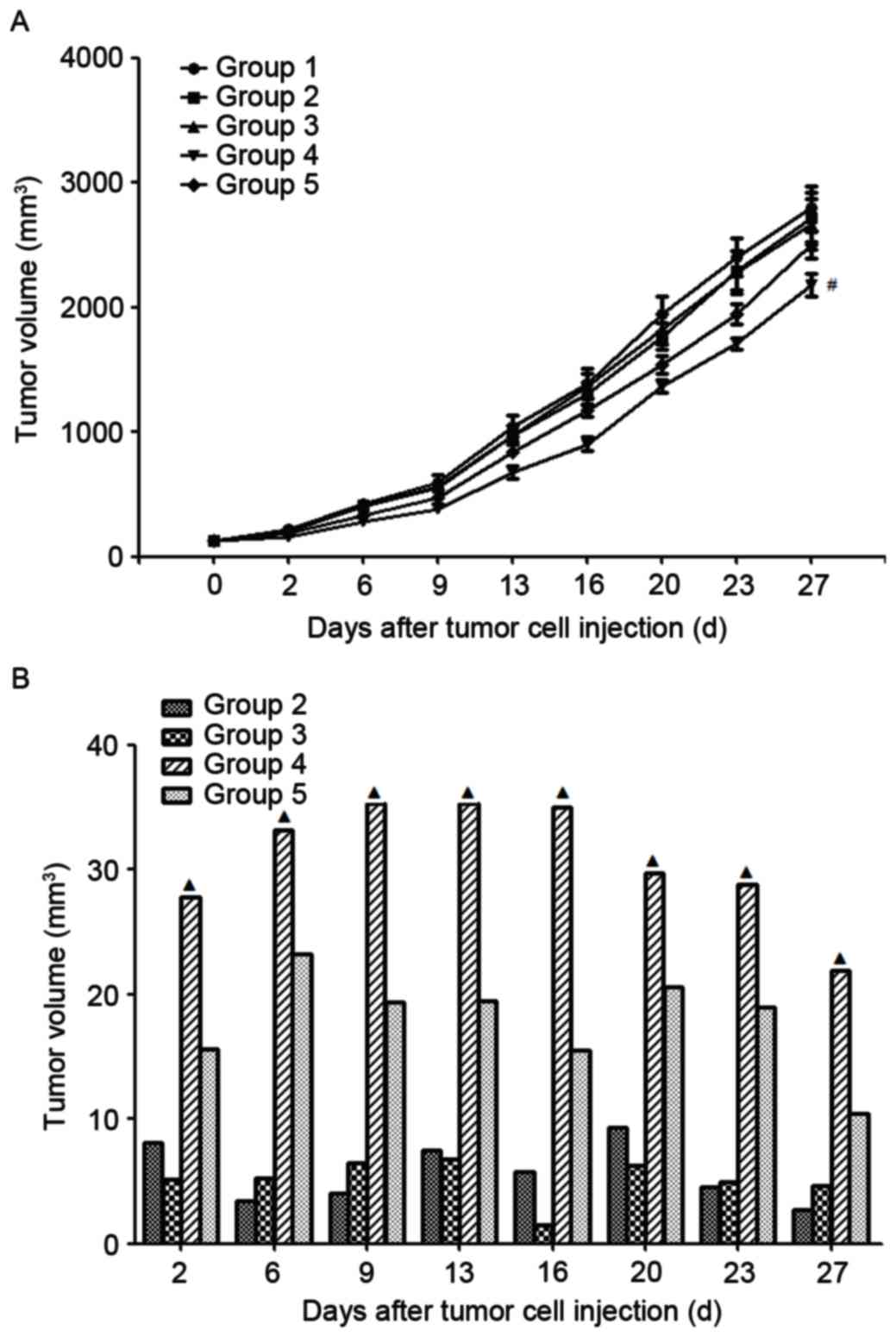 | Figure 5.(A) Tumor growth curve and (B)
inhibition rates. Group 1, treated with vehicle; Group 2, treated
with TRAIL-Mu3 at a concentration of 5 mg/kg; group 3, treated with
TRAIL-Mu3 at a concentration of 15 mg/kg; group 4, treated with
TRAIL-Mu3 at a concentration of 45 mg/kg; and group 5, treated with
TRAIL at a concentration of 45 mg/kg. #P<0.05 vs.
group 1, 2, 3, 5; ▲P<0.05 vs. group 2, 3, 5. TRAIL,
tumor necrosis factor ligand superfamily member 10. |
Side effects, survival quality and
analysis
All the mice in the TRAIL and TRAIL-Mu3 groups began
to manifest slight syndromes such a stunt responses, bad appetite,
and little activity on the 8th day after the treatment. However,
there were no significant differences between TRAIL and TRAIL-Mu3
groups in mental status, weight, appetite and so on.
Discussion
TRAIL, which is able to selectively induce apoptosis
in cancer cells, is a potential targeted drug for cancer therapy
(20). However, its clinical use
is limited by cellular resistance to cell death which occurs in
~50% of cancer cells (21).
Various methods have been developed to overcome TRAIL resistance
(4,22).
In recent years, CPPs have become one of the most
popular techniques for intracellular access (6). CPPs are typically short cationic
sequences and may be derived from natural sources or be
synthetically designed constructs. It was previously demonstrated
that this sequence was able to be shortened to a few amino acids,
without altering its translocation capacity (23). In addition, this highly efficient
translocation capacity has been observed in a variety of cell lines
with minimal toxicity, overcoming challenges frequently associated
with other delivery methods (24,25).
In the present study, 5 amino acids of the
extracellular region (114-281aa) of wild-type TRAIL were
selectively altered to form a continuous sequence of 8 Arg residues
which approximately maintained the conformation of TRAIL protein,
and additionally had a penetrating peptide-like structure. This
TMPPA was termed TRAIL-Mu3.
Following PCR amplification with designed primers, a
target gene of ~500 bp was obtained. The gene was ligated with
pMD19-T vector and pET32a plasmid, successively. Following
transformation into bacteria, culturing, enzyme digestion,
electrophoresis and sequencing, it was confirmed that TRAIL-Mu3 was
successfully synthesized. Positively-transformed bacteria with
plasmid pET32a/TRAIL-Mu3 were obtained. The following SDS-PAGE
electrophoresis demonstrated that the positively-transformed
bacteria with plasmid pET32a/TRAIL-Mu3 were able to express
TRAIL-Mu3 successfully. Subsequently, cation exchange purification,
hydroxyapatite purification and anion exchange purification methods
were used to purify the TRAIL-Mu3 proteins. The results
demonstrated that the purity of TRAIL-Mu3 protein was high. Western
blot analysis further confirmed that the TRAIL-Mu3 protein was
successfully obtained.
TRAIL-Mu3 has a penetrating peptide-like structure;
therefore, the affinity of TRAIL-Mu3 to the cancer cell membrane
was detected. In the immunofluorescence analysis, it was observed
that TRAIL-Mu3 exhibited significantly stronger affinity to the
SW620 colorectal cancer cell membrane compared with TRAIL. It was
hypothesized that TRAIL-Mu3 may enhance the affinity to the cancer
cell membrane. TRAIL-Mu3 was able to aggregate on the cancer cell
membrane, and exerted increased signal transduction and antitumor
effects.
The present study aimed to investigate the antitumor
effects of TRAIL-Mu3 on tumor cells in vitro and in
vivo. The antitumor effects of TRAIL-Mu3 and TRAIL were
detected in 32 different tumor cell lines through CCK-8 method. It
was observed that TRAIL-Mu3 exhibited markedly stronger antitumor
effects compared with TRAIL in these 32 tumor cell lines (data not
shown), including 5 colorectal tumor cell lines. Additionally,
TRAIL-Mu3 was able to reverse the resistance of TRAIL-resistant
tumor cell lines (data not shown) including HT-29 tumor cell line.
The antitumor effects of TRAIL-Mu3 were further detected in HT-29
tumor-bearing mice. Similarly, TRAIL-Mu3 at the same concentration
exhibited significantly better antitumor effects compared with
TRAIL on HT-29 tumor-bearing nude mice.
To the best of our knowledge, the present study was
the first to introduce the concept of mutations in TRAIL and to
obtain the novel drug TRAIL-Mu3. In addition, RAIL-Mu3 exerted
stronger antitumor effects compared with TRAIL in colorectal cancer
in vitro and in vivo. However, there remain certain
limitations to the present study. The detailed mechanisms
underlying the antitumor effects of TRAIL-Mu3 were not completely
elucidated. Future studies are required to complete the
investigation into the mechanism.
The present study constructed a novel drug TMPPA
termed TRAIL-Mu3. TRAIL-Mu3 exerted significantly stronger
antitumor effects compared with TRAIL on colorectal cancer in
vitro and in vivo. TRAIL-Mu3 was able to reverse the
resistance of TRAIL-resistant HT-29 colorectal cancer cells
successfully.
Acknowledgements
The present study was supported by grants from the
National Natural Scientific Foundation of China (grant nos.
81301962 and 81372444).
References
|
1
|
Fulda S: Safety and tolerability of TRAIL
receptor agonists in cancer treatment. Eur J Clin Pharmacol.
71:525–527. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wiley SR, Schooley K, Smolak PJ, Din WS,
Huang CP, Nicholl JK, Sutherland GR, Smith TD, Rauch C, Smith CA,
et al: Identification and characterization of a new member of the
TNF family that induces apoptosis. Immunity. 3:673–682. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ashkenazi A: Targeting the extrinsic
apoptosis pathway in cancer. Cytokine Growth Factor Rev.
19:325–331. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dimberg LY, Anderson CK, Camidge R,
Behbakht K, Thorburn A and Ford HL: On the TRAIL to successful
cancer therapy? Predicting and counteracting resistance against
TRAIL-based therapeutics. Oncogene. 32:1341–1350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Adamik P, Emmenegger T, Briedis M,
Gustafsson L, Henshaw I, Krist M, Laaksonen T, Liechti F, Procházka
P, Salewski V and Hahn S: Barrier crossing in small avian migrants:
Individual tracking reveals prolonged nocturnal flights into the
day as a common migratory strategy. Sci Rep. 6:215602016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koren E and Torchilin VP: Cell-penetrating
peptides: Breaking through to the other side. Trends Mol Med.
18:385–393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wagstaff KM and Jans DA: Protein
transduction: Cell penetrating peptides and their therapeutic
applications. Curr Med Chem. 13:1371–1387. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin W, Xie X, Yang Y, Fu X, Liu H and Deng
J: Thermosensitive magnetic liposomes with doxorubicin
cell-penetrating peptides conjugate for enhanced and targeted
cancer therapy. Drug Deliv. 23:3436–3443. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sugawara K, Shinohara H, Kadoya T and
Kuramitz H: Sensing lymphoma cells based on a
cell-penetrating/apoptosis-inducing/electron-transfer peptide
probe. Anal Chim Acta. 924:106–113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoo J, Lee D, Gujrati V, Rejinold NS,
Lekshmi KM, Uthaman S, Jeong C, Park IK, Jon S and Kim YC:
Bioreducible branched poly(modified nona-arginine) cell-penetrating
peptide as a novel gene delivery platform. J Control Release.
246:142–154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu H, Li Z, Mao S, Ma B, Zhou S, Deng L,
Liu T, Cui D, Zhao Y, He J, et al: Antitumor effect of sFlt-1 gene
therapy system mediated by Bifidobacterium Infantis on Lewis lung
cancer in mice. Cancer Gene Ther. 18:884–896. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li ZJ, Zhu H, Ma BY, Zhao F, Mao SH, Liu
TG, He JP, Deng LC, Yi C and Huang Y: Inhibitory effect of
Bifidobacterium infantis-mediated sKDR prokaryotic expression
system on angiogenesis and growth of Lewis lung cancer in mice. BMC
Cancer. 12:1552012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gan HT, Lee J, Latiff SM, Chuah C, Toh P,
Lee WY and Gagnon P: Characterization and removal of aggregates
formed by nonspecific interaction of IgM monoclonal antibodies with
chromatin catabolites during cell culture production. J Chromatogr
A. 1291:33–40. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng H, Yuan Z, Zhu H, Li L, Shi H, Wang
Z, Fan Y, Deng Q, Zeng J, He Y, et al: Expression of hPNAS-4
radiosensitizes Lewis lung cancer. Int J Radiat Oncol Biol Phys.
84:e533–e540. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu H, Zhang Y, Hu X, Yi C, Zhong S, Wang
Y and Yang F: The effects of high-dose qinggan huoxue recipe on
acute liver failure induced by d-galactosamine in rats. Evid Based
Complement Alternat Med. 2013:9057152013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang C, Yan X, Meng T, Hu T and Pan J:
Immunofluorescence analysis of membrane-associated proteins for
clathrin-mediated endocytosis in plant root cells. Methods Mol
Biol. 1662:151–157. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Li D, Hu Z, Zhao S, Zheng Z and Li
W: Protective effects of green tea polyphenol against renal injury
through ros-mediated JNK-MAPK pathway in lead exposed rats. Mol
Cells. 39:508–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu H, Zhao F, Yu S, He J, Deng L, Yi C
and Huang Y: The synergistic effects of low-dose irinotecan and
TRAIL on TRAIL-resistant HT-29 colon carcinoma in vitro and in
vivo. Int J Mol Med. 30:1087–1094. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu H, Huang M, Ren D, He J, Zhao F, Yi C
and Huang Y: The synergistic effects of low dose fluorouracil and
TRAIL on TRAIL-resistant human gastric adenocarcinoma AGS cells.
Biomed Res Int. 2013:2938742013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui DD, Huang Y, Mao SH, Chen SC, Qiu M,
Ji LL and Yi C: Synergistic antitumor effect of TRAIL and
adriamycin on the human breast cancer cell line MCF-7. Braz J Med
Biol Res. 42:854–862. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Zhao J, Zhu W, Gou H, Cao D, Yang
Y, Huang Y and Yi C: Synergistic effect of subtoxic-dose cisplatin
and TRAIL to mediate apoptosis by down-regulating Decoy receptor 2
and up-regulating caspase-8, caspase-9 and bax expression on
NCI-H460 and A549 cells. Iran J Basic Med Sci. 16:710–718.
2013.PubMed/NCBI
|
|
22
|
Jiang Q, Zhu H, Liang B, Huang Y and Li C:
Apoptosis-inducing effect of the DR5 monoclonal antibody, D-6,
alone or in combination with cisplatin, on A2780 ovarian cancer
cells. Mol Med Rep. 6:316–320. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Falanga A, Galdiero M and Galdiero S:
Membranotropic cell penetrating peptides: The outstanding Journey.
Int J Mol Sci. 16:25323–25337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xie X, Yang Y, Lin W, Liu H, Liu H, Yang
Y, Chen Y, Fu X and Deng J: Cell-penetrating peptide-siRNA
conjugate loaded YSA-modified nanobubbles for ultrasound triggered
siRNA delivery. Colloids Surf B Biointerfaces. 136:641–650. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Parnaste L, Arukuusk P, Zagato E,
Braeckmans K and Langel U: Methods to follow intracellular
trafficking of cell-penetrating peptides. J Drug Target.
24:508–519. 2016. View Article : Google Scholar : PubMed/NCBI
|