Introduction
Tongue squamous cell carcinoma (TSCC) has a
particularly high mortality rate among malignancies, predominantly
due to the difficulty of early detection, and its incidence is
increasing rapidly according to the American Cancer Society
(1,2). Studies have shown that TSCC has a
poorer prognosis and higher recurrence rate, compared with several
other types of cancer due to metastasis being common (3). Clinical results suggest that a lack
of available biomarkers result in the majority of patients with
TSCC not being diagnosed until an advanced stage. Therefore,
understanding the molecular basis of tongue carcinogenesis may
provide potential molecular markers for more rapid diagnosis of
TSCC. Several biological molecules have been reported in the saliva
and serum, and histological analysis has indicated that they are
useful for screening and early detection of several types of cancer
(4–6).
Serum is important for supporting cell viability and
pH maintenance (7), whereas saliva
is important in oral hygiene, food digestion and the repair of
mucosal damage, as demonstrated by animal behavioural experiments
and in vivo analysis of patients (8). Studies have suggested that saliva and
serum are functionally similar and reflect the physiological state
of the body (9–11), and that saliva is a non-invasive
sample for potential clinical diagnosis. In the present study,
serum deprivation experiments were performed on the Tca-8113 TSCC
cell line to screen for potential biomarkers suitable for TSCC
diagnosis. This was based on previous observations that serum
deprivation synchronises proliferating cells at the G0 phase,
induces apoptosis and stimulates the fibrotic response (12).
The results of the present study showed that the
Tca-8113 cells resisted serum deprivation-induced apoptosis.
Two-dimensional gel electrophoresis (2-DE) and mass
spectrometry-based protein identification were used for the
proteomic analysis of serum-deprived Tca-8113 cells. The results
provided insight for potential diagnostic biomarkers or drug
targets for the treatment of TSCC.
Materials and methods
Cell culture and MTT assay
The cells were routinely cultured, and MTT
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) assays were
performed as previously described (13). Briefly, the TSCC Tca-811 cell line,
provided by the Ninth People's Hospital of Shanghai (Shanghai,
China) was maintained by routine culturing in RPMI-1640 medium with
10% fetal bovine serum (FBS) (both from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in a 37°C incubator with 5%
CO2. The cells were digested with 0.25% trypsin and
seeded into 96-well plates (cat no. 3524; Corning Incorporated,
Corning, NY, USA) at a density of 1×104 cells/well and
cultured for 18–24 h to 90% confluence. Serum deprivation was
performed following washing twice with PBS. The cells were cultured
for 12, 24, 36, 48 and 72 h following serum deprivation in the
media at 37°C, with 10% serum as a control. Following cell culture,
the culture medium were discarded, 200 µl of 0.5 mg/ml MTT in PBS
was added to wells, and the cells were cultured for 4 h at 37°C.
Following removal of the supernatants, 200 µl of DMSO was added and
cells were shaken lightly for 15 min. The optical density value was
measured using a microplate spectrophotometer (POWERWAVE 340;
BioTek Instruments, Inc., Winooski, VT, USA) at 490 nm. All
experiments were repeated four times and the mean ± standard
deviation of results were calculated.
Proteomic analysis
The proteomic experiments were performed as
described previously (14). The
steps were as follows: i) At least 1.0×108 Tca-8113
cells were harvested and lysed for sample preparation, and the
bicinchoninic acid method was used for measurement of protein
concentration; ii) each 80 µg sample was subjected to 2-DE on an
Ettan IPGphor II apparatus (GE Healthcare Life Sciences, Chalfont,
UK) in triplicate. Silver-stained gels were scanned at 300 dpi
resolution, and differential analysis was performed to assign
spots. Student's t-test was used for filtering, and spots with a
fold-change >1.2 were considered differentially expressed; iii)
selected spots were subjected to MALDI-TOF mass spectrometry and
tandem TOF/TOF mass spectrometry. The identified peptides were used
for Basic Local Alignment Search Tool searches of the International
Protein Index rat sequence database (http://www.matrixscience.com). The protein number,
protein name, sequence coverage, score and Mr/pI were obtained and
included in the analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
The serum-deprivation treated Tca-8113 cells were
prepared for one-step RT-qPCR SYBR Green assay as previous
described (15). Briefly, RNA was
extracted using the TRIzol LS reagent and the cDNA Synthesis kit
(both from Gibco; Thermo Fisher Scientific, Inc.) was used for cDNA
synthesis. A total of 1 µl reverse transcriptase product was used
to amplify the six genes (Annexin A1, peroxiredoxin 6, heat shock
protein 27, heat shock protein 70, caspase-7 and GAPDH). Primers
(Table I) were designed based on
the sequences of the five differentially expressed proteins
identified in the proteomic analysis and GAPDH gene was used as
reference. The primers were synthesized by Sangon Biotech
(Shanghai) Co., Ltd. (Shanghai, China). The SYBR Green Real-Time
PCR Master Mix (TOYOBO; Toyobo Life Science, Inc., Osaka, Japan)
was used and the PCR cycling parameters were as follows: 94°C for
30 sec, followed by 35 cycles of 60°C for 30 sec and 72°C for 30
sec. A MyCycler thermal cycler (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was used for the PCR analysis. Each gene was
quantified by 2−ΔΔCq method (16) and the relative expression level was
calculated by the ratio of each targeted gene to GAPDH gene.
 | Table I.Reverse transcription-polymerase chain
reaction primer sequences for the six genes. |
Table I.
Reverse transcription-polymerase chain
reaction primer sequences for the six genes.
| Gene | Primer sequence | Length (bp) |
|---|
| Annexin A1 | F:
5′-AAGACTTGGCTGATTCAGATGC-3′ | 124 |
|
| R:
5′-AACACTCTGCGAAGTTGTGGATA-3′ |
|
| Peroxiredoxin 6 | F:
5′-CGCATCCGTTTCCACGACTT-3′ | 271 |
|
| R:
5′-TGGCAAGCTCCCGATTCCTAT-3′ |
|
| Heat shock protein
27 | F:
5′-AGTGGTCGCAGTGGTTAGGC-3′ | 186 |
|
| R:
5′-GGTTGACATCCAGGGACACG-3′ |
|
| Heat shock protein
70 | F:
5′-CTGCTGCGACAGTCCACTACC-3′ | 177 |
|
| R:
5′-TCGGCTCCGCTCTGAGATT-3′ |
|
| Caspase-7 | F:
5′-GAAGAGGCTCCTGGTTTGTG-3′ | 106 |
|
| R:
5′-CTGGCAACTCTGTCATTCACC-3′ |
|
| GAPDH | F:
5′-GGGAAACTGTGGCGTGAT-3′ | 299 |
|
| R:
5′-GAGTGGGTGTCGCTGTTGA-3′ |
|
Statistical analysis
Data are reported as the mean ± standard deviation,
and Student's t-tests were performed using the SPSS 16.0 software
package (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
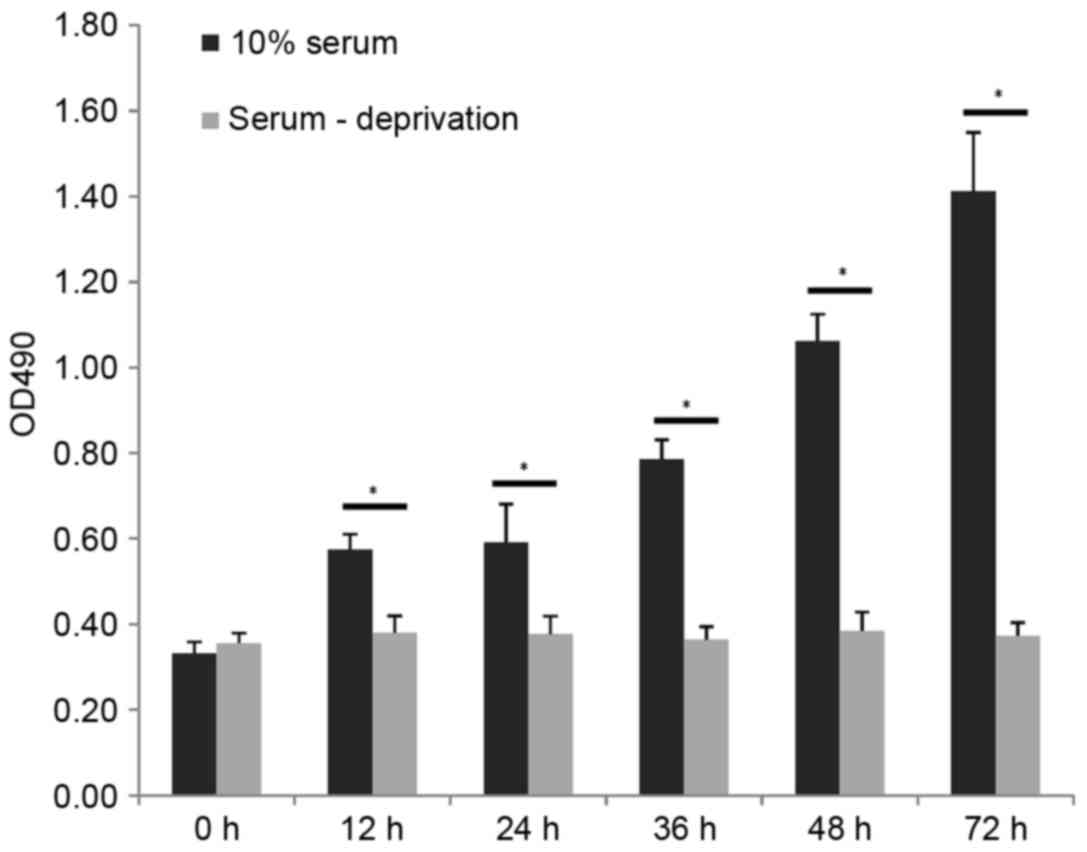
Serum deprivation inhibits cell
proliferation
Serum deprivation inhibited the proliferation of
cells, as exhibited in Fig. 1.
When the Tca-8113 cells were subjected to serum deprivation for 12,
24, 36, 48 or 72 h, the proliferative activity was measured using
an MTT assay. No significant differences were observed over time in
the serum-deprived group (P>0.05), however, proliferation
continued to increase in the cells cultured in 10% FBS medium
(P<0.05).
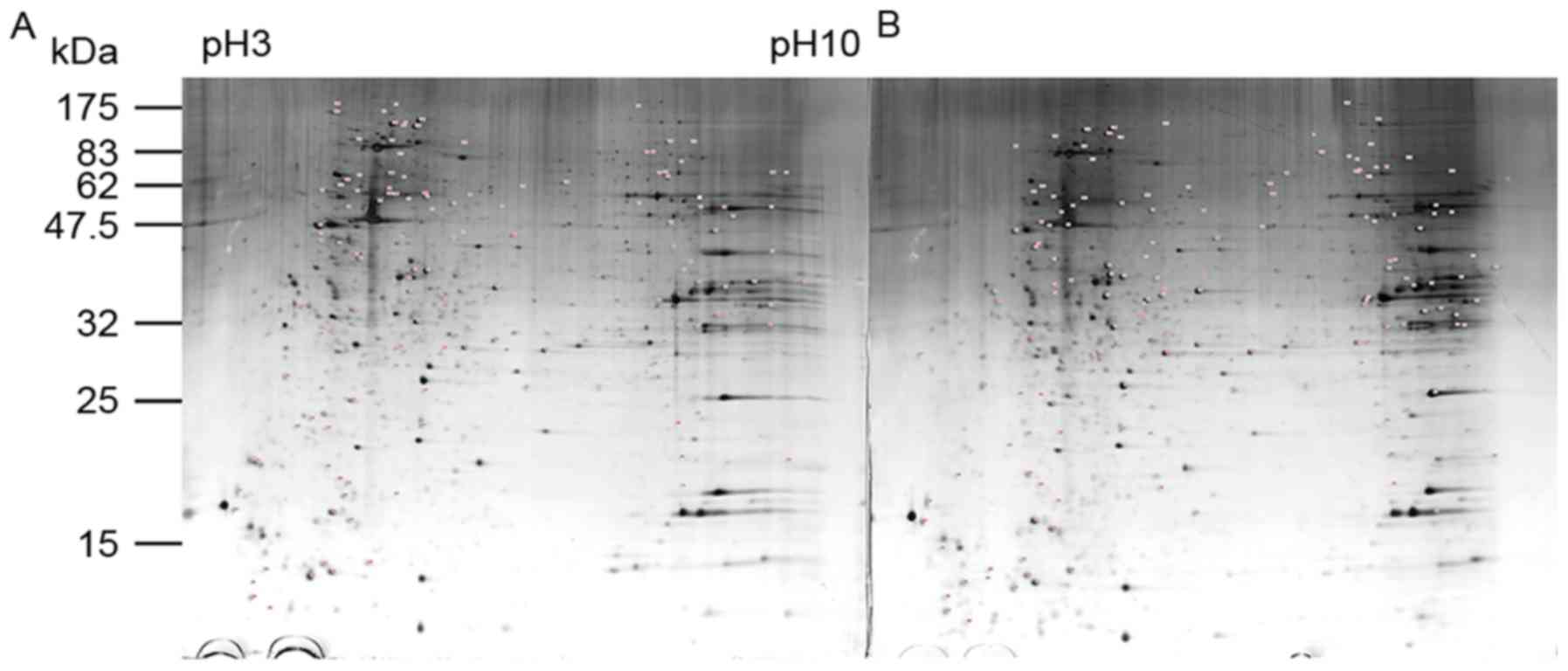
Identification of differentially
expressed proteins using proteomic analysis
As proliferation was inhibited in the Tca-8113 cells
subjected to serum deprivation for 24 h, the time-points of 0 h
(control) and 24 h were selected for the comparison of triplicate
samples using 2-DE and mass spectrometry. Representative 2-DE
images are shown in Fig. 2. A
total of 1,237 protein spots were detected on the 2-DE gel, with 43
significantly upregulated and 45 significantly downregulated at 24
h, compared with levels at 0 h (P<0.05; average spot intensity
difference >1.2-fold). Of these 88 potential differentially
expressed proteins, 35 were identified using MALDI-TOF/MS, whereas
the remaining 53 did not yield suitably complete polypeptide
fragments or were too low in abundance for reliable identification.
The results of the spot ID, average spot intensity, protein number,
protein name, sequence coverage, score and Mr/pI are exhibited in
Table II. The present study
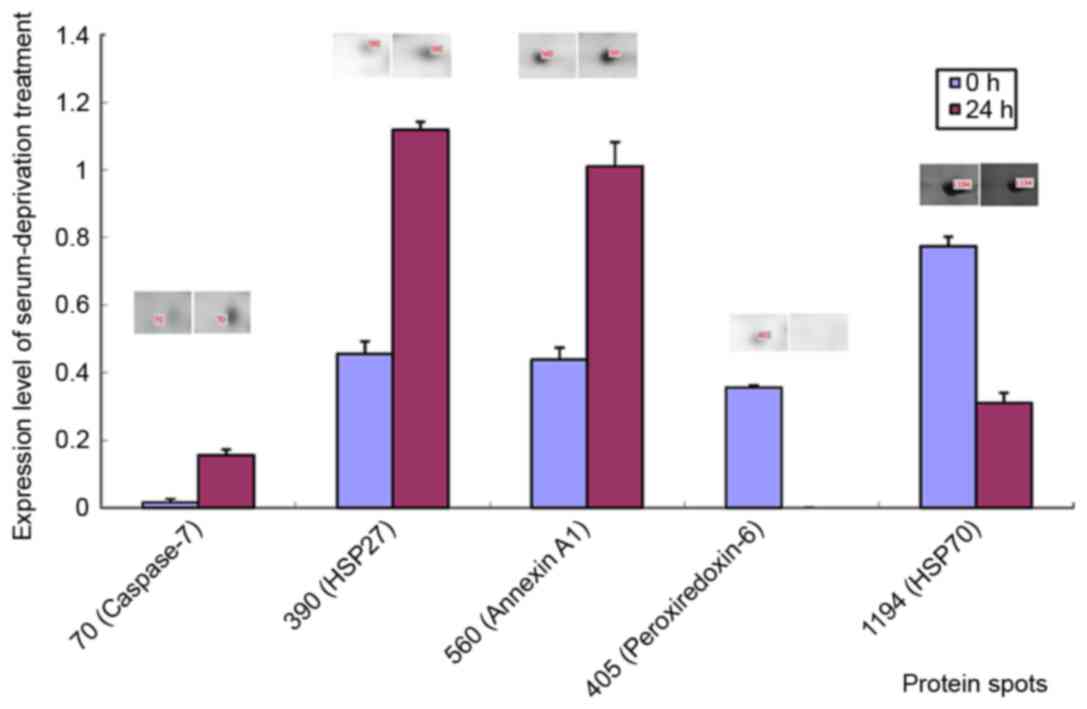
focussed on five differentially expressed proteins: Caspase-7 (spot
70), heat shock protein 27 (HSP 27; spot 390), Annexin A1 (spot
560), peroxiredoxin-6 (spot 405) and HSP 70 (spot 1194; Fig. 3). Notably, all are involved in
mitochondrial oxidative stress, consistent with a previous
conclusion that serum deprivation can induce apoptosis (7).
 | Table II.List of differentially expressed
proteins during serum-deprivation treatment. |
Table II.
List of differentially expressed
proteins during serum-deprivation treatment.
|
| Percentage of total
volume of all spotsa |
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|---|
| Spot ID | Control (0 h) | Serum-deprivation
(24 h) | Protein number | Protein name | Sequence coverage
(%) | Score | Mr/pI |
|---|
| Upregulated |
|
|
|
|
|
|
|
| 59 | 0.000±0.000 | 0.086±0.005 | gi|2250700 | KNP-I α
protein | 62 | 138 | 23,806/8.61 |
|
489 | 0.000±0.000 | 0.002±0.001 | gi|4505773 | Prohibitin | 58 | 197 | 29,843/5.57 |
|
497 | 0.000±0.000 | 0.153±0.005 | gi|28931 | β-subunit (AA
1–312) | 42 | 95 | 34,026/4.90 |
|
566 | 0.001±0.001 | 0.110±0.015 | gi|194376310 | Actin, β,
partial | 34 | 174 | 38,950/5.19 |
|
343 | 0.007±0.004 | 0.214±0.030 | gi|194375974 |
Peroxiredoxin-2 | 57 | 193 | 20,209/8.90 |
| 70 | 0.016±0.010 | 0.156±0.016 | gi|20150093 | Chain B, crystal
structure of caspase-7 complexed with xiap | 63 | 143 | 12,244/5.73 |
|
220 | 0.010±0.003 | 0.090±0.004 | gi|55959240 | Nicotinamide
nucleotide adenylyltransferase 1 | 66 | 132 | 18,413/8.99 |
|
103 | 0.078±0.023 | 0.417±0.035 | gi|1362965 | T cell receptor
Mb11 β chain | 48 | 109 | 15,697/6.56 |
|
376 | 0.076±0.024 | 0.399±0.017 | gi|5822016 | Chain C,
erythropoietin complexed with extracellular domains of
erythropoietin receptor | 66 | 165 | 18,655/9.28 |
|
549 | 0.065±0.019 | 0.250±0.024 | gi|21667979 | Oxysterol binding
protein-related protein 3 isoform 2d | 40 | 123 | 35,786/5.11 |
|
390 | 0.456±0.037 | 1.119±0.024 | gi|662841 | Heat shock protein
27 | 39 | 111 | 22,427/7.83 |
|
560 | 0.439±0.035 | 1.011±0.071 | gi|4502101 | Annexin A1 | 52 | 215 | 38,918/6.57 |
|
392 | 0.856±0.067 | 1.734±0.087 | gi|1871210 | T-complex protein
1, β subunit (TCP-1-β) | 63 | 154 | 22,924/5.88 |
| Downregulated |
|
|
|
|
|
|
|
|
358 | 1.744±0.043 | 1.021±0.067 | gi|2392338 | Glyoxalase-I | 42 | 121 | 20,861/5.12 |
|
775 | 0.852±0.030 | 0.456±0.067 | gi|301030821 | c-Myc
promoter-binding protein 1 | 42 | 100 | 36,874/6.17 |
|
1184 | 0.416±0.007 | 0.174±0.012 | gi|194388088 | Heat shock 70 kDa
protein 1A/1B | 34 | 205 | 64,170/5.39 |
|
1162 | 0.120±0.004 | 0.050±0.011 | gi|194384236 | Heat shock 70 kDa
protein 1A/1B | 19 | 87 | 60,091/5.39 |
|
1194 | 0.774±0.028 | 0.311±0.029 | gi|62897129 | Heat shock 70 kDa
protein 8 isoform 1 variant | 17 | 78 | 71,083/5.28 |
|
885 | 1.450±0.087 | 0.476±0.039 | gi|92911770 |
XTP3TPA-transactivated protein 1 | 50 | 146 | 23,896/6.42 |
|
714 | 0.822±0.156 | 0.233±0.076 | gi|296211572 | Tubulin α-1B chain
isoform 2 | 30 | 110 | 46,797/4.96 |
|
204 | 0.327±0.035 | 0.057±0.018 | gi|56605994 | CDGSH iron-sulfur
domain-containing protein 2 | 82 | 177 | 15,497/9.66 |
|
1077 | 0.055±0.005 | 0.005±0.003 | gi|32189394 | ATP synthase
subunit β, mitochondrial precursor | 46 | 208 | 56,525/5.26 |
|
198 | 0.161±0.022 | 0.000±0.000 | gi|57162210 | CDC14 cell division
cycle 14 homolog B (S. cerevisiae) | 47 | 126 | 31,655/8.33 |
|
206 | 0.107±0.012 | 0.000±0.000 | gi|51479152 | ATP synthase
subunit d, mitochondrial isoform b | 48 | 89 | 15,820/6.60 |
|
245 | 0.052±0.010 | 0.000±0.000 | gi|119595056 | Glutathione
S-transferase pi, isoform CRA_c | 24 | 41 | 19,577/4.84 |
|
314 | 0.054±0.009 | 0.000±0.000 | gi|306517833 | MHC class I
antigen | 48 | 109 | 21,098/6.64 |
|
405 | 0.356±0.006 | 0.000±0.000 | gi|4758638 |
Peroxiredoxin-6 | 41 | 133 | 25,133/6.00 |
|
419 | 0.267±0.034 | 0.000±0.000 | gi|3329384 | Translation
initiation factor 4e | 55 | 131 | 27,481/10.00 |
|
772 | 0.076±0.004 | 0.000±0.000 | gi|7705592 | Mediator of RNA
polymerase II transcription subunit 31 | 61 | 201 | 15,966/8.72 |
|
845 | 0.131±0.003 | 0.000±0.000 | gi|89574029 | Mitochondrial ATP
synthase, H+ transporting F1 complex β subunit | 39 | 134 | 48,083/4.95 |
|
1000 | 0.235±0.047 | 0.000±0.000 | gi|48146259 | CCT2 | 31 | 159 | 57,766/6.01 |
|
1006 | 0.173±0.035 | 0.000±0.000 | gi|220702506 | Chain A,
TapasinERP57 HETERODIMER | 34 | 174 | 54,541/5.61 |
|
1080 | 0.052±0.003 | 0.000±0.000 | gi|169404695 | Chain A, pyruvate
kinase M2, phosphotyrosine binding protein | 29 | 153 | 57,091/8.00 |
|
1189 | 0.063±0.010 | 0.000±0.000 | gi|14718539 | HIC-3 | 15 | 82 | 65,103/5.89 |
|
1222 | 0.186±0.005 | 0.000±0.000 | gi|386758 | GRP78 precursor,
partial | 32 | 169 | 72,185/5.03 |
|
1230 | 0.035±0.001 | 0.000±0.000 | gi|15293595 | Seven transmembrane
helix receptor | 43 | 85 | 24,812/8.87 |
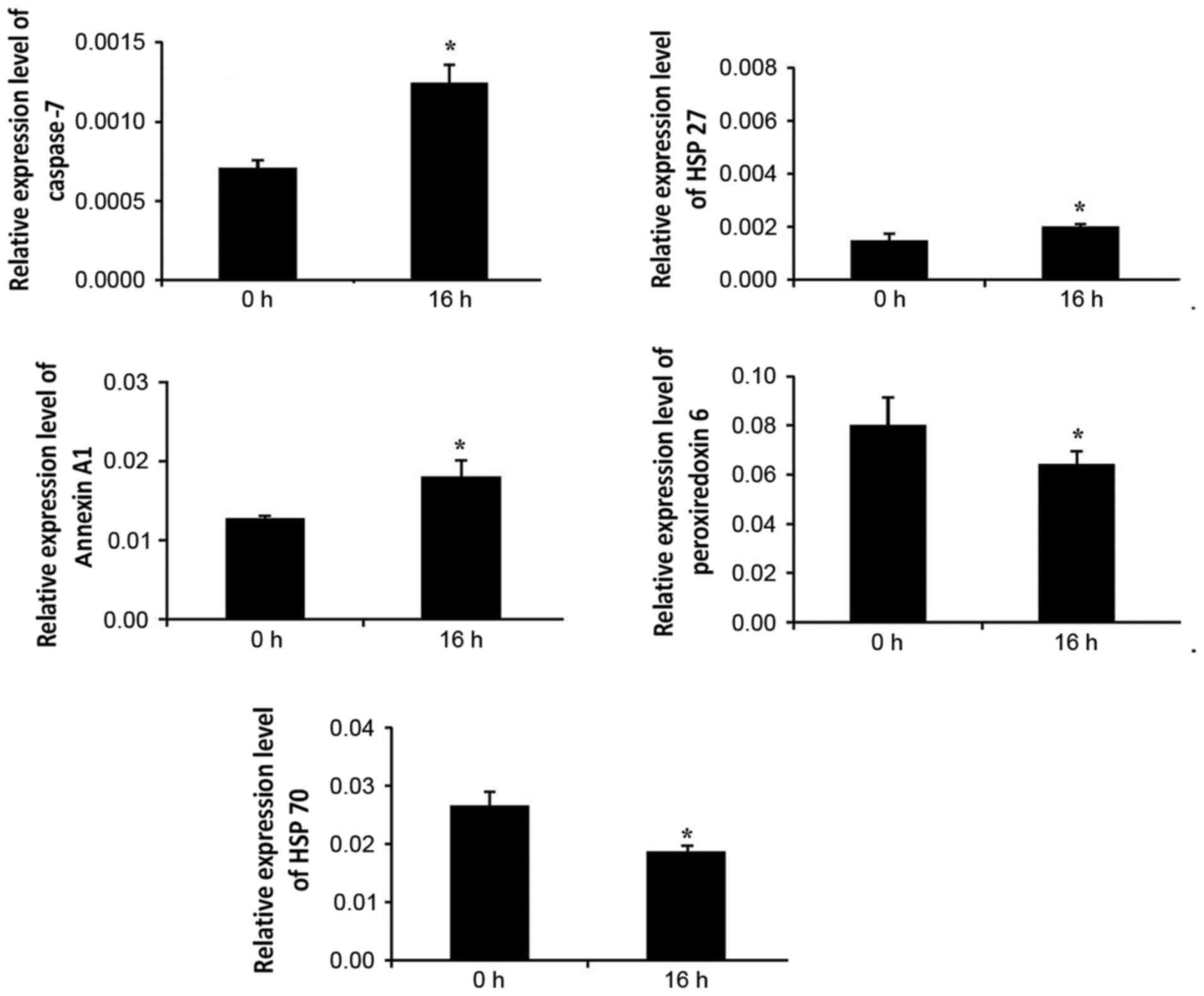
RT-PCR verification
The present study performed RT-PCR analysis to
investigate the expression of caspase-7, HSP 27, Annexin A1,
peroxiredoxin-6 and HSP 70 in the Tca-8113 cells subjected to serum
deprivation at two time-points (0 and 16 h). The stably expressed
GAPDH mRNA was selected as an internal reference gene as the
expression of cytoskeletal proteins was significantly disrupted.
For example, the expression of β-actin was increased at the protein
(Table II) and mRNA levels (data
not shown), whereas the expression of tubulin was decreased
(Table II). As shown in Fig. 4, the expression patterns of the
five proteins were similar to the results obtained using 2-DE.
Discussion
Serum deprivation is widely used for cell cycle
arrest and apoptosis, and various molecular mechanisms may be
involved in the modulation of serum deprivation-induced apoptosis
(17). However, in the present
study, the effects of serum deprivation on cell proliferation and
apoptosis were not observed in the Tca-8113 cells, even following a
long duration (72 h) of treatment (Fig. 1). The proteomic analysis showed
that a 24 h period of serum deprivation induced significant
expression changes of numerous mitochondrial proteins, including
prohibitin, peroxiredoxin, glyoxalase, caspase and ATP synthase
(Table II). This is consistent
with a previous study, in which mitochondrial function was readily
disrupted by alterations of the cultivation environment through
serum deprivation, which affects mitochondrial viability in several
fibroblast lines (18). However,
the detailed molecular mechanism linking culture conditions and
mitochondrial activity remains to be fully elucidated. The results
of the present study provided novel insight into the mechanism by
which Tca-8113 cells subjected to serum deprivation avoid
apoptosis.
Although the mechanism by which serum deprivation
stimulates reactive oxygen species (ROS) production remains to be
elucidated, studies have shown that ROS resulting from serum
deprivation can induce programmed cell death (7,19).
Oxidative stress has a wide range of effects on cellular
proliferation, differentiation, migration and other physiological
processes (20). Several
antioxidant proteins are involved in alleviating oxidative stress
by removing ROS. The present study found that peroxiredoxin-2
(Prx2) was upregulated, whereas the associated Prx6 was
downregulated following serum deprivation. Prx enzymes have six
isoforms, and they are located at sites of energy production and
cellular motion, where ROS can be produced in abundance,
particularly in mitochondria and at the plasma membrane. The
peroxiredoxins are known for their function in removing endogenous
peroxides. Therefore, the upregulation of peroxiredoxins is linked
to high levels of oxidative stress (21). Prx2 is the major Prx enzyme in the
cytoplasm, where it reduces peroxynitrite and hydroperoxides via a
rapid, diffusion-controlled reaction (22). Prx2 is also important in the
mechanism of H2O2-induced redox signalling
(23). The overexpression of Prx2
has been associated with the progression of squamous cell
carcinoma, and it may be an underlying biomarker for radiotherapy
in colorectal cancer (24) lung
cancer (25) and bladder cancer
(26). Similarly, the expression
of Prx6 has been associated with high rates of metastasis of breast
cancer cell lines, and the overexpression of Prx6 occurs in
malignant mesothelioma, breast cancer, oesophageal carcinoma and
oligodendroglioma (27). A
previous study revealed a marked upregulation of Prx2 following
demethylation in gefitinib-resistant A549 cells (28). The present results suggested that
Tca-8113 cells may resist the serum deprivation-induced apoptosis
by upregulating Prx2, which may serve as a potent target for
assisting in anti-angiogenic drug treatment in human TSCC.
Annexin A1 (ANXA1) is a well-characterised
apoptosis-associated membrane protein, which is regulated by the
epidermal growth factor receptor, insulin receptor, TRPM7 channel
kinase 1, protein kinase C and protein kinase A. ANXA1 has also
been linked with carcinogenesis and metastasis in various types of
tumour (29). As an essential
protein in tumourigenesis and apoptosis, the overexpression of
ANXA1 is linked to carcinogenesis and high pathological
differentiation grade (30,31).
There is accumulating evidence suggesting that ANXA1 may be
regulated by pro-inflammatory proteins, including
lipopolysaccharide and interleukin-6, and it is reported to act as
a brake to control the inflammatory response (32). Kang et al found that ANXA1
was positively upregulated during serum deprivation-induced
autophagic degradation in HCT116 cells, indicating a potential role
in autophagy by downregulating the inflammatory response (33). The overexpression of ANXA1 can also
inhibit the aberrant dysregulation of cytoskeletal proteins and
proliferation of pulmonary microvascular endothelial cells
(34), which is consistent with
the observed morphology, static proliferation and upregulation of
actin in the serum deprived Tca-8113 cells in the present study.
The results of the present study showed that ANXA1 was upregulated
at the mRNA and protein levels following serum deprivation,
suggesting that autophagy, and not apoptosis, was a factor in
Tca-8113 cells; autophagy protected the cells against serum
deprivation by promoting cell survival.
Autophagy and apoptosis are the two major cell death
pathways, and are tightly regulated at the genetic level to ensure
that tissue and organ development proceeds correctly, in addition
to other pathophysiological processes (35). A hallmark of autophagy is lysosomal
degradation, which performs an adaptive function to protect
organisms against infections, cancer and ageing (36). Apoptosis is a programmed process
for killing and removing certain cells during morphological
development, which also assists in maintaining tissue homeostasis
in multicellular organisms. The aberrant regulation of apoptosis
has been consistently linked with human proliferative diseases,
including malignant tumours (37).
Apoptosis involves a large number of molecules, including cysteine
proteases in the form of caspases, which are synthesised as
pro-forms and become activated by cleavage at aspartate residues.
Initiator caspases, including caspase-1, 2, 4, 5, 8, 9, 10, 11 and
12, integrate molecular signals and activate downstream effector
caspases, including caspase-3, 6, 7 and 14. As caspases can cleave
and activate each other, triggering of the caspase cascade
initiates a rapidly amplified signal, which ensures accurate
apoptotic cell death. Caspases are also able to cleave several
other substrates, including nuclear lamins and cytoskeletal
proteins, thereby inducing typical morphological characteristics
associated with apoptosis. Caspase-7 is most closely linked with
caspase-3, and these enzymes are activated by mitochondria-induced
apoptosis.
Effector caspases are in charge of initiating DNA
fragmentation, cell shrinkage and membrane blebbing (38). Brentnall et al (39) showed that caspase-9 and caspase-3
are involved in remodeling of mitochondria and ROS production.
Caspase-7 has no significant effect on sensitivity to intrinsic
cell death, but is involved in ROS production and cell detachment.
In the present study, serum deprivation upregulated the level of
caspase-7 in Tca-8113 cells, which underwent shrinkage and membrane
blebbing, suggesting that serum deprivation stimulated TSCC
differentiation and inhibited cancer growth.
HSPs are also closely linked with stress responses,
including the response to serum deprivation. HSP70 is a major
stress-inducible protein, which can fulfil multiple roles depending
on its location. The upregulation of intracellular HSP70 is
involved in the stress associated with recovery from clinical
treatment, including radiotherapy or chemotherapy, whereas
extracellular HSP70 stimulates the innate immune system to initiate
antitumour immunity (40). In the
present study, the serum deprivation-induced downregulation of
HSP70 may assist TSCCs to escape and evade attacks from natural
killer (NK) cells, however, the downregulation of major
histocompatibility complex-I can compensate for this and ensure
activation of the NK cells. HSP27 is primarily involved in
proteasome-mediated protein degradation and the regulation of
apoptosis. A previous study revealed that HSP27 mediates apoptotic
programming in pancreatic cancer cells (41), and clinical immunohistochemical
evidence demonstrates that the overexpression of HSP27 causes
high-grade differentiation in TSCCs (42). Therefore, the serum
deprivation-induced upregulation of HSP27 may be correlated with
the morphology and differentiation of TSCCs, consistent with the
expression of caspase-7.
In conclusion, the present investigative preclinical
study found that Tca-8113 TSCC cells resisted apoptosis induced by
serum deprivation. The changes in proteins were primarily involved
in the oxidative stress process in the mitochondrion, which
indicated that ROS may be important in serum deprivation-induced
oxidative stress. In the future, further detailed investigations on
the signalling pathway and clinical verification are required.
Acknowledgements
The authors would like to thank the Natural Science
Foundation of China (grant nos. 81473593 and 81473458), the Jiangsu
Qing Lan Project (grant no. QL-2014) for financial support, and the
Priority Academic Program Development of Jiangsu Higher Education
Institutions (Integration of Chinese and Western Medicine; grant
no. PAPD-2014) for funding.
References
|
1
|
Ching CT, Sun TP, Huang SH, Hsiao CS,
Chang CH, Huang SY, Chen YJ, Cheng CS, Shieh HL and Chen CY: A
preliminary study of the use of bioimpedance in the screening of
squamous tongue cancer. Int J Nanomed. 5:213–220. 2010. View Article : Google Scholar
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Layland MK, Sessions DG and Lenox J: The
influence of lymph node metastasis in the treatment of squamous
cell carcinoma of the oral cavity, oropharynx, larynx, and
hypopharynx: N0 versus N+. Laryngoscope. 115:629–639. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Korostoff A, Reder L, Masood R and Sinha
UK: The role of salivary cytokine biomarkers in tongue cancer
invasion and mortality. Oral Oncol. 4:282–287. 2011. View Article : Google Scholar
|
|
5
|
Faratzis G, Tsiambas E, Rapidis AD,
Machaira A, Xiromeritis K and Patsouris E: VEGF and ki 67
expression in squamous cell carcinoma of the tongue: An
immunohistochemical and computerized image analysis study. Oral
Oncol. 45:584–588. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Patel RS, Clark JR, Dirven R, Wyten R, Gao
K and O'Brien CJ: Prognostic factors in the surgical treatment of
patients with oral carcinoma. Anz J Surg. 79:19–22. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee SB, Kim JJ, Kim TW, Kim BS, Lee MS and
Yoo YD: Serum deprivation-induced reactive oxygen species
production is mediated by Romo1. Apoptosis. 15:204–218. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Grossman N, Binyamin LA and Bodner L:
Effect of rat salivary glands extracts on the proliferation of
cultured skin cells-a wound healing model. Cell Tissue Bank.
4:205–212. 2004. View Article : Google Scholar
|
|
9
|
De Giuseppe R, Cossellu G, Vigna L,
Dicorato F, De Vita C, Venturelli G, Bamonti F, Maiavacca R and
Farronato G: Correlation between salivary and serum oxidized LDL
levels: A pilot study on overweight/obese subjects. J Oral Pathol
Med. 44:884–887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hayes LD, Sculthorpe N, Herbert P, Baker
JS, Hullin DA, Kilduff LP and Grace FM: Poor levels of agreement
between serum and saliva testosterone measurement following
exercise training in aging men. Aging Male. 18:67–70. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Seethalakshmi C, Koteeswaran D and
Chiranjeevi V: Correlation of serum and salivary biochemical
parameters in end stage renal disease patients undergoing
hemodialysis in pre and post-dialysis state. J Clin Diagn Res.
12:CC12–CC14. 2014.
|
|
12
|
Boraldi F, Annovi G, Paolinelli-Devincenzi
C, Tiozzo R and Quaglino D: The effect of serum withdrawal on the
protein profile of quiescent human dermal fibroblasts in primary
cell culture. Proteomics. 8:66–82. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li WZ, Wang XY, Li ZG, Zhang JH and Ding
YQ: Celecoxib enhances the inhibitory effect of cisplatin on
Tca8113 cells in human tongue squamous cell carcinoma in vivo and
in vitro. J Oral Pathol Med. 7:579–584. 2010.
|
|
14
|
Wu J, Wang F, Gong Y, Li D, Sha J, Huang X
and Han X: Proteomic analysis of changes induced by nonylphenol in
Sprague-Dawley rat Sertoli cells. Chem Res Toxicol. 22:668–675.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheuk BL and Cheng SW: Annexin A1
expression in atherosclerotic carotid plaques and its relationship
with plaque characteristics. Eur J Vasc Endovasc Surg. 41:364–371.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan F, Cheng Q, Li G and Tong T:
Nucleostemin knockdown sensitizes hepatocellular carcinoma cells to
ultraviolet and serum starvation-induced apoptosis. PLoS One.
10:e01416782015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takeda K, Akagi S, Takahashi S, Onishi A,
Hanada H and Pinkert CA: Mitochondrial activity in response to
serum starvation in bovine (Bos taurus) cell culture. Cloning Stem
Cells. 4:223–229. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu J and Du L: PERK pathway is involved
in oxygen-glucose-serum deprivation-induced NF-kB activation via
ROS generation in spinal cord astrocytes. Biochem Biophys Res
Commun. 467:197–203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gerrits EG, Alkhalaf A, Landman GW, van
Hateren KJ, Groenier KH, Struck J, Schulte J, Gans RO, Bakker SJ,
Kleefstra N and Bilo HJ: Serum peroxiredoxin 4: A marker of
oxidative stress associated with mortality in type 2 diabetes
(ZODIAC-28). PLoS One. 2:e897192014. View Article : Google Scholar
|
|
21
|
Ding Y, Yamada S, Wang KY, Shimajiri S,
Guo X, Tanimoto A, Murata Y, Kitajima S, Watanabe T, Izumi H, et
al: Overexpression of peroxiredoxin 4 protects against high-dose
streptozotocin-induced diabetes by suppressing oxidative stress and
cytokines in transgenic mice. Antioxid Redox Signal. 13:1477–1490.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Manta B, Hugo M, Ortiz C, Ferrer-Sueta G,
Trujillo M and Denicola A: The peroxidase and peroxynitrite
reductase activity of human erythrocyte peroxiredoxin 2. Arch
Biochem Biophys. 484:146–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martinez A, Peluffo G, Petruk AA, Hugo M,
Piñeyro D, Demicheli V, Moreno DM, Lima A, Batthyány C, Durán R, et
al: Structural and molecular basis of the peroxynitrite-mediated
nitration and inactivation of trypanosoma cruzi iron-superoxide
dismutases (Fe-SODs) A and B: Disparate susceptibilities due to the
repair of Tyr35 radical by Cys83 in Fe-SODB through intramolecular
electron transfer. J Biol Chem. 289:12760–12778. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cerda MB, Lloyd R, Batalla M, Giannoni F,
Casal M and Policastro L: Silencing peroxiredoxin-2 sensitizes
human colorectal cancer cells to ionizing radiation and
oxaliplatin. Cancer Lett. 388:312–319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rostila A, Puustinen A, Toljamo T, Vuopala
K, Lindström I, Nyman TA, Oksa P, Vehmas T and Anttila SL:
Peroxiredoxins and tropomyosins as plasma biomarkers for lung
cancer and asbestos exposure. Lung Cancer. 77:450–459. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shiota M, Yokomizo A, Kashiwagi E,
Takeuchi A, Fujimoto N, Uchiumi T and Naito S: Peroxiredoxin 2 in
the nucleus and cytoplasm distinctly regulates androgen receptor
activity in prostate cancer cells. Free Radic Biol Med. 51:78–87.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang B, Wang Y and Su Y: Peroxiredoxins,
a novel target in cancer radiotherapy. Cancer Lett. 286:154–160.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kwon T, Jin KR, Lee JC, Park YH, Shin HJ,
Cho S, Kang YK, Kim BY, Yoon DY and Yu DY: An important role for
peroxiredoxin II in survival of A549 lung cancer cells resistant to
gefitinib. Exp Mol Med. 47:e1652015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sobral-Leite M, Wesseling J, Smit VT,
Nevanlinna H, van Miltenburg MH, Sanders J, Hofland I, Blows FM,
Coulson P and Patrycja G: Annexin A1 expression in a pooled breast
cancer series: association with tumor subtypes and prognosis. BMC
Med. 13:1562015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin CY, Jeng YM, Chou HY, Hsu HC, Yuan RH,
Chiang CP and Kuo MY: Nuclear localization of annexin A1 is a
prognostic factor in oral squamous cell carcinoma. J Surg Oncol.
97:544–550. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Faria PC, Sena AA, Nascimento R, Carvalho
WJ, Loyola AM, Silva SJ, Durighetto AF, Oliveira AD, Oliani SM and
Goulart LR: Expression of annexin A1 mRNA in peripheral blood from
oral squamous cell carcinoma patients. Oral Oncol. 46:25–30. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vago JP, Nogueira CR, Tavares LP, Soriani
FM, Lopes F, Russo RC, Pinho V, Teixeira MM and Sousa LP: Annexin
A1 modulates natural and glucocorticoid-induced resolution of
inflammation by enhancing neutrophil apoptosis. J Leukoc Biol.
92:249–258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kang JH, Li M, Chen X and Yin XM:
Proteomics analysis of starved cells revealed Annexin A1 as an
important regulator of autophagic degradation. Biochem Biophys Res
Commun. 407:581–586. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yi B, Jing Z, Wang G, Qian G and Lu K:
Annexin A1 protein regulates the expression of PMVEC cytoskeletal
proteins in CBDL rat serum-induced pulmonary microvascular
remodeling. J Transl Med. 11:982013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thorburn A: Apoptosis and autophagy:
Regulatory connections between two supposedly different processes.
Apoptosis. 1:1–9. 2008. View Article : Google Scholar
|
|
36
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
PLOS ONE Staff, . Correction: Association
of genetic markers in the BCL-2 family of apoptosis-related genes
with endometrial cancer risk in a Chinese population. PLoS One.
10:e01176322015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Coutinho-Camillo CM and Soares FA: CASP7
(caspase 7, apoptosis-related cysteine peptidase). Atlas Genetics
Cytogenetics Oncology Haematology. 3:160–163. 2015.
|
|
39
|
Brentnall M, Rodriguez-Menocal L, Guevara
RL, Cepero E and Boise LH: Caspase-9, caspase-3 and caspase-7 have
distinct roles during intrinsic apoptosis. BMC Cell Biol.
14:322013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Multhoff G, Pockley AG, Schmid TE and
Schilling D: The role of heat shock protein 70 (Hsp70) in
radiation-induced immunomodulation. Cancer Lett. 368:179–184. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang G, Ziesch A, Hocke S, Kampmann E,
Ochs S, De Toni EN, Göke B and Gallmeier E: Overexpression of heat
shock protein 27 (HSP27) increases gemcitabine sensitivity in
pancreatic cancer cells through S-phase arrest and apoptosis. J
Cell Mol Med. 2:340–350. 2015.
|
|
42
|
Zhang NN, Sheng SH, Chen D, et al:
Expression and significance of HSP27 in tongue squamous cell
carcinoma. Beijing J Stomatol. 3:146–148. 2010.
|