Introduction
Neonatal respiratory distress syndrome (NRDS), also
termed hyaline membrane disease, is one of the most prevalent
causes of morbidity and mortality in newborns (1). The oxygen diffusion efficiency
through the alveoli-capillary exchange barrier is impacted by the
dysregulation of numerous factors, leading to the pathophysiology
of NRDS (2). Cellular stress at
the gas-blood level is associated with alterations in alveolar
surfactant proteins (3) and the
upregulation of numerous innate immune responses of
pro-inflammatory cytokines to foreign challenges (4). A lack of surfactant may drive the
pathogenesis towards NRDS, while surfactant replacement therapy may
mitigate symptoms of the disease by decreasing the surface tension
of alveoli and facilitating their inflation (5,6).
However, surfactant serves an additional role in immunological
processes. A recent study demonstrated that applying clinical
surfactant for patients led to a stronger response to challenges
from foreign microbiota (7).
Additionally, animal-derived surfactant applied for clinical use in
humans has been demonstrated to be of use against lung pathogens
and to mediate excess host damage from neutrophils (8). Therefore, it may be hypothesized that
mechanisms that mechanisms of NRDS involving surfactant exert
additional functions to promoting alveolar inflation.
Acute respiratory distress syndrome (ARDS) is a
clinical phenotype precipitated by the manifestation of a severe
form of lung injury due to numerous lung insults (9). The pathophysiological manifestation
of ARDS is derived from a cascade that is triggered by the complex
combination of risk factors, including asthma, sepsis, pneumonia,
increased neutrophil presence in the lungs and other variables;
accumulation of these numerous systemic factors forms the
pathological endpoint of ARDS (10).
The present study used peripheral blood obtained
from infants with (n=4) or without (n=2) NRDS in tandem with
mRNA-sequencing (mRNA-seq) analysis to reveal various factors
involved in the mechanisms of NRDS, and to compare onset mechanisms
between NRDS and ARDS.
Materials and methods
Samples
The Ethics Committee of the Affiliated Hospital of
Inner Mongolia Medical University (Huhehaote, China) approved the
present study. All study participants provided written informed
consent and were recruited at the Affiliated Hospital of Inner
Mongolia Medical University between January 1 and June 30,
2016.
A total of four infants with severe NRDS and two
healthy infants were recruited for the present study. Subject
characteristics are presented in Table
I. The subjects were not related, and all of their relatives in
the three most recent generations were residents in Inner Mongolia
province and self-identified as Chinese Inner Mongolia race. NRDS
was diagnosed according to the European Consensus Guidelines on the
Diagnosis of NRDS in Preterm Infants 2010 (11). The criteria were as follows:
Progressive deterioration of shortness of breath commencing at 12 h
post-birth, with arterial partial pressure of oxygen
(PaO2) <50 mmHg during inspiration accompanied by
central cyanosis. Oxygen therapy was required to maintain
PaO2 >50 mmHg. Chest X-ray (CXR) indicated NRDS
specific signs, including ground glass opacity and air-filled
bronchi. The control groups were defined by CXR demonstrating clear
lung field without pulmonary infection signs. White blood cell
counts and C-reactive protein values were normal, to exclude the
possibility of any infection. Subjects with any of the following
were excluded from the present study: i) Any serious congenital
diseases, including complex congenital cardiac anomaly or diaphragm
hernia; ii) inherited metabolic disorders, including
phenylketonuria, congenital hypothyroidism or diabetes mellitus;
iii) confirmed maternal infection history during the third
trimester of pregnancy; iv) multigestation; v) maternal diabetes
mellitus; or vi) neonatal asphyxia.
 | Table I.Subject information. |
Table I.
Subject information.
| Sample ID | Sex | Mother | Gestational age,
weeks |
|---|
| Control 1 | Male | G2P2 | 38+3 |
| Control 2 | Female | G3P3 | 38+3 |
| NRDS1 | Male | G2P1 | 37+6 |
| NRDS2 | Male | G3P1 | 37+4 |
| NRDS3 | Female | G3P2 | 37 |
| NRDS4 | Female | G2P1 | 37+3 |
mRNA extraction and sequencing
Total RNAs from 4 NRDS and 2 control subjects were
isolated and quality controlled using protocols from Illumina, Inc.
(San Diego, CA, USA). A total amount of 3 µg RNA/sample was used as
the input material for library construction. Ribosomal RNA (rRNA)
was removed using the Epicentre Ribo-zero rRNA Removal kit
(Illumina, Inc.), according to the manufacturer's protocol.
Strand-specific sequencing libraries were generated via the dUTP
method using the resulting RNA with the NEB Next Ultra Directional
RNA Library Prep kit for Illumina (New England BioLabs, Inc.,
Ipswich, MA, USA), according to the manufacturer's protocol.
RNA-seq was performed on an Illumina Hiseq 2000 platform (Illumina,
Inc.) and 100-bp paired-end reads were generated, according to the
manufacturer's protocol.
RNA-seq data analysis
The adapter sequences were removed from the raw
sequencing data and the individual libraries were converted to the
FASTQ format. Sequence reads were aligned to the human genome
(hg19) using TopHat2 (v2.0.9; ccb.jhu.edu/software/tophat/index.shtml) and the
resulting alignment files were reconstructed using Cufflinks
(v2.1.1; cole-trapnell-lab.github.io/cufflinks) (12) and Scripture (beta2; software.broadinstitute.org/software/scripture).
The transcripts assembled by Cufflinks and Scripture were used for
the identification of differentially expressed genes (DEGs). For
mRNA analyses, the RefSeq database (build 37.3; www.ncbi.nlm.nih.gov/refseq) was selected for the
annotation references. The read counts of each transcript were
normalized to the length of the individual transcript and to the
total mapped fragment counts in each sample, and expressed as
fragments per kilobase of exon per million fragments mapped of
mRNAs in each sample. mRNA differential expression analyses were
applied to the NRDS and control groups. The P-value was calculated
using R3.2.5 software (www.r-project.org). Adjusted P<0.05 (Student's
t-test with Benjamini-Hochberg false discovery rate adjustment) was
used as the threshold for significant DEGs. DEGs were analyzed by
enrichment analyses to detect over-represented functional terms
present in the genomic background. Gene ontology (GO) analysis was
performed using the GO-seq package (bioconductor.org/packages/release/bioc/html/goseq.html)
in R3.2.5 software, in which gene length bias was corrected. Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway analysis was
performed using KOBAS 3.0 software (kobas.cbi.pku.edu.cn).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNAs were isolated and quality controlled
using protocols from Illumina, Inc. All cDNAs were synthesized
using Superscript III (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The top five over- and underexpressed genes were
selected for the RT-qPCR assay. All RT-qPCR primers are presented
in Table II. All qPCR reactions
were performed on a Roche Lightcycler 480 PCR system (Roche Applied
Science, Penzberg, Germany) using Toyobo Thunderbird SYBR RT-qPCR
Mix (Toyobo Life Science, Osaka, Japan), with three technical
repeats. The amplification procedure was as follows: 95°C for 5
min, followed by 40 cycles of 95°C for 10 sec and 60°C for 20 sec.
Relative quantification of target genes was performed using the
2−ΔΔCq method with GAPDH as a reference gene (13). Pearson correlation was used to
calculate the association between RNA sequence and RT-qPCR
(14).
 | Table II.Primers used for RT-qPCR. |
Table II.
Primers used for RT-qPCR.
| Gene | Primer |
|---|
| PIP4K2B | Forward:
ATTGCACTGGAGACCAGCAA |
|
| Reverse:
GTACGCACAAAAGACTGGCG |
| FCAR | Forward:
GACCACCCTCCTGTGTCTTG |
|
| Reverse:
GGATGGAGTCGTAGCACCTG |
| MLLT6 | Forward:
GTGGGCCATGGCAGAAGTAG |
|
| Reverse:
CCCTGATTCAAAGCCCCGAA |
| DDX52 | Forward:
TGCGATCAACTACTTCGGCA |
|
| Reverse:
ATCGCAGCCATGGACGTTTA |
| AC010970 | Forward:
TGTCAGCACTCCCAACAGAC |
|
| Reverse:
GTCCCTCATGGCCACAAGTT |
| APBA2 | Forward:
ACTGGGACCGCTACAGTACA |
|
| Reverse:
ATCCAGACTGTCAGCATCGC |
| BRD2 | Forward:
GTCTACCGATTCCCACCTCG |
|
| Reverse:
GCCAAGATGGCTGTAGGTGT |
| HLA-DQA1 | Forward:
CTGACAAACATCGCTGTGCT |
|
| Reverse:
GAAGCACCAACTGAACGCAG |
| MDGA1 | Forward:
CTCCGAGTACCCCACAGCTA |
|
| Reverse:
CATGGATCCCCAAAGTTGCAG |
| WASF3 | Forward:
CTTTTAGGGAACCCGCTGGA |
|
| Reverse:
TAGAGCGAACATGGACGACAG |
| GAPDH | Forward:
TGCACCACCAACTGCTTAGC |
|
| Reverse:
GGCATGGACTGTGGTCATGAG |
Results
RNA-seq libraries
To replicate the results, control samples were
obtained from two patients and NRDS samples from four patients. In
total, >300 million reads were acquired from the six samples.
The average percentage of uniquely mapped paired reads was 50.45%
and the average GC percentage was 53.7%. Detailed alignment data
are presented in Table III.
 | Table III.Results of alignment from RNA
sequencing libraries. |
Table III.
Results of alignment from RNA
sequencing libraries.
| Sample ID | Total reads | Uniquely mapped
paired read (%) | Uniquely mapped
unpaired read (%) | GC rate, % |
|---|
| Control 1 | 45805780 | 24193674
(52.8180) | 551484
(1.2040) | 55.8 |
| Control 2 | 53239568 | 36388990
(68.3495) | 995799
(1.8704) | 48.2 |
| NRDS1 | 49799542 | 19819046
(39.7977) | 479564
(0.9630) | 55.7 |
| NRDS2 | 53005504 | 26210282
(49.4482) | 573098
(1.0812) | 56.2 |
| NRDS3 | 53700026 | 23724822
(44.1803) | 514729
(0.9585) | 56.3 |
| NRDS4 | 50728662 | 24525002
(48.3455) | 580637
(1.1446) | 50.1 |
Identification of DEGs
Compared with the control group, 80 genes were
differentially expressed in the NRDS group. Among them, 69 genes
were upregulated and 11 were downregulated. Fig. 1 illustrates all significant DEGs in
an MA plot. The top 10 upregulated and downregulated genes (in
terms of log2FC) are presented in Table IV.
 | Table IV.Top 10 differentially expressed genes
in the neonatal respiratory distress syndrome group compared with
the control group. |
Table IV.
Top 10 differentially expressed genes
in the neonatal respiratory distress syndrome group compared with
the control group.
| Gene |
Log2FC | P-value | Description |
|---|
| Upregulated |
| PIP4K2B | 4.83 |
1.96×10−24 |
Phosphatidylinositol-5-phosphate 4-kinase
type 2β |
| FCAR | 4.43 |
5.43×10−19 | Fc fragment of IgA
receptor |
| MLLT6 | 3.87 |
1.19×10−13 | PHD finger domain
containing |
| DDX52 | 3.52 |
2.82×10−11 | DExD-box helicase
52 |
| AC010970.2 | 3.09 |
4.95×10−10 | AC010970.2 |
| HBD | 2.69 |
1.49×10−08 | Hemoglobin subunit
δ |
| GPR84 | 2.58 |
2.65×10−07 | G protein-coupled
receptor 84 |
| MTRNR2L12 | 2.47 |
1.05×10−07 | MT-RNR2-like
12 |
| LRRC37A2 | 2.46 |
4.07×10−07 | Leucine rich repeat
containing 37 member A2 |
| Downregulated |
| APBA2 | −3.56 |
4.91×10−11 | Amyloid β precursor
protein binding family A member 2 |
| BRD2 | −3.19 |
2.45×10−13 | Bromodomain
containing 2 |
| HLA-DQA1 | −2.61 |
1.04×10−06 | Major
histocompatibility complex, class II, DQ α 1 |
| MDGA1 | −2.08 |
1.10×10−04 | MAM domain
containing glycosylphosphatidylinositol anchor 1 |
| WASF3 | −1.94 |
1.89×10−04 | WAS protein family
member 3 |
| COL10A1 | −1.88 |
1.84×10−05 | Collagen type X α 1
chain |
| GAN | −1.78 |
8.67×10−11 | Gigaxonin |
| LRRN3 | −1.39 |
2.48×10−06 | Leucine rich repeat
neuronal 3 |
| GATA3 | −1.36 |
9.47×10−05 | GATA binding
protein 3 |
| CSF1R | −1.13 |
1.4×10−04 | Colony stimulating
factor 1 receptor |
In addition, the log2 fold change of 10 genes
obtained by RNA-seq and RT-qPCR analysis was compared to validate
the DEGs. There was a statistically significant Pearson correlation
(R2=0.897) between the expression levels measured by
RNA-seq and RT-qPCR (Figs. 2 and
3).
 | Figure 3.Expression of the top five over- and
underexpressed genes, detected by RNA-seq (blue) and RT-qPCR (red).
RNA-seq, RNA sequencing; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; PIP4K2B,
phosphatidylinositol-5-phosphate 4-kinase type 2 β; FCAR, Fc
fragment of IgA receptor; MLLT6, MLLT6 PHD finger containing;
DDX52, DExD-box helicase 52; AC070970, AC010970.1; APBA2, amyloid β
precursor protein binding family A member 2; BRD2, bromodomain
containing 2; HLA-DQA1, major histocompatibility complex class II
DQ α 1; MDGA1, MAM domain containing glycosylphosphatidylinositol
anchor 1; WASF3, WAS protein family member 3. |
GO and KEGG analysis
The mRNA-seq data set comparison to KEGG (12
pathways; Fig. 4) and GO (105 gene
sets; top 20 presented in Table V)
pathways highlighted the up- and downregulation of three groupings
of genes associated with biological function: Cellular processes;
innate immune response; and pathogen recognition response. Of the
12 pathways, five of them were associated with immunological
mechanisms: The tumor necrosis factor (TNF) signaling pathway
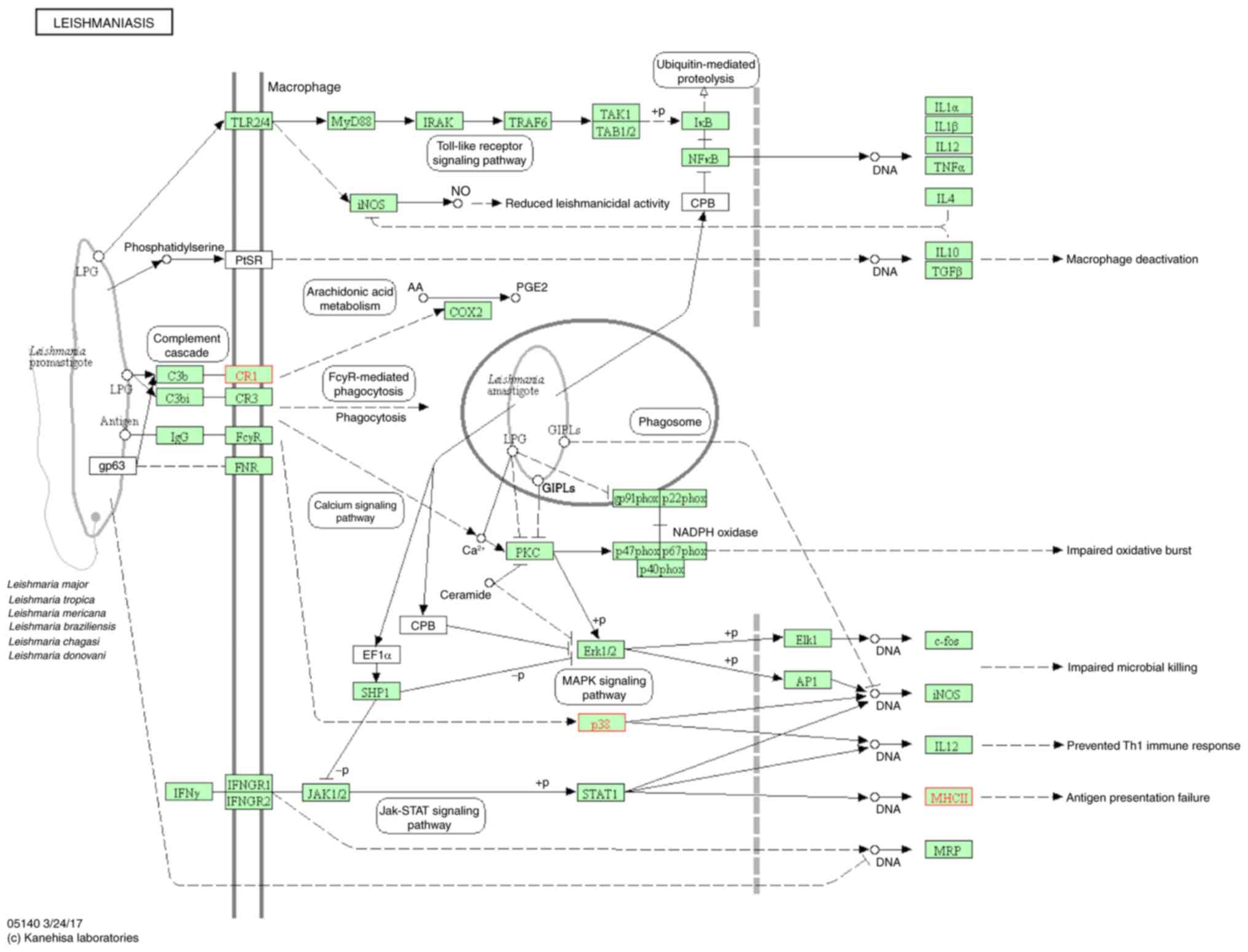
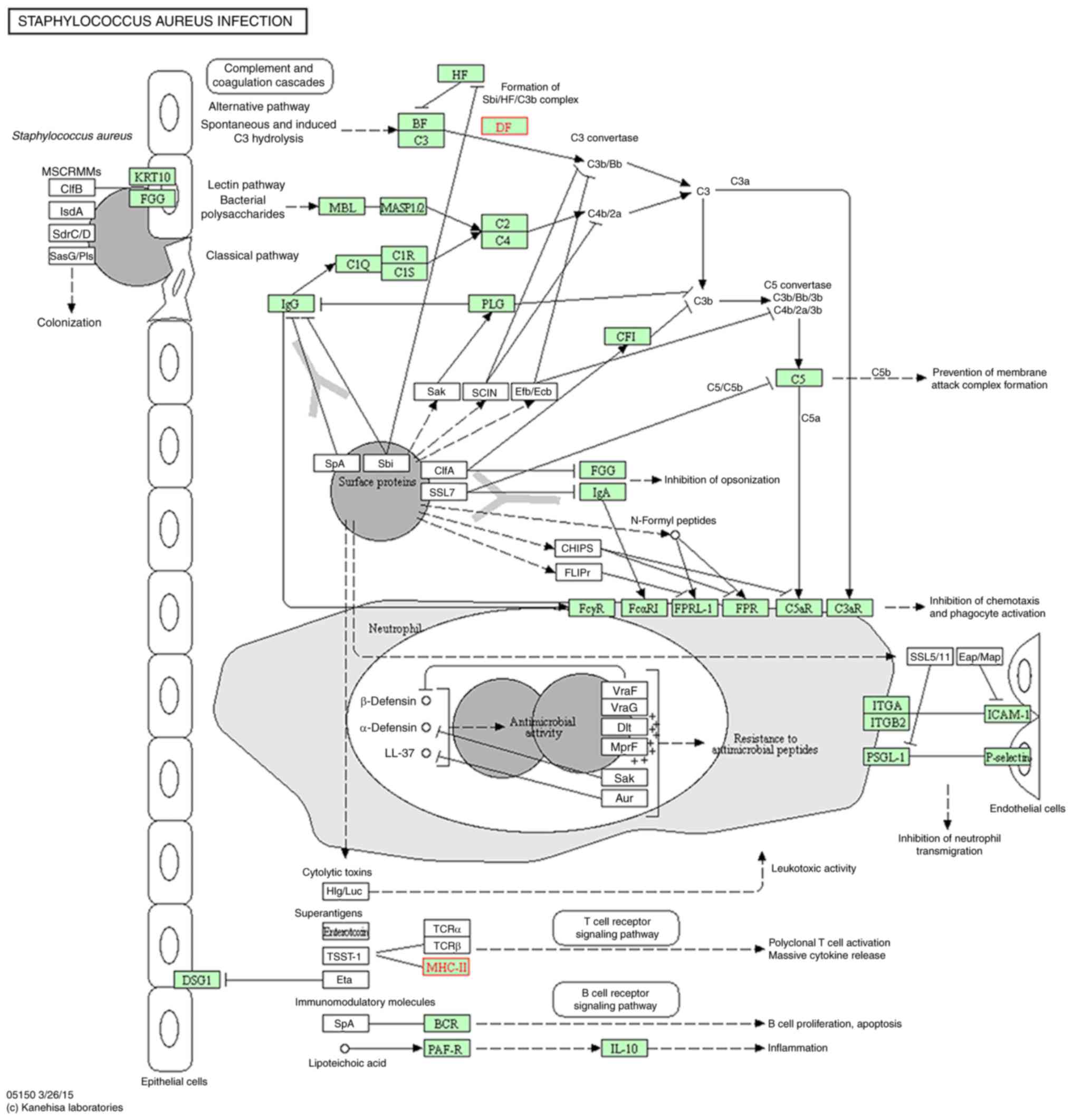
(Fig. 5); influenza A (Fig. 6); leishmaniasis (Fig. 7); Staphylococcus aureus
infection (Fig. 8); and
tuberculosis (Fig. 9). Suppressor
of cytokine signaling 3, mitogen-activated protein kinase 14 and
major histocompatibility complex II are overexpressed in these
pathways; all three of these genes are involved in immune
processes.
 | Table V.Top 20 enriched GO terms among
significantly differentially expressed genes. |
Table V.
Top 20 enriched GO terms among
significantly differentially expressed genes.
| GO ID | Title | Description | No. of genes | P-value |
|---|
| GO:0006260 | DNA
replication | The cellular
metabolic process in which a cell duplicates one or more molecules
of DNA. | 5 | 0.000278 |
| GO:0001878 | Response to
yeast | Any process that
results in a change in state or activity of a cell or an organism
as a result of a stimulus from a yeast species. | 2 | 0.000873 |
| GO:0060045 | Positive regulation
of cardiac muscle cell proliferation | Any process that
activates or increases the frequency, rate or extent of cardiac
muscle cell proliferation. | 2 | 0.001500 |
| GO:0032094 | Response to
food | Any process that
results in a change in state or activity of a cell or an organism
as a result of a food stimulus. | 2 | 0.001750 |
| GO:0051726 | Regulation of cell
cycle | Any process that
modulates the rate or extent of progression through the cell
cycle. | 4 | 0.001760 |
| GO:0042246 | Tissue
regeneration | The regrowth of
lost or destroyed tissues. | 2 | 0.002900 |
| GO:0042742 | Defense response to
bacterium | Reactions triggered
in response to the presence of a bacterium that act to protect the
cell or organism. | 3 | 0.003520 |
| GO:0060333 |
Interferon-gamma-mediated signaling
pathway | A series of
molecular signals initiated by the binding of interferon-gamma to a
receptor on the surface of a cell, and ending with regulation of a
downstream cellular process. | 3 | 0.004290 |
| GO:0032201 | Telomere
maintenance via semi-conservative replication | The process in
which telomeric DNA is synthesized semi-conservatively by the
conventional replication machinery and telomeric accessory factors
as part of cell cycle DNA replication. | 2 | 0.005140 |
| GO:0045003 | Double-strand break
repair via synthesis-dependent strand annealing | Synthesis-dependent
stand annealing is a major mechanism of double-strand break repair
in mitosis that allows for the error-free repair of a double-strand
break without the exchange of adjacent sequences. | 2 | 0.006020 |
| GO:0000077 | DNA damage
checkpoint | A cell cycle
checkpoint that regulates progression through the cell cycle in
response to DNA damage. | 2 | 0.006490 |
| GO:0006513 | Protein
monoubiquitination | Addition of a
single ubiquitin group to a protein. | 2 | 0.006490 |
| GO:0006953 | Acute-phase
response | An acute
inflammatory response that involves non-antibody proteins whose
concentrations in the plasma increase in response to infection or
injury of homeothermic animals. | 2 | 0.006490 |
|
|
| |
| GO:0000722 | Telomere
maintenance via recombination | Any recombinational
process that contributes to the maintenance of proper telomeric
length. | 2 | 0.007460 |
| GO:0050829 | Defense response to
Gram-negative bacterium | Reactions triggered
in response to the presence of a Gram-negative bacterium that act
to protect the cell or organism. | 2 | 0.007970 |
| GO:0006955 | Immune
response | Any immune process
that functions in the calibrated response of an organism to a
potential internal or invasive threat. | 5 | 0.008730 |
| GO:0097192 | Extrinsic apoptotic
signaling pathway in absence of ligand | A series of
molecular signals in which a signal is conveyed from the cell
surface to trigger the apoptotic death of a cell. | 2 | 0.011300 |
| GO:0009408 | Response to
heat | Any process that
results in a change in state or activity of a cell or an organism
(in terms of movement, secretion, enzyme production, gene
expression, etc.) as a result of a heat stimulus, a temperature
stimulus above the optimal temperature for that organism. | 2 | 0.017400 |
| GO:0032508 | DNA duplex
unwinding | The process in
which interchain hydrogen bonds between two strands of DNA are
broken or ‘melted’, generating a region of unpaired single
strands. | 2 | 0.018100 |
| GO:0001504 | Neurotransmitter
uptake | The directed
movement of neurotransmitters into neurons or glial cells. | 1 | 0.022300 |
Within the groupings of cellular processes, the DNA
replication and Fanconi anemia pathways were downregulated by 93
and 318 nuclear receptor (NR) genes, respectively, in response to
NRDS pathogenesis compared with the control group. By contrast, the
cellular proteolysis by ubiquitination and hormone signaling
pathways were upregulated by 104 and 82 NR genes, respectively.
Discussion
The phenotype of NRDS is not concluded from a single
symptom; rather it is derived from numerous cumulative factors
which precipitate NRDS. RNA-seq is an useful tool to illustrate the
physiological responses to multiple factors which contribute to the
manifestation of NRDS. The following identifies a number of
mechanisms observed in the RNA-seq results, which are consistent
with those reported in the process of ARDS.
In a case-control study reported by Molnar et
al (15), increased levels of
ubiquitin specific peptidase 10 (USP10) were observed in the ARDS
group. In the present study, USP1 was observed to be increased in
the NRDS group (data not shown). As USP proteins are involved in
autophagy, it may be hypothesized that ARDS and NRDS exhibit
similar processes of autophagy.
A recent study by Qiao et al (16) illustrated that the
ubiquitin-proteasome pathway (UPP) targeted signal transducer and
activator of transcription 6 and further regulated T helper cells 2
and 9; this regulation may serve as an ideal target for mitigating
the underlying pathology of allergic diseases, particularly asthma.
Dysregulation and chronic overexpression of the UPP ligase systems
and other ubiquitinated kinase protein components may cause
inflammation of the bronchi and promote the pathophysiology of
NRDS. Majetschak et al (17) demonstrated that ubiquitin-activated
20S core proteasome particles in bronchoalveolar lavage fluid were
markedly increased following lung contusions. The proteolytic
cleavage of functional proteasome complexes and additional
post-protein processing observed within the alveolar space
following lung injury may be associated with the pathogenesis of
NRDS. It is unknown how cleavage and post-regulation of proteins
involved in lung edema clearance interact with essential surfactant
proteins, which may further enhance the NRDS pathological
cascade.
Inhibition of the ubiquinated protein kinase
myristoylated alanine rich protein kinase C substrate (MARCKS)
serves a role in mitigating excess pro-inflammatory cytokines
release from neutrophils following lipopolysaccharide (LPS) from
Escherichia coli and the activation of Toll-like receptor
(TLR)2, TLR4 and TLR9 (18). The
results of this previous study illustrated correct and incorrect
post-transcriptional regulation. When organisms were treated with
the protein myristoylated N-terminal sequence, an inhibitor of
MARCKS, neutrophil migration was mediated; marked alterations were
observed in the reduction of mRNA expression associated with
LPS-challenged interleukin-8 and TNF-α.
The assessment of various lung cellular responses to
numerous environmental factors, by assessing mRNA-seq associated
with NRDS, is a viable and sophisticated way to characterize NRDS
and ARDS. Assessing the progression of various symptoms towards the
onset of diseases may help to further characterize cellular
responses associated with disease and to further elucidate the
underlying pathogenesis of diseases. A previous study assessed how
alterations at the mRNA level affect how transcriptional gene
regulation is moderated prior to the production and translation of
proteins (15). Gao and Barnes
(19) performed a meta-analysis
using RNA-seq data of 83 lung-specific genes; only 62 of the genes
were required to code for proteins necessary for lung development
and pathogenesis. Specifically, in ARDS there are 21 principal
genes of interest to target that are associated with the
physiological onset of ARDS and similar acute lung injuries.
Due to the hydrophobic nature of surfactants, they
are able to keep the alveoli dry and allow for optimal oxygen-blood
exchange. Dysregulation of surfactant phospholipids has been
previously demonstrated to be associated with alveolar instability
and an increased patient risk of developing NRDS (20). Translational regulation of gene
families has essential roles in maintaining lung homeostasis,
including surfactant-associated proteins (SFTPs; SFTPA1, SFTPA2,
SFTPC and SFTPD) which serve a role in understanding the pathology
of NRDS (21,22).
In a study by D'Ovidio et al (23), clinicians assessed the mortality
associated with lung transplants and various mRNA levels among
different experimental groups and their surfactant protein
polymorphism (SP-A, SP-B, SP-C and SP-D) expression levels. The
pilot study demonstrated notable mortality rates in patients;
however, those with increased mRNA expression of SP generally
exhibited a decreased incidence of allograft rejection by the
innate immune system, thus illustrating a marked difference in lung
transplant mortality associated with surfactants. Additionally,
Silva et al (24) observed
that the application of the anti-inflammatory agent LASSBio596 may
be applied to ARDS mouse models to increase survival rates by
mitigating macrophages, neutrophils and transforming growth
factor-β by increasing surfactant yield, which in turn reduced
surface tension. By maintaining elasticity and surfactant
alterations in lung mechanics, the authors were able to counteract
degradation immediately following lung injury leading to ARDS.
The removal of surfactant proteins leads to
substantial alterations in the alveoli-capillary barrier, leading
to the serious phenotype of sepsis. Pathogenesis-induced sepsis of
the peripheral lung is denoted by inflammation associated with the
presence of the microbial metabolite lactate, which is a standard
lung biomarker of sepsis (25). In
response to a microbial presence in the form of adhesion or
byproducts, the innate immune system is the first to mount a
response. In order to further assess the biomarkers of NRDS, Ware
et al (26) demonstrated
that SP-D overexpression is an effective marker of sepsis
prognosis; increased expression of this protein from alveolar
epithelial type II cells is associated with increased
mortality.
In the grouping of gene families associated with the
innate immune response, it has been reported that all four
pathways, complement and coagulation cascades, tumor TNF signaling,
inflammatory bowel disease and prolactin signaling, were
upregulated by 50–67 genes in response to NRDS. In the case of
pneumonia, pro-inflammatory cytokines and systemic inflammation,
the host-associated response will lead to increased TNF expression
followed by dysregulation of lung-induced apoptosis by neutrophils
(27). Proteinase-activated
cascades following lung injury promote the activation of the
coagulation response that begins the innate inflammatory response
and the additional pro-inflammatory factors, including TNF
(28). Macrophage-derived TNF has
been demonstrated to further provoke lung injury by stimulating p55
receptor-mediated death signaling; this further exacerbates injury
by increasing lung neutrophils and leads to the deleterious
phenotype of ARDS (29).
Upstream activation of TNF-α within the lung
periphery is an essential mediator of neutrophils, and the
pathology of NRDS and ARDS. Plasma TNF-α levels have been observed
to be increased in patients with NRDS compared with controls
(30). A study performed in mice
by Lomas-Neira et al (31)
demonstrated that the transfer of TNF-α to mice lacking neutrophils
did not stimulate the progression of lung injury. By comparison,
mice lacking neutrophils expressing TNF-α displayed reduced acute
lung injury. These previous results suggested that TNF-α expression
in neutrophils is an underlying factor the promotes the progression
of the pathogenesis from the initial lung-injury into a
shock-primed innate immune response.
In response to colonization by pathogenic organisms,
including Streptococcus pneumoniae, proteinase-activated
receptor 1 serves a role in splicing for the activation of
coagulation pathways leading to inflammation; thus, beginning the
cascade of neutrophilic pro-inflammatory responses to invasive
pathogens, excess neutrophil recruitment contributes to lung injury
through alveolar surfactant barrier disruption and alveolar leakage
(18).
The pathogen recognition response for NRDS KEGG
pathways all had upregulated gene families in the present study;
this was the case for all except Staphylococcus aureus
infection, which was downregulated by 10 NR genes. Upregulated gene
families included leishmaniasis, tuberculosis, and influenza A, and
the number of genes involved in these three groups were 3, 4 and 3,
respectively. Microbial proliferation and sepsis following acute
lung injury is a primary contributor to the cascade and severity of
NRDS. Lv et al (32)
demonstrated through transcriptomic analysis that genes associated
with foreign pathogens (NLR family pyrin domain containing 3 and
S100 calcium binding protein A8/A9) were markedly upregulated in
response to microbial LPS.
In addition to sepsis, other factors not involved
with the innate immune response or surfactants may contribute to
NRDS and ARDS. For example, Lv et al (32) demonstrated, through systematic
mRNA-seq of ARDS, a significant upregulation of 122 genes and a
downregulation of 91 genes associated with essential protein
functions, including the mitotic cell cycle. Specifically, the
investigators noted that cyclin (CCNB)1 and CCNB2, which serve a
role in cell cycle regulation, may be associated with the onset of
ARDS.
A previous study of the human microbiome indicated
dysbiosis within the human system, typically leading to a
dysregulation of host responses to various systemic challenges; in
particular, these challenges threaten lung homeostasis and provoke
the perpetual inflammation associated with invasive pathogenic
microbiota (33). Invasion,
persistence and colonization of foreign microbiota challenge the
host immune system to further provoke ARDS; the proliferation of
microbiota and heightened responses to challenge further provoke
the accumulated lung injury to develop into sepsis (34). Infections with and the persistence
of numerous viral strains and microbiota, including influenza A
(35), Mycobacterium
tuberculosis (36),
Staphylococcus aureus/H1N1 strain of the influenza virus
(37) and multi-drug resistant
Acinetobacter baumannii (38).
Dysregulation of TNF and the coagulation cascade
pathways may be associated with ubiquitin-mediated kinase
activation in the case of proteasome activated lysis, which may
activate cascades leading to neutrophil over-stimulation and
eventual dysregulated lung damage. In conclusion, the mechanisms
leading to a specific NRDS phenotype are complex, although mRNA-seq
as a tool may be used to elucidate genomic response to lung-induced
stress. Consequently, cascading immune mechanisms associated with
the dysregulation of one aspect of the immune response may
accumulate progressively towards NRDS.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. gjzr81260107) and
the Inner Mongolia Natural Science Foundation (grant no.
2011MS1111).
References
|
1
|
Angus DC, Linde-Zwirble WT, Clermont G,
Griffin MF and Clark RH: Epidemiology of neonatal respiratory
failure in the United States: Projections from California and New
York. Am J Respir Crit Care Med. 164:1154–1160. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Santos C, Ferrer M, Roca J, Torres A,
Hernández C and Rodriguez-Roisin R: Pulmonary gas exchange response
to oxygen breathing in acute lung injury. Am J Respir Crit Care
Med. 161:26–31. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Doyle IR, Bersten AD and Nicholas TE:
Surfactant proteins-A and -B are elevated in plasma of patients
with acute respiratory failure. Am J Respir Crit Care Med.
156:1217–1229. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dolinay T, Kim YS, Howrylak J, Hunninghake
GM, An CH, Fredenburgh L, Massaro AF, Rogers A, Gazourian L,
Nakahira K, et al: Inflammasome-regulated cytokines are critical
mediators of acute lung injury. Am J Respir Crit Care Med.
185:1225–1234. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Speer CP, Robertson B, Curstedt T,
Halliday HL, Compagnone D, Gefeller O, Harms K, Herting E, McClure
G, Reid M, et al: Randomized European multicenter trial of
surfactant replacement therapy for severe neonatal respiratory
distress syndrome: Single versus multiple doses of Curosurf.
Pediatrics. 89:13–20. 1992.PubMed/NCBI
|
|
6
|
Enhorning G, Shennan A, Possmayer F, Dunn
M, Chen CP and Milligan J: Prevention of neonatal respiratory
distress syndrome by tracheal instillation of surfactant: A
randomized clinical trial. Pediatrics. 76:145–153. 1985.PubMed/NCBI
|
|
7
|
Thawer S, Auret J, Schnoeller C, Chetty A,
Smith K, Darby M, Roberts L, Mackay RM, Whitwell HJ, Timms JF, et
al: Surfactant protein-D is essential for immunity to helminth
infection. PLoS Pathog. 12:e10054612016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
de Guevara YL, Hidalgo OB, Santos SS,
Brunialti MK, Faure R and Salomao R: Effect of natural porcine
surfactant in Staphylococcus aureus induced pro-inflammatory
cytokines and reactive oxygen species generation in monocytes and
neutrophils from human blood. Int Immunopharmacol. 21:369–374.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bernard GR, Artigas A, Brigham KL, Carlet
J, Falke K, Hudson L, Lamy M, Legall JR, Morris A and Spragg R: The
American-European Consensus Conference on ARDS. Definitions,
mechanisms, relevant outcomes and clinical trial coordination. Am J
Respir Crit Care Med. 149:818–824. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Khadaroo RG and Marshall JC: ARDS and the
multiple organ dysfunction syndrome. Common mechanisms of a common
systemic process. Crit Care Clin. 18:127–141. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sweet DG, Carnielli V, Greisen G, Hallman
M, Ozek E, Plavka R, Saugstad OD, Simeoni U, Speer CP, Halliday HL,
et al: European consensus guidelines on the management of neonatal
respiratory distress syndrome in preterm infants-2010 update.
Neonatology. 97:402–417. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Trapnell C, Williams BA, Pertea G,
Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ and Pachter
L: Transcript assembly and quantification by RNA-Seq reveals
unannotated transcripts and isoform switching during cell
differentiation. Nat Biotechnol. 28:511–515. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Häne BG, Jäger K and Drexler HG: The
Pearson product-moment correlation coefficient is better suited for
identification of DNA fingerprint profiles than band matching
algorithms. Electrophoresis. 14:967–972. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Molnar CS, Sárvári M, Vastagh C, Maurnyi
C, Fekete C, Liposits Z and Hrabovszky E: Altered gene expression
profiles of the hypothalamic arcuate nucleus of male mice suggest
profound developmental changes in peptidergic signaling.
Neuroendocrinology. 103:369–382. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qiao G, Ying H, Zhao Y, Liang Y, Guo H,
Shen H, Li Z, Solway J, Tao E, Chiang YJ, et al: E3 ubiquitin
ligase Cbl-b suppresses proallergic T cell development and allergic
airway inflammation. Cell Rep. 6:709–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Majetschak M, Sorell LT, Patricelli T,
Seitz DH and Knoferl MW: Detection and possible role of proteasomes
in the bronchoalveolar space of the injured lung. Physiol Res.
58:363–372. 2009.PubMed/NCBI
|
|
18
|
José RJ, Williams AE, Mercer PF,
Sulikowski MG, Brown JS and Chambers RC: Regulation of neutrophilic
inflammation by proteinase-activated receptor 1 during bacterial
pulmonary infection. J Immunol. 194:6024–6034. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gao L and Barnes KC: Recent advances in
genetic predisposition to clinical acute lung injury. Am J Physiol
Lung Cell Mol Physiol. 296:L713–L725. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gregory TJ, Longmore WJ, Moxley MA,
Whitsett JA, Reed CR, Fowler AA III, Hudson LD, Maunder RJ, Crim C
and Hyers TM: Surfactant chemical composition and biophysical
activity in acute respiratory distress syndrome. J Clin Invest.
88:1976–1981. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Melton KR, Nesslein LL, Ikegami M,
Tichelaar JW, Clark JC, Whitsett JA and Weaver TE: SP-B deficiency
causes respiratory failure in adult mice. Am J Physiol Lung Cell
Mol Physiol. 285:L543–L549. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bridges JP, Wert SE, Nogee LM and Weaver
TE: Expression of a human surfactant protein C mutation associated
with interstitial lung disease disrupts lung development in
transgenic mice. J Biol Chem. 278:52739–52746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
D'Ovidio F, Kaneda H, Chaparro C, Mura M,
Lederer D, Di Angelo S, Takahashi H, Gutierrez C, Hutcheon M,
Singer LG, et al: Pilot study exploring lung allograft surfactant
protein A (SP-A) expression in association with lung transplant
outcome. Am J Transplant. 13:2722–2729. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Silva JD, de Oliveira GP, Cdos Samary S,
Araujo CC, Padilha Gde A, Filho Costa e Silva F, da Silva RT,
Einicker-Lamas M, Morales MM, Capelozzi VL, et al: Respiratory and
systemic effects of LASSBio596 plus surfactant in experimental
acute respiratory distress syndrome. Cell Physiol Biochem.
38:821–835. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, D'Annibale-Tolhurst MA, Adler KB,
Fang S, Yin Q, Birkenheuer AJ, Levy MG, Jones SL, Sung EJ, Hawkins
EC, et al: A myristoylated alanine-rich C kinase substrate-related
peptide suppresses cytokine mRNA and protein expression in
LPS-activated canine neutrophils. Am J Respir Cell Mol Biol.
48:314–321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ware LB, Koyama T, Zhao Z, Janz DR,
Wickersham N, Bernard GR, May AK, Calfee CS and Matthay MA:
Biomarkers of lung epithelial injury and inflammation distinguish
severe sepsis patients with acute respiratory distress syndrome.
Crit Care. 17:R2532013. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bordon J, Aliberti S, Fernandez-Botran R,
Uriarte SM, Rane MJ, Duvvuri P, Peyrani P, Morlacchi LC, Blasi F
and Ramirez JA: Understanding the roles of cytokines and neutrophil
activity and neutrophil apoptosis in the protective versus
deleterious inflammatory response in pneumonia. Int J Infect Dis.
17:e76–e83. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chambers RC and Scotton CJ: Coagulation
cascade proteinases in lung injury and fibrosis. Proc Am Thorac
Soc. 9:96–101. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Patel BV, Wilson MR, O'Dea KP and Takata
M: TNF-induced death signaling triggers alveolar epithelial
dysfunction in acute lung injury. J Immunol. 190:4274–4282. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meng Q, Xiao DX, Zhang Z and Fu WH:
Changes and clinical significance of the peripheral blood
mononuclear cell NF-κB activity and TNF-α in neonatal respiratory
distress syndrome. Chin J Child Health Care. 4:411–412. 2009.(In
Chinese).
|
|
31
|
Lomas-Neira J, Perl M, Venet F, Chung CS
and Ayala A: The role and source of tumor necrosis factor-alpha in
hemorrhage-induced priming for septic lung injury. Shock.
37:611–620. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lv X, Zhang Y, Lu W, Wang Q, Li Sy, Guo L,
Qian Gs, Zhou Sw and Li Yy: Digital gene expression analysis of
transcriptomes in lipopolysaccharide-induced acute respiratory
distress syndrome. Clin Chim Acta. 453:182–189. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dickson RP, Martinez FJ and Huffnagle GB:
The role of the microbiome in exacerbations of chronic lung
diseases. Lancet. 384:691–702. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kitsios GD, Morowitz MJ, Dickson RP,
Huffnagle GB, McVerry BJ and Morris A: Dysbiosis in the intensive
care unit: Microbiome science coming to the bedside. J Crit Care.
38:84–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Short KR, Kroeze EJBV, Fouchier RAM and
Kuiken T: Pathogenesis of influenza-induced acute respiratory
distress syndrome. Lancet Infect Dis. 14:57–69. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Arevalo-Lorido JC, Bureo-Dacal JC,
Romero-Requena J, Ramos-Salado JL and Bureo-Dacal P: Tuberculosis
complicated by ARDS. Ir J Med Sci. 170:2092001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lees NJ, Rosenberg A, Hurtado-Doce AI,
Jones J, Marczin N, Zeriouh M, Weymann A, Sabashnikov A, Simon AR
and Popov AF: Combination of ECMO and cytokine adsorption therapy
for severe sepsis with cardiogenic shock and ARDS due to
Panton-Valentine leukocidin-positive Staphylococcus aureus
pneumonia and H1N1. J Artif Organs. 19:399–402. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin CY, Chen YM, Lin MC, Chang YP, Chao
TY, Wang CC, Tsai YC, Shen LS, Li CL and Lin AS: Risk factors of
multidrug-resistant Acinetobacter baumannii recurrence after
successful eradication in ventilated patients. Biomed J.
39:130–138. 2016. View Article : Google Scholar : PubMed/NCBI
|