Introduction
Nasopharyngeal carcinoma (NPC) is a malignant cancer
derived from epithelial cells located in the nasopharynx (1). Radiotherapy is one of the most common
treatment options for patients with NPC (2). However, due to distant metastases and
local recurrence in some patients with NPC (3), radioresistance can be a serious
obstacle to therapy success. Therefore, there is an urgent medical
need to understand the molecular mechanisms of NPC progression and
radioresistance, to develop novel diagnostic and therapeutic
strategies that can potentially enhance tumor cell
radiosensitivity.
MicroRNAs (miRNAs) are a class of endogenous single
stranded non-coding RNAs of 18–25 nucleotides in length that
regulate gene expression by binding to the 3′untranslated regions
(3′UTRs) of target mRNAs (4,5).
Accumulating evidence has demonstrated that miRNAs regulate
numerous physiological processes, including cell proliferation,
cell cycle stage, apoptosis, migration, invasion and
differentiation (6,7). Growing evidence supports the critical
role of miRNAs in the progression of human cancers, where they
function as either oncogenic miRNAs (oncomirs) or tumor suppressors
through the regulation of cellular proliferation, differentiation
and apoptosis (8–10). A number of studies have
demonstrated that miRNAs are involved in cellular ionizing
radiation (IR) responses through cell cycle regulation and the
apoptosis signal pathway in several types of cancer, including NPC
(11–13).
miRNA-222 (miR-222) has been demonstrated to
function as an oncomir in various types of human cancer, with
effects on cell growth, oncogenesis, invasion, migration and drug
resistance of tumor cells (14–18).
A recent report revealed that miR-222 confers radioresistance in
glioblastoma cells through the activation of the protein kinase B
(AKT) signaling pathway, independent of phosphatase and tensin
homolog (PTEN) status (17).
However, the detailed function of miR-222 in NPC remains unclear.
The present study investigated the role of miR-222 in NPC
carcinogenesis, particularly in NPC radioresistance.
Materials and methods
Cell culture and tissue samples
The three human NPC cell lines (HONE-1, C666-1 and
TWO3) and the NP69 human immortalized nasopharyngeal epithelial
cell line were obtained from The Cell Bank of Type Culture
Collection of Chinese Academy of Science (Shanghai, China) were
cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal
bovine serum (Thermo Fisher Scientific, Inc.), 100 U/ml penicillin
(Sigma Aldrich; Merck KGaA, Darmstadt, Germany) and 100 mg/ml
streptomycin at 37°C in a humidified atmosphere of 5%
CO2.
Human NPC and adjacent normal tissue samples (n=30
each) were harvested at The Tumor Hospital of Jilin Province
(Changchun, China) between July 2014 and July 2015. All tissue
samples were immediately snap-frozen in liquid nitrogen following
surgery, and stored in liquid nitrogen until use. The study was
approved by the Ethic Committee of the Tumor Hospital of Jilin
Province (Changchun, China) and written informed consent was
obtained from every patient
Cell transfection
miR-222 mimic, 5′-AGCUACAUCGGCUACUGGGUUU-3′ and the
corresponding negative control, miR-NC, UUC UCC GAA CGU GUG UCA CGU
TT, miR-222 inhibitors, anti-miR-222, 5′-AGCUACAUCUGGCUACUGGGU-3′
and the corresponding miRNA negative control, anti-miR-NC,
5′-UCUACUCUUUCUAGGAGGUUGUGA-3′, were obtained from Qiagen Sciences,
Inc. (Frederick, MD, USA). A PTEN overexpression plasmid was
obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China).
Plasmids (100 ng) and miRNAs (100 nM) were transiently transfected
into C666-1 cells using Lipofectamine 3000 Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Transfected cells were cultured for 1–3 days until
subsequent analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA of cell lines was extracted with TRIzol
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and the purity
and concentration of the RNA was determined by a dual-beam
ultraviolet spectrophotometer (Eppendorf, Hamburg, Germany). The
total RNA was then reverse transcribed into cDNA using a Universal
cDNA synthesis kit (Exiqon, Inc., Woburn, MA, USA) according to the
manufacturer's protocol. cDNA was amplified and quantified using
the Strotatagene Mx3005P real-time PCR System (Agilent
Technologies, Inc., Santa Clara, CA, USA) with the Taqman Universal
PCR Master Mix (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Specific primer sequences were: Mature miR-222 forward, ACA
CTC CAG CTG GGA GCT ACA TCT GGC TAC TG and reverse, CTC AAC TGG TGT
CGT GGA) and U6 (control) forward, CTC GCT TCG GCA GCA CA and
reverse, AAC GCT TCA CGA ATT TGC GT (Applied Biosystems; Thermo
Fisher Scientific, Inc. PTEN, forward, TTG TGG TCT GCC AGC TAA A
and reverse, CGC TCT ATA CTG CAA ATG CT and GAPDH forward, GCA CCG
TCA AGG CTG AGA AC and reverse-TGG TGA AGA CGC CAG TGG A. Primers
were used in this study as described previously (17). qPCR was performed in triplicate
consisting of 40 cycles of a denaturation step at 95°C for 10 sec,
annealing at 58°C for 30 sec and extension at 72°C for 40 sec
following a cycle of a pre-denaturation step at 95°C for 40 sec. U6
and GAPDH were used as endogenous controls for the detection of
miR-222 and PTEN, respectively. For data analysis, the
2−∆∆Cq method (19) was
used to calculate fold change using the Rotor-Gene 6000 Series
Software 1.7 (Qiagen Sciences, Inc.).
Cell viability assay
For the cell viability assay, transfected cells were
seeded in 96-well plates at a density of 2×103
cells/well. Cells were continually cultured for 72 h prior to the
addition of 10 µl 0.5 mg/ml MTT to each well. Cells were incubated
for 4 h at 37°C. The medium was subsequently removed and 100 µl
dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA) was added to each
well. Following 20 min of agitation, absorbance was detected at 490
nm with an iMark Microplate Absorbance Reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Colony formation assay
Transfected cells (1×103 per well) were
seeded in 6-well plates and cultured for 10 days. Colonies were
washed with phosphate-buffered saline (pH, 7.2) and subsequently
fixed with 4% paraformaldehyde (Sigma-Aldrich; Merck KGaA) at room
temperature for 10 min and stained with 1% crystal violet
(Sigma-Aldrich; Merck KGaA) at room temperature for 5 min. Colony
numbers were counted under an IX71 inverted light microscope
(Olympus Corporation, Tokyo, Japan).
Cell apoptosis assay
Cell apoptosis was examined using flow cytometry in
transfected cells. At 48 h post-transfection, cells were harvested
and the apoptosis assay was performed using an Annexin V/Propidium
Iodide Detection kit (Nanjing KeyGen Biotech Co., Ltd., Nanjing,
China) in a FACS Calibur flow cytometer (BD Biosciences, Franklin
Lakes, NJ, USA), according to the manufacturer's protocol. The
apoptosis ratio was calculated using CellQuest software 3.4 (BD
Biosciences).
In vitro radiosensitivity assay
Cells transfected with the miR-222 mimic were
treated with 0, 1, 2, 4, 6 and 8 Gy X-ray radiation, using a
Faxitron RX-650 (Faxitron Bioptics, Lincolnshire, IL, USA) with 100
kVp. The dose was administered at a rate of 3 Gy/min at room
temperature. A colony formation assay was then performed on the
irradiated cells., survival curve parameters were determined using
a Kaplan-Meier plot. Additionally, cells transfected with miR-222
mimic or miR-NC were exposed to 4 Gy of X-ray radiation. Cells were
harvested at 48 h after IR and a cell apoptosis assay was
subsequently performed as described above.
Vector construction and luciferase
activity assay
The 3′UTR of PTEN containing the putative miR-222
binding site was synthesized (GenePharma Co., Ltd) and inserted
into the pGL3 control vector (Promega Corporation, Madison, WI,
USA). Mutations in the miR-222 binding site of the PTEN 3′UTR were
introduced with the QuikChange Site-Directed Mutagenesis kit
(Stratagene; Aligent Technologies, Inc., Santa Clara, CA, USA)
according to the manufacturer's protocol.
For the luciferase reporter assay, C666-1 cells were
cultured in 96-well plates and co-transfected with 100 ng pGL3 with
wild type PTEN-3′UTR (Wt-PTEN-3′UTR) or pGL3 with mutant PTEN-3′UTR
(Mut-PTEN-3′UTR). Additionally, 80 ng luciferase co-reporter vector
pRL-SV40 was added, alone or in combination with miR-NC (100 nM) or
miR-222 mimic (100 nM), using Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Luciferase activity was measured using the Dual
Luciferase Reporter Assay System (Promega Corporation, Madison, WI,
USA) 48 h post-transfection. Firefly luciferase activity was
normalized to Renilla luciferase activity for each
transfected well.
Western blotting
Protein extracts were obtained from cultured cells
using radioimmunoprecipitation lysis buffer (Beyotime Institute of
Biotechnology, Beijing, China). The total concentration of protein
was measured using a bicinchoninic acid protein assay kit (Boster
Biological Technology, Pleasanton, CA, USA). Protein lysates (30 µg
per lane) were separated by 10% sodium dodecyl sulfate
polyacrylamide gel electrophoresis (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA), prior to transfer onto nitrocellulose membranes
(Bio-Rad Laboratories, Inc.). Membranes were blocked with 5%
non-fat milk in Tris-buffered saline for 1 h at room temperature
and incubated with the following antibodies overnight at 4°C:
Anti-PTEN (1:1,000; sc-133197; Santa Cruz Biotechnology Inc.,
Dallas, TX, USA), anti-phosphoinositide 3-kinase (PI3K; 1:1,000;
sc-293172; Santa Cruz), anti-AKT (1:500; sc-8312; Santa Cruz
Biotechnology Inc.) and anti-phosphorylated-AKT (p-AKT; Ser473;
sc-271966; 1:500; Santa Cruz Biotechnology Inc.). Anti-GAPDH
(1:1,000; sc-47724; Santa Cruz Biotechnology Inc.) was used as an
internal control for protein loading. The membrane was subsequently
incubated with horseradish peroxidase conjugated goat anti-mouse
immunoglobulin G (IgG; 1:5,000; sc-516102; Santa Cruz Biotechnology
Inc.) or goat anti-rabbit IgG (1:5,000; sc-2040; Santa Cruz
Biotechnology Inc.) secondary antibodies for 2 h at room
temperature. Proteins were visualized using a chemiluminescent
detection system (Thermo Fisher Scientific, Inc.) and exposed on
X-ray film.
Statistical analysis
All data are expressed as the mean ± standard
deviation of at least three independent experiments or samples. The
statistical difference was determined using a two-tailed Student
t-test or one-way analysis of variance followed by a Bonferroni
post-hoc test. All data analyses were performed using SPSS 19.0
software (IBM Corp., Armonk, NY, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-222 expression is upregulated in
NPC tissues and cells
The expression levels of miR-222 in NPC and adjacent
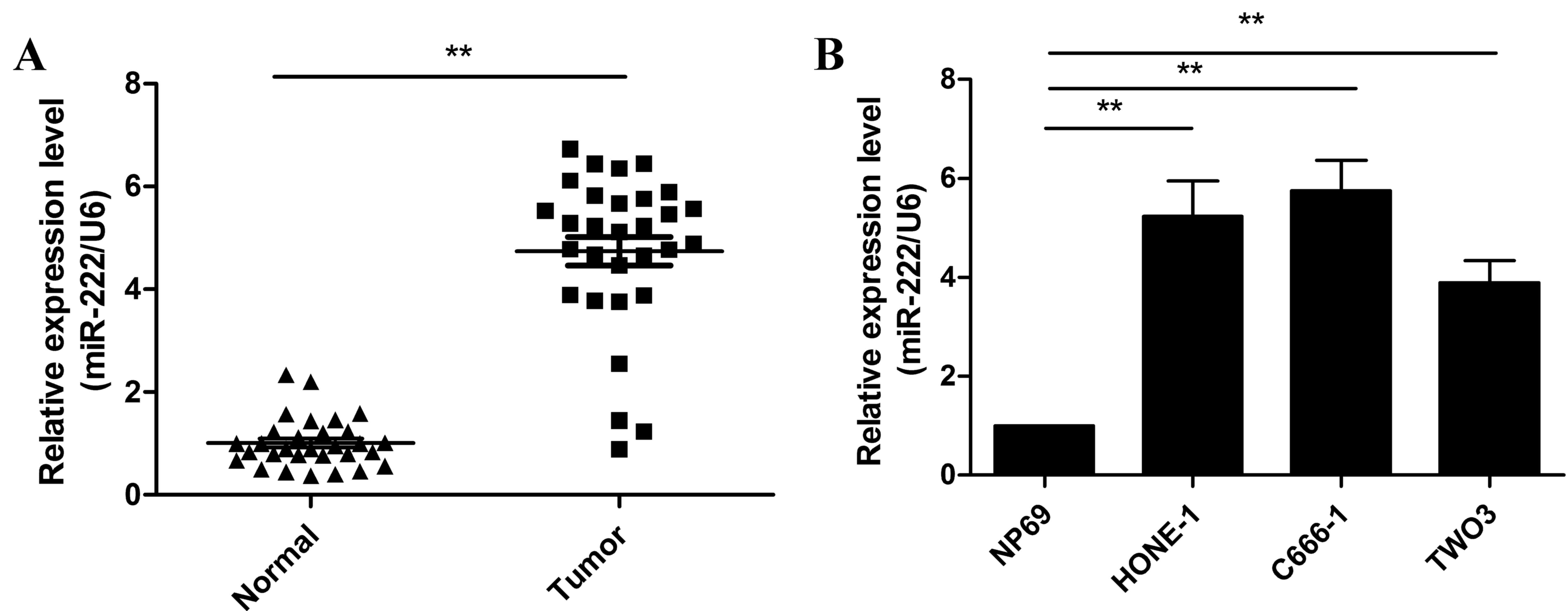
normal tissues was measured by RT-qPCR. As presented Fig. 1A, the level of miR-222 expression
in NPC tissues was significantly higher than that of adjacent
normal tissues (P<0.01). The expression of miR-222 in three NPC
cell lines (C666-1, HONE-1 and TWO3) and the NP69 nasopharyngeal
epithelial cell line was subsequently examined. miR-222 expression
was significantly upregulated in the three NPC cell lines compared
with NP69 cells (P<0.05; Fig.
1B). This data suggests that miR-222 may be involved in the
initiation and progression of NPC.
miR-222 overexpression increased cell
viability and colony formation, and inhibits the apoptosis of NPC
cells
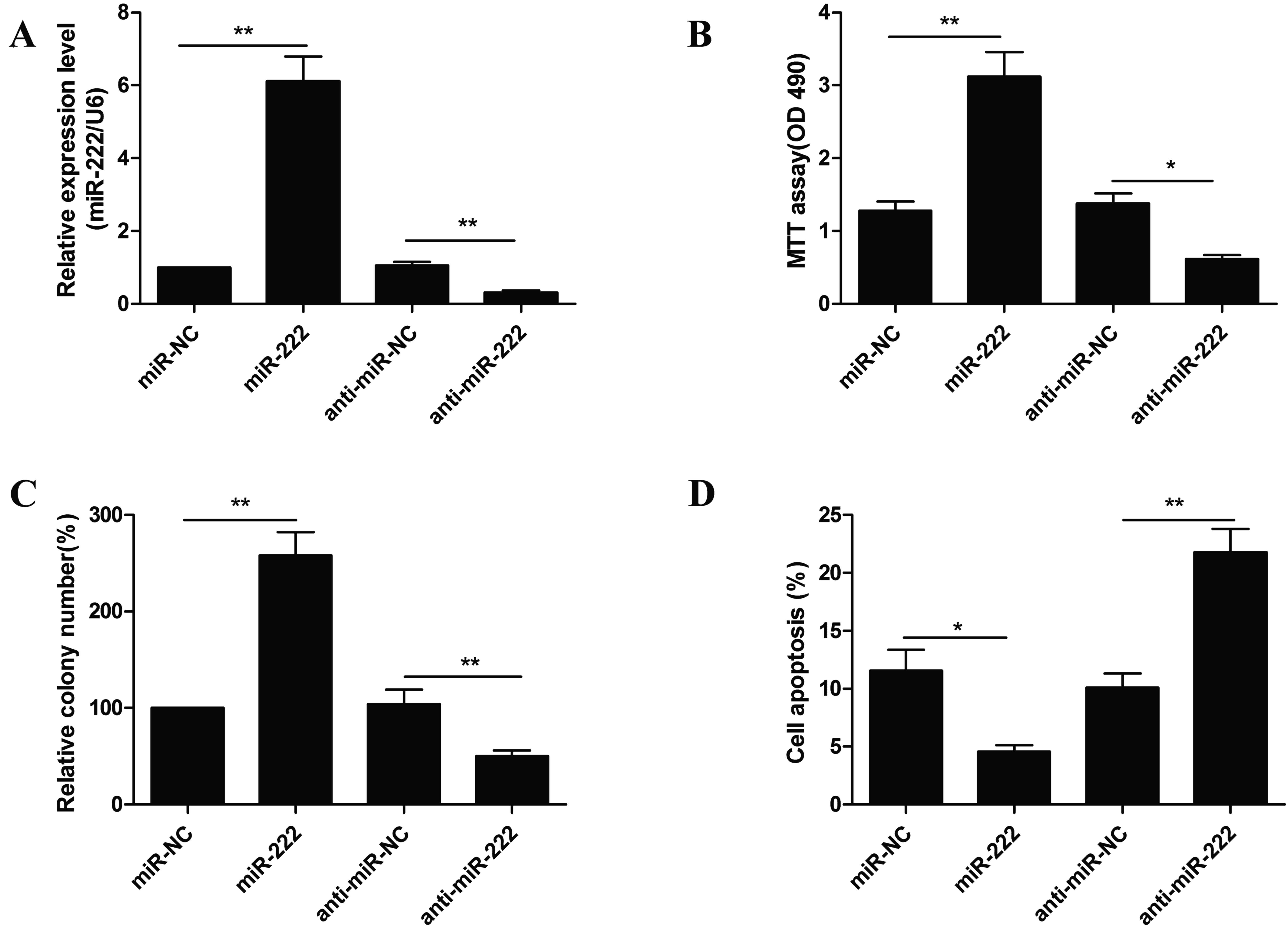
To examine the biological effects of miR-222 in NPC
cells, C666-1 cells were transfected with miR-222 mimic or
anti-miR-222 and the biological function of miR-222 in the C666-1
cells was subsequently evaluated. RT-qPCR analysis confirmed that
C666-1 cells transfected miR-222 mimic had an upregulation of
miR-222 expression, whereas transfection with anti-miR-222 resulted
in a downregulation in miR-222 expression compared with NCs
(Fig. 2A). MTT assay indicated
that miR-222 mimic transfected C666-1 cells had increased cell
proliferation compared with miR-NC. Conversely, a significant
decrease in proliferation was observed in cells transfected with
anti-miR-222 compared with anti-miR-NC (Fig. 2B). Similar results were obtained in
the colony formation assay. As presented in Fig. 2C, upregulated miR-222 expression
promoted colony formation, whereas downregulated miR-222 expression
inhibited colony formation in C666-1 cells compared with the
negative controls. Flow cytometry was subsequently used to examine
the role of miR-222 in apoptosis (data not presented). It was
demonstrated that downregulated miR-222 expression induced
apoptosis, whereas upregulated miR-222 expression decreased cell
apoptosis compared with the negative control groups (Fig. 2D). These results suggest that
miR-222 functions as an oncomir in NPC cells.
miR-222 confers radioresistance in NPC
cells
It is well established that certain miRNAs can
regulate the radioresistance of cancer cells (11–13,17,18).
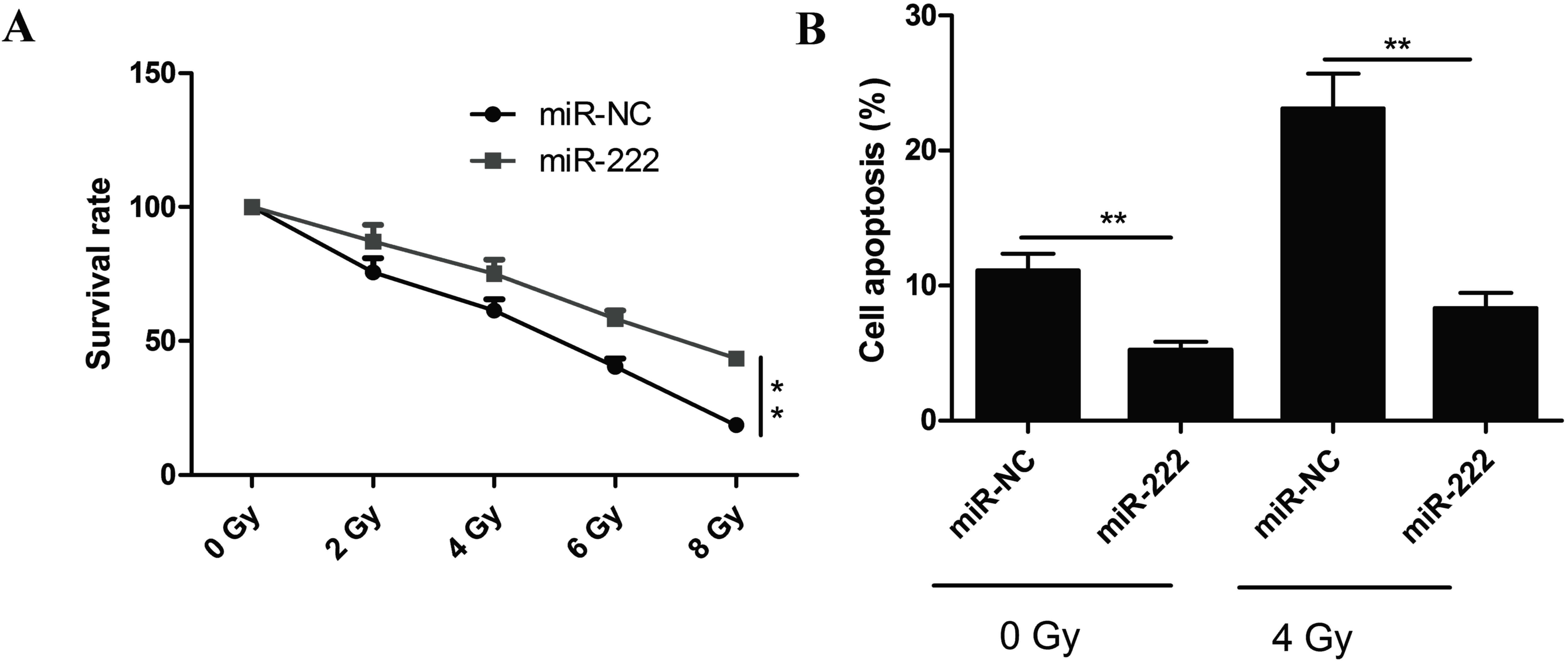
Thus, the effect of miR-222 on NPC cell radioresistance was
investigated. C666-1 cells were transfected with miR-222 mimic
prior to treatment with different doses of radiation. Colony
formation assays were subsequently performed and survival curve
parameters were counted. The survival rate of miR-222 mimic
transfected cells significantly increased, whereas the survival
rate of miR-NC transfected cells significantly decreased (Fig. 3A). The apoptotic rate in cells
overexpressing miR-222 following IR was also examined by flow
cytometry (data not presented). miR-222 overexpression
significantly decreased radiation-induced apoptosis in C666-1 cells
at 0 Gy and 4 Gy (Fig. 3B),
suggesting that miR-222 may increase radioresistance in NPC
cells.
PTEN is a direct target of miR-222 in
NPC cells
PTEN has been previously identified as a direct
target of miR-222 in several types of cancer (17,18).
However, the link between miR-222 and PTEN in NPC remains unclear.
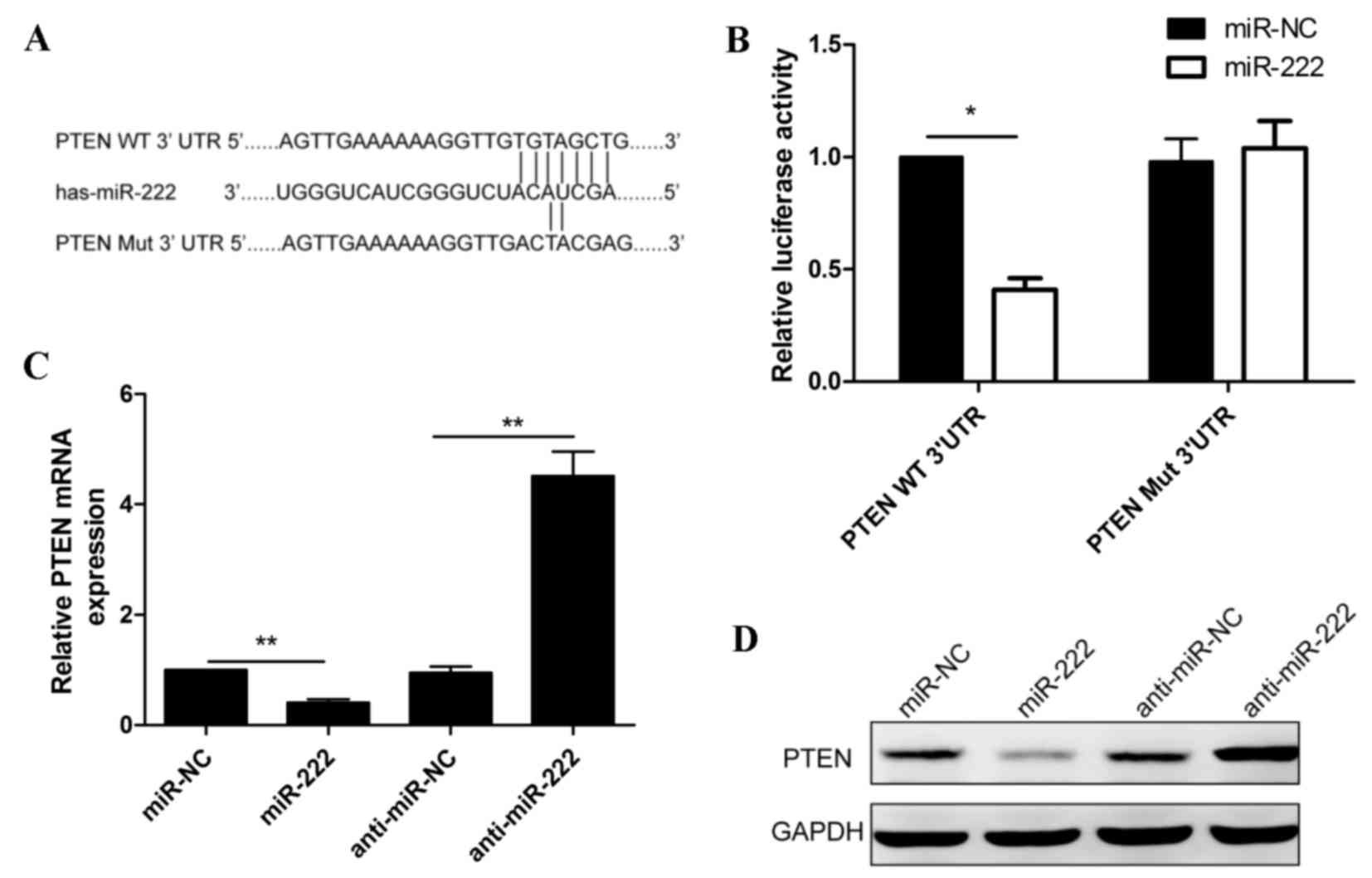
To verify if PTEN is a direct target of miR-222 in NPC, a human
PTEN 3′UTR fragment containing the binding sites or mutated binding
sites of miR-222 (Fig. 4A) was
fused to a luciferase reporter vector and co-transfected with
miR-222 mimic or miR-NC into C666-1 cells. The luciferase reporter
assay was subsequently performed. As presented in Fig. 4B, the relative luciferase activity
was reduced following co-transfection with miR-222 mimic and
Wt-PTEN-3′UTR, compared with co-transfection with miR-222 mimic and
Mut-PTEN-3′UTR. To determine whether miR-222 expression affected
endogenous PTEN mRNA and protein expression, miR-222 mimic, miR-NC,
anti-miR-222 and anti-miR-NC were transfected into C666-1 cells for
48 h and were subsequently analyzed with RT-qPCR and western
blotting. The results revealed that miR-222 overexpression
inhibited PTEN mRNA and protein expression in C666-1 cells, whereas
reduced miR-222 expression increased PTEN mRNA and protein
expression in C666-1 cells (Fig. 4C
and D), suggesting that PTEN is a target of miR-222 in NPC
cells.
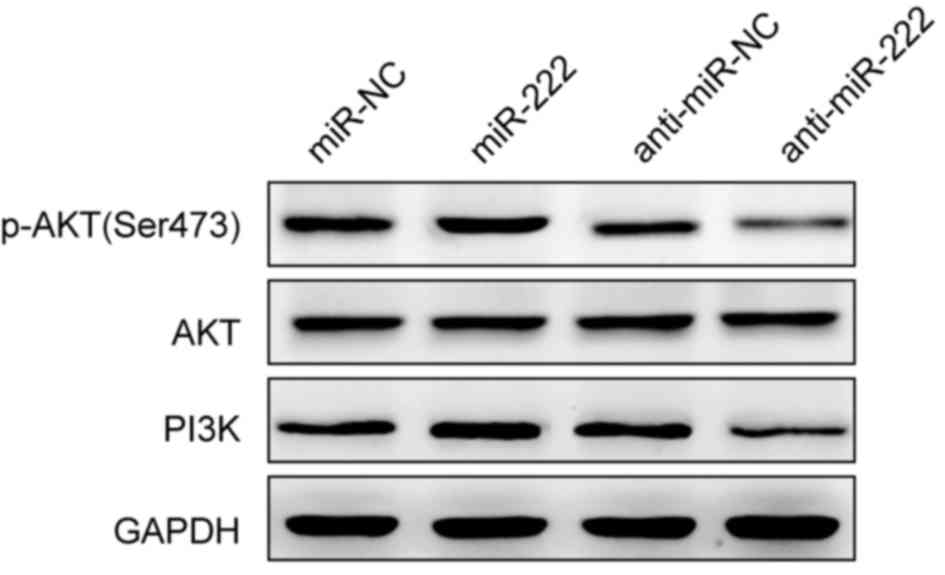
miR-222 regulates the PI3K/AKT
signaling pathway in NPC cells
PTEN has been reported to act as a negative
regulator of the PI3K/AKT pathway, via the dephosphorylation of
phosphatidylinositol (3,4,5)-triphosphate (20). To investigate if miR-222 could
regulate the PI3K/AKT pathway, PI3K, AKT and p-AKT protein levels
were detected by western blot in miR-222 mimic or anti-miR-222
transfected C666-1 cells. PI3K and p-AKT protein expression levels
were increased in miR-222 mimic transfected cells, and decreased in
anti-miR-222-transfected cells compared with the negative controls
(Fig. 5). AKT protein expression
was unchanged among the groups (Fig.
5). These findings suggest that the promotion of cell
proliferation, migration and invasion by miR-222 may be mediated
via the activation of the PI3K/AKT signaling pathway.
Discussion
Recent studies have demonstrated the involvement of
several miRNAs in NPC initiation and development, through the
regulation of target gene expression (11–13,21).
The present study revealed that miR-222 expression was increased in
clinical NPC tissues and cell lines, compared to adjacent normal
tissues and cell lines. miR-222 was demonstrated to be involved in
NPC progression through the regulation of proliferation, colony
formation and cell apoptosis. Furthermore, miR-222 was observed to
increase radioresistance in NPC cells. PTEN was verified as a
direct, functional target of miR-222 in NPC cells. These findings
contribute to the understanding of NPC development and
radioresistance, and suggest miR-222 as a potential target in NPC
therapy.
As an oncomir, miR-222 has been widely reported to
be upregulated in numerous types of human cancer, including
non-small cell lung cancer (22),
bladder cancer (23), breast
cancer (24), glioblastoma
(25) and hepatocellular carcinoma
(26). Accumulating evidence
suggests that miR-222 contributes to tumor development,
progression, metastasis, and may be an effective biomarker or
therapy target (15–18,23–26).
Previous studies have demonstrated that miR-222 is involved in the
radioresistance of glioblastoma and gastric cancer cells (17,18).
However, the role of miR-222 in NPC remains elusive. In the present
study, miR-222 expression was upregulated in NPC tissues and cell
lines, and miR-222 overexpression promoted tumor growth and the
radioresistance of NPC cells, suggesting that miR-222 functions as
an oncomir in NPC cells.
Located on chromosome 10q23.3, PTEN is downregulated
in numerous types of human cancer and functions as a tumor
suppressor (27). Recent studies
have revealed that PTEN expression is downregulated in NPC tissues,
and that normal PTEN expression may suppress NPC development
(28,29). It has been reported that PTEN can
regulate radioresistance in cancer cells (17,18)
and the PTEN/PI3K/AKT pathway (30). PTEN has previously been identified
as a direct target gene of miR-222 in glioblastoma cells and
gastric cancer cells (17,18). Consistent with these results, the
present study identified PTEN as a potential target of miR-222 in
NPC cells. miR-222 was also demonstrated to regulate the PI3K/AKT
pathway. These results suggest that miR-222 can promote tumor
growth and confer radioresistance in NPC cells by directly
targeting PTEN and thus indirectly regulating the PI3K/AKT
signaling pathway.
In conclusion, the present study provides evidence
that miR-222 expression is upregulated in NPC tissues and cell
lines, and that miR-222 may promote cell proliferation and colony
formation, decrease cell apoptosis and confer radioresistance in
NPC cells. Furthermore, PTEN was identified as a crucial target
gene of miR-222. It was demonstrated that mir-222 inhibited PTEN
expression and positively regulated the PI3K/AKT signaling pathway,
suggesting that miR-222 functions as an oncomir in NPC and may be a
potential therapeutic target in NPC.
Acknowledgements
The present study was supported by the Project of
the Health and Family Planning Commission (2014ZC030).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lai SZ, Li WF, Chen L, Luo W, Chen YY, Liu
LZ, Sun Y, Lin AH, Liu MZ and Ma J: How does intensity-modulated
radiotherapy versus conventional two-dimensional radiotherapy
influence the treatment results in nasopharyngeal carcinoma
patients? Int J Radiat Oncol Biol Phys. 80:661–668. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xiao WW, Huang SM, Han F, Wu SX, Lu LX,
Lin CG, Deng XW, Lu TX, Cui NJ and Zhao C: Local control, survival,
and late toxicities of locally advanced nasopharyngeal carcinoma
treated by simultaneous modulated accelerated radiotherapy combined
with cisplatin concurrent chemotherapy: Long-term results of a
phase 2 study. Cancer. 117:1874–1883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brennecke J and Cohen SM: Towards a
complete description of the microRNA complement of animal genomes.
Genome Biol. 4:2282003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Carthew RW and Sontheimer EJ: Origins and
Mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kang M, Xiao J, Wang J, Zhou P, Wei T,
Zhao T and Wang R: MiR-24 enhances radiosensitivity in
nasopharyngeal carcinoma by targeting SP1. Cancer Med. 5:1163–1173.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lin T, Zhou F, Zhou H, Pan X, Sun Z and
Peng G: MicroRNA-378g enhanced radiosensitivity of NPC cells
partially by targeting protein tyrosine phosphatase SHP-1. Int J
Radiat Biol. 91:859–866. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao L, Tang M, Hu Z, Yan B, Pi W, Li Z,
Zhang J, Zhang L, Jiang W, Li G, et al: miR-504 mediated
down-regulation of nuclear respiratory factor 1 leads to
radio-resistance in nasopharyngeal carcinoma. Oncotarget.
6:15995–16018. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miller TE, Ghoshal K, Ramaswamy B, Roy S,
Datta J, Shapiro CL, Jacob S and Majumder S: MicroRNA-221/222
confers tamoxifen resistance in breast cancer by targeting p27Kip1.
J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rao X, Di Leva G, Li M, Fang F, Devlin C,
Hartman-Frey C, Burow ME, Ivan M, Croce CM and Nephew KP:
MicroRNA-221/222 confers breast cancer fulvestrant resistance by
regulating multiple signaling pathways. Oncogene. 30:1082–1097.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee JC, Zhao JT, Clifton-Bligh RJ, Gill A,
Gundara JS, Ip JC, Glover A, Sywak MS, Delbridge LW, Robinson BG
and Sidhu SB: MicroRNA-222 and microRNA-146b are tissue and
circulating biomarkers of recurrent papillary thyroid cancer.
Cancer. 119:4358–4365. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li W, Guo F, Wang P, Hong S and Zhang C:
miR-221/222 confers radioresistance in glioblastoma cells through
activating AKT independent of PTEN status. Curr Mol Med.
14:185–195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F,
Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z and Chun-Sheng
K: MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamada KM and Araki M: Tumor suppressor
PTEN: Modulator of cell signaling, growth, migration and apoptosis.
J Cell Sci. 114:2375–2382. 2001.PubMed/NCBI
|
|
21
|
Wang Y, Guo Z, Shu Y, Zhou H, Wang H and
Zhang W: BART miRNAs: An unimaginable force in the development of
nasopharyngeal carcinoma. Eur J Cancer Prev. 26:144–150. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yamashita R, Sato M, Kakumu T, Hase T,
Yogo N, Maruyama E, Sekido Y, Kondo M and Hasegawa Y: Growth
inhibitory effects of miR-221 and miR-222 in non-small cell lung
cancer cells. Cancer Med. 4:551–564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang DQ, Zhou CK, Jiang XW, Chen J and
Shi BK: Increased expression of miR-222 is associated with poor
prognosis in bladder cancer. World J Surg Oncol. 12:2412014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hwang MS, Yu N, Stinson SY, Yue P, Newman
RJ, Allan BB and Dornan D: miR-221/222 targets adiponectin receptor
1 to promote the epithelial-to-mesenchymal transition in breast
cancer. PLoS One. 8:e665022013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Quintavalle C, Garofalo M, Zanca C, Romano
G, Iaboni M, del Basso De Caro M, Martinez-Montero JC, Incoronato
M, Nuovo G, Croce CM and Condorelli G: miR-221/222 overexpession in
human glioblastoma increases invasiveness by targeting the protein
phosphate PTPu. Oncogene. 31:858–868. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong QW, Ching AK, Chan AW, Choy KW, To
KF, Lai PB and Wong N: MiR-222 overexpression confers cell
migratory advantages in hepatocellular carcinoma through enhancing
AKT signaling. Clin Cancer Res. 16:867–875. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang S and Yu D: PI(3)king apart PTEN's
role in cancer. Clin Cancer Res. 16:4325–4330. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cai LM, Lyu XM, Luo WR, Cui XF, Ye YF,
Yuan CC, Peng QX, Wu DH, Liu TF, Wang E, et al: EBV-miR-BART7-3p
promotes the EMT and metastasis of nasopharyngeal carcinoma cells
by suppressing the tumor suppressor PTEN. Oncogene. 34:2156–2166.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou XM, Sun R, Luo DH, Sun J, Zhang MY,
Wang MH, Yang Y, Wang HY and Mai SJ: Upregulated TRIM29 promotes
proliferation and metastasis of nasopharyngeal carcinoma via
PTEN/AKT/mTOR signal pathway. Oncotarget. 7:13634–13650. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hafsi S, Pezzino FM, Candido S, Ligresti
G, Spandidos DA, Soua Z, McCubrey JA, Travali S and Libra M: Gene
alterations in the PI3K/PTEN/AKT pathway as a mechanism of
drug-resistance (review). Int J Oncol. 40:639–644. 2012.PubMed/NCBI
|