Introduction
Perthes' disease is a self-limiting disease of
children, and causes interrupted blood supply to the capital
femoral epiphysis (1). As one of
the major outcomes of the deficient blood supply associated with
Perthes' disease, a large proportion of the patients will end up
with steroid-induced avascular necrosis of femoral head (SANFH),
which is characterized by degeneration of the femoral head and
death of the dynamics component of bone (2). Since the 1960s, scientists have been
attempting to reveal the pathogenesis of SANFH and have proposed
multiple theories. Mechanisms including fat coagulation, increased
intraosseous pressure and osteoporosis have been investigated for
their potential to identify the pathogenesis of SANFH (3,4).
However, even with the investigation of SANFH, there are a number
of questions that need to be answered concerning the pathological
alterations associated with osteocytes and osteoblasts in SANFH. In
previous years, several studies have inferred the strong
association of osteoblastogenesis with SANFH (5,6). A
comprehensive understanding of the pattern of the death of
osteocytes and osteoblasts, and factors involved in the process,
will provide a novel insight into the pathogenesis of SANFH.
Several studies have already been performed to validate the key
role of the fate of osteoblasts in determining the onset of SANFH
(7,8). It is also been demonstrated that a
deficiency in the number and function of osteoblasts results in
impaired osteogenic capacity, and therapies based on osteoblast
grafts can alleviate the symptom of femoral head necrosis (9). Therefore, identification of
biomarkers predicating or modulating the proliferation of
osteoblasts will open novel therapeutic avenues for SANFH.
MicroRNAs (miRs) have important roles in determining
the growth, differentiation and function of bone cells (10). By binding to 3′ untranslated
regions (3′UTRs) of target mRNAs, miR family members can repress
the function of the targeted genes. More than 600 miR members have
been proved to have an effect on bone and cartilaginous tissues,
influencing the fate of osteoblasts and osteoclasts as well as
chondrocytes and other mesenchymal cell types (11–15).
Inose et al (16)
demonstrated that the level of miR-206 is markedly downregulated
during differentiation of C2C12 cells into osteoblasts, while
overexpression of miR impairs bone formation by targeting the gap
junction a-1 protein (9,16). In addition, the association between
the effect of miR-206 on osteogenic differentiation with the onset
of SANFH is further explained by Liu et al (17), by connecting the function of the
miR to the Wnt/β-catenin signaling pathway. Results of the two
studies confirmed the crucial role of miR-206 in determining the
differentiation potential of osteoblasts. Therefore, it is
reasonable to investigate the signaling pathways involved in the
function of miR-206 in osteoblasts.
Although hundreds of targets of miR-206 have been
predicated by bioinformatics analysis, the present study focused on
the expression of programmed cell death 4 (PDCD4) in osteoblasts.
Expression of the PDCD4 gene influences the activity of the
transcription factor AP-1 directly (18,19),
and diminished PDCD4 level allows initiation of the
osteoclastogenic program by releasing proto oncogene c-Fos from
inhibition (10). Given the
function of miR-206 and PDCD4 in bone metabolic processes, a clear
explanation of the interaction between the two factors will further
highlight the process underlying the pathological alterations of
SANFH. In the present study, the expression status of miR-206 and
PDCD4 were first investigated with clinical SANFH samples.
Subsequently, the expression levels of miR-206 and PDCD4 were
modulated to assess their exact roles in the proliferation and
apoptosis of osteoblasts. The results of the present study
suggested that miR-206 promoted the onset of SANFH, by inducing
osteoblast apoptosis via inhibition of PDCD4.
Materials and methods
Chemicals and agents
Antibodies against PDCD4 (cat. no. ab45263),
alkaline phosphatase (ALP; cat. no. ab83259), Bcl-2-associated X
protein (Bax; cat. no. ab32503) and B-cell lymphoma 2 (Bcl-2; cat.
no. ab32124) were purchased from Abcam (Cambridge, UK). The
antibody against GAPDH (KC-5G5) was purchased from Zhejiang
Kangchen Biotech Co., Ltd. (Wuhan, China). The secondary goat
anti-rabbit (cat. no. BA1054) immunoglobulin (Ig)G-horseradish
peroxidase-conjugated antibody was purchased from Boster Biological
Technology (Pleasanton, CA, USA). A mimic
(5′-UGGAAUGUAAGGAAGUGUGUGG-3′) and inhibitor
(5′-CCACACACUUCCUUACAUUCCA-3′) for miR-206 was obtained from Chang
Jing Bio-Tech, Ltd. (Changsha, China). A normal control (NC) mimic
(5′-UUCUCCGAACGUGUCACGUT-3′) was purchased from Chang Jing
Bio-Tech, Ltd. The Annexin V/propidium iodide (PI) apoptosis kit
(cat. no. CCS012) was purchased from MultiSciences Biotech Co.,
Ltd. (Susteren, The Netherlands). Lipofectamine™ 2000
(cat. no. 52887) reagent was obtained from Invitrogen; Thermo
Fisher Scientific, Inc. (Waltham, MA, USA). The
Cell-Light™ 5-ethynyl-2′-deoxyuridine (EdU)
Apollo®488/567 in vitro Imaging kit was purchased
from Guangzhou RiboBio Co., Ltd. (Guangzhou, China; cat. no.
C10327). RNAiso Plus (cat. no. 9109) was from Takara Bio, Inc.
(Otsu, Japan). The Reverse Transcription (RT) kit and quantitative
polymerase chain reaction (qPCR) agents were purchased from
DBI®Bioscience (www.xinghanbio.com/). A protein Concentration
Determination kit (cat. no. 23227) was purchased from Thermo Fisher
Scientific, Inc. A Dual Luciferase Assay kit (cat. no. E1980) was
purchased from Promega Corporation (Madison, WI, USA).
Cell cultures
Human osteoblast lineage hFOB1.19 and 293T cells
were purchased from the American Type Culture Collection (Manassas,
VA, USA) and maintained in media consisting of 45% Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.),
45% F12 medium (Gibco; Thermo Fisher Scientific, Inc.), 10% fetal
bovine serum and 1% mixed antibiotics (v/v)
(penicillin/streptomycin) (R&D Systems, Inc., Minneapolis, MN,
USA) at 37°C in an atmosphere of 5% CO2 and 95% air.
Cells from passage three to five were used for assays in the
present study.
Specimen collection
SANFH specimens were collected from 15 SANFH
patients from February 2015 to March 2017 in the Department of
Orthopedic Pathology in Baoan People's Hospital of the Southern
Medical University (Shenzhen, China). A total of 15 bone specimens
from patients undergoing total hip replacement for femoral neck
fracture were collected as controls. All the cases involved in the
present study had detailed information on the clinicopathological
and prognostic characteristics. The present study was approved by
Baoan People's Hospital of the Southern Medical University Ethics
Committee. The Ethics Committee approved the associated screening,
inspection and data collection of the patients, and all subjects
signed a written informed consent form. All experiments were
undertaken following the provisions of the Declaration of
Helsinki.
Plasmid construction and
transfection
Specific small interfering (si)RNA (50 nM) targeting
PDCD4 (5′-UCAGUAUCUGCUCAUUUUCUATT-3′) and negative control
(NC) siRNA (50 nM) (5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased
from Chang Jing Bio-Tech, Ltd. For the dual luciferase assay, wild
type and mutant sequences of PDCD4 3′UTR were synthesized by
Sangon Biotech Co., Ltd. (Shanghai, China) and ligated into the
psiCHECK-2 plasmid (Promega Corporation, Madison, WI, USA).
Transfection was performed using Lipofectamine™ 2000
according to the manufacturer's protocol. Twenty-four hours after
transfection, cells were collected and subjected to subsequent
experiments.
RNA extraction and RT-qPCR
Total RNA from SANFH specimens or hFOB1.19 cells
(normal, NC siRNA transfected, and PDCD4 siRNA transfected)
was extracted using an RNA extraction kit according to the
manufacturers' protocol. cDNA templates were obtained via RT using
RNA according to the manufacturer's protocol. The final reaction
mixture of volume 20 µl contained 10 µl Bestar®
SYBR-Green qPCR Master Mix and 0.5 µl each primer. PDCD4 forward,
5′-ACAGGTGTATGATGTGGAGGA-3′ and reverse,
5′-TTCTCAAATGCCCTTTCATCCAA-3′; miR-206 forward,
5′-ACACTCCAGCTGGGTGGAATGTAAGGAAGTGT-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; ALP forward, 5′-ACTGGGGCCTGAGATACCC-3′
and reverse, 5′-TCGTGTTGCACTGGTTAAAGC-3′. GAPDH was used as an
internal reference, the primers were as follows: Forward,
5′-ATCGTGCGTGACATTAAGGAGAAG-3′ and reverse,
5′-AGGAAGGAAGGCTGGAAGAGTG-3′. U6 was used as an internal reference,
the primers were as follows: Forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′. A total of 1 µl cDNA template
and 8 µl RNase free H2O reaction volume was used. The
thermocycling conditions for the amplification were as follows: A
denaturation step at 94°C for 2 min, followed by 40 cycles at 94°C
for 20 sec, 58°C for 20 sec and 72°C for 30 sec, and the reaction
was stopped by a step at 25°C for 5 min. The melting curve was
analyzed between 62°C and 95°C, and the relative expression levels
of the target genes were calculated by a Real-Time PCR Detection
system (Mx3000P; Agilent Technologies, Inc., Santa Clara, CA, USA)
using the formula of 2−ΔΔCq (20).
Western blot assay
Total proteins were extracted using the Total
Protein Extraction kit according to the manufacturer's protocol.
The concentrations of the protein samples were determined according
to the Bicinchoninic Acid assay. 30 µg protein from different
samples were subjected to 10% sodium dodecyl sulfate polyacrylamide
gel electrophoresis at 80 V for 2.5 h. The proteins were
transferred onto polyvinylidene difluoride membranes and rinsed
with Tris base buffer solution with Tween-20 (TBST). Following
this, membranes were blocked with skimmed milk solution for 1 h at
room temperature. Subsequently, membranes were incubated with one
of the primary antibodies against PDCD4 (1:2,000) for 50 min,
anti-Bax (1:1,000) for 25 min, anti-ALP (1:1,000) for 40 min,
anti-Bcl-2 (1:1,000) for 25 min, and anti-GAPDH (internal
reference; 1:1,000) for 30 min at 300 mA at room temperature and
then incubated with peroxidase-conjugated secondary antibodies
(1:20,000) for 40 min at room temperature. Blots were developed
using the Beyo ECL Plus reagent (Beyotime Institute of
Biotechnology, Haimen, China) and analyzed with the Gel Imaging
system (WD-9413B; Beijing Liuyi Biotechnology Co., Ltd., Beijing,
China) and the relative expression levels of different proteins
were calculated with Bio-Rad Quantity One software version 4.6.3
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Dual luciferase assay
The modulation effect of miR-206 on PDCD4 was
determined with dual luciferase assay in 293T cells
(2×105/ml): luciferase activity was detected by Dual
Luciferase Assay kit according to the manufacturers' instruction 48
h following transfection of different combinations of plasmid and
mimic. psi-CHECK2-PDCD4 and miR-206 mimic co-transfection was
performed with the with Lipofectamine 2000. The fluorescence
intensity was detected using a Microplate Reader (GloMax; Promega
Corporation) and normalized in comparison with Renilla
luciferase.
Cell viability and proliferation
detection
Exponentially growing hFOB1.19 cells
(5×105 cells/ml) were seeded into a 96-well plate. Then
cells were incubated for 72 h (each treatment was administered nine
times) and every 24 h, 10 µl/ml CCK-8 (Beyotime Insitute of
Biotechnology) solution was added to selected wells and incubated
at 37°C for 60 min. Optical density values at 450 nm were measured
with a microplate reader (GloMax; Promega Corporation) and used to
give a representative assessment of cell viability. hFOB1.19 cells
were harvested following a 48-h culture and assessed using
5-ethynyl-2′-deoxyuridine (EdU) assay according to the
manufacturer's protocol and the result was detected under a
fluorescent microscope, magnification, 200x.
Immunofluorescence detection
Immunofluorescence detection was conducted as
described in a previous study (21). Briefly, paraformaldehyde-fixed
cells were permeabilized with 0.1% Triton X-100 for 30 min.
Following blocking in 10% goat serum (Thermo Fisher Scientific,
Inc.) for 15 min, primary rabbit polyclonal antibodies against
different proteins of interest (PDCD4) (1:500) were then added, and
the cells were incubated overnight at 4°C. Subsequently, cells were
incubated with Cy3-labeled secondary antibody (1:1,000) (cat. no.
ab6939; Abcam) for 1 h in the dark. Thereafter, the cells were
stained with 4,6-diamino-2-phenyl indole for 5 min at room
temperature. The results were imaged with a fluorescent microscope
(BX53; Olympus Corporation, Tokyo, Japan) at magnification,
×40.
Hoechst 33258 staining
The morphological alterations of the cellular nuclei
due to apoptosis were detected using a Hoechst staining kit (20 µM)
according to the manufacturer's protocol. The results were observed
under a fluorescence microscope under 460 nm; magnification,
×120.
Flow cytometry
Apoptotic process was detected using an Annexin V/PI
apoptosis kit according to the manufacturer's protocol. The
apoptotic rates were analyzed using a FACScan flow cytometer
(Accuri C6; BD Biosciences). The total apoptotic rate was equal to
the sum of the late apoptotic rate (UR, upper right
quadrant-advanced stage apoptosis) and the early apoptotic rate
(LR, lower right quadrant-prophase apoptosis).
Statistical analysis
All the data were expressed as the mean ± the
standard deviation (n=3). The association between expression of
miR-206 and PDCD4 at mRNA level was analyzed with Pearson
correlation analysis. The difference between groups was analyzed
using Student's t-test or one-way analysis of variance followed by
the Least Significant Difference. All the statistical analyses were
conducted using SPSS version 19.0 (IBM Corp., Armonk, NY, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
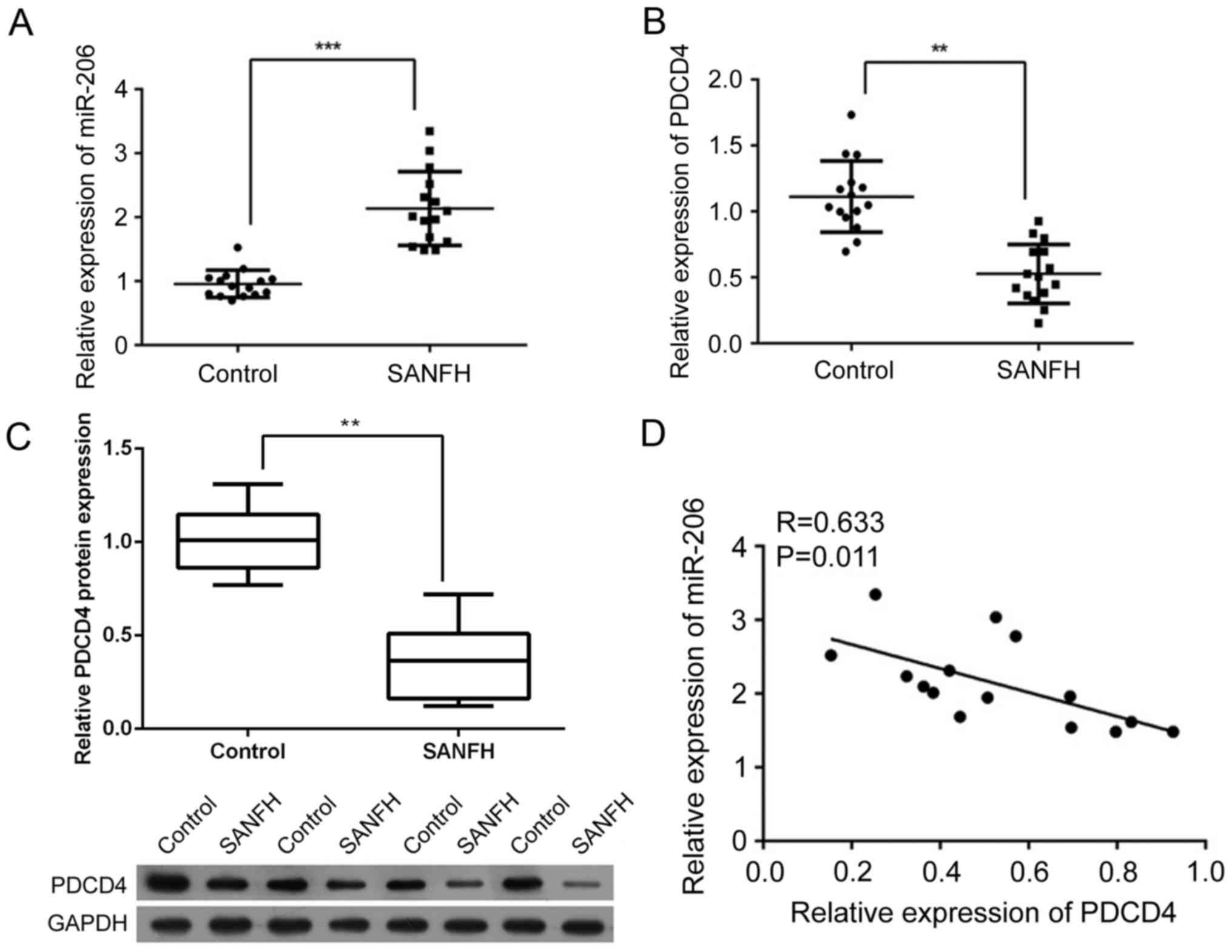
Expression level of miR-206 increases
while expression of PDCD4 is suppressed in SANFH samples
A total of 15 SANFH samples were collected to
investigate the association between the expression of miR-206 and
PDCD4. As presented in Fig. 1A,
the expression of miR-206 was significantly increased in SANFH
samples compared with the control samples (P<0.001). However,
the expression of PDCD4 was significantly inhibited at the mRNA and
protein level (P<0.01; Fig. 1B and
C, respectively). In addition, it was demonstrated that miR-206
was negatively correlated with PDCD4 at the mRNA level (R=0.633,
P<0.01; Fig. 1D), indicating a
potential interaction between the two factors.
miR-206 directly binds to the promoter
of the PDCD4 gene and inhibits the transcription of PDCD4
The interaction between miR-206 and PDCD4 was
analyzed with a dual luciferase assay. As presented in Fig. 2, the co-transfection of miR-206 and
the wild type promoter sequence of PDCD4 significantly decreased
the relative luciferase activity in 293T cells (P<0.05). Given
that miR-206 and the mutant promoter sequence of PDCD4 had no
effect on relative luciferase activity, the results of the dual
luciferase assay indicated the that miR-206 bound directly and
specifically to the promoter sequence of the PDCD4 gene, which
exerted a negative effect on the expression of PDCD4 at the
transcriptional level.
miR-206 contributes to suppressing
cell viability and proliferation, and inducing apoptosis in
hFOB1.19 cells
The function of miR-206 in osteoblastogenesis was
assessed by altering the expression of miR-206 in a bilateral
pattern. Both a specific mimic and inhibitor of miR-206 were stably
introduced into the human osteoblast lineage hFOB1.19. Based on the
results of the CCK-8 assay, the cell viability of miR-206
overexpressing cells was significantly decreased, while the
viability of the cells treated with an miR-206 inhibitor
significantly increased compared with the control (P<0.05;
Fig. 3A). Consistent with the
results of the CCK-8 assay, a greater number of EdU positive cells
(stained red) were detected in the cells treated with the miR-206
inhibitor while fewer was detected with miR-206 overexpressing
cells (Fig. 3B). The cells
overexpressing-miR-206 demonstrated a significantly increased level
of apoptosis as illustrated by Hoechst33258 staining and flow
cytometry: A larger proportion of Hoechst33258 positive cells
(bright blue), and significantly increased apoptotic rate were
detected (P<0.01; Fig. 3C and
D). However, inhibition of miR-206 in hFOB1.19 cells had no
influence on the cell apoptotic process compared with the NC group
(Fig. 3C and D).
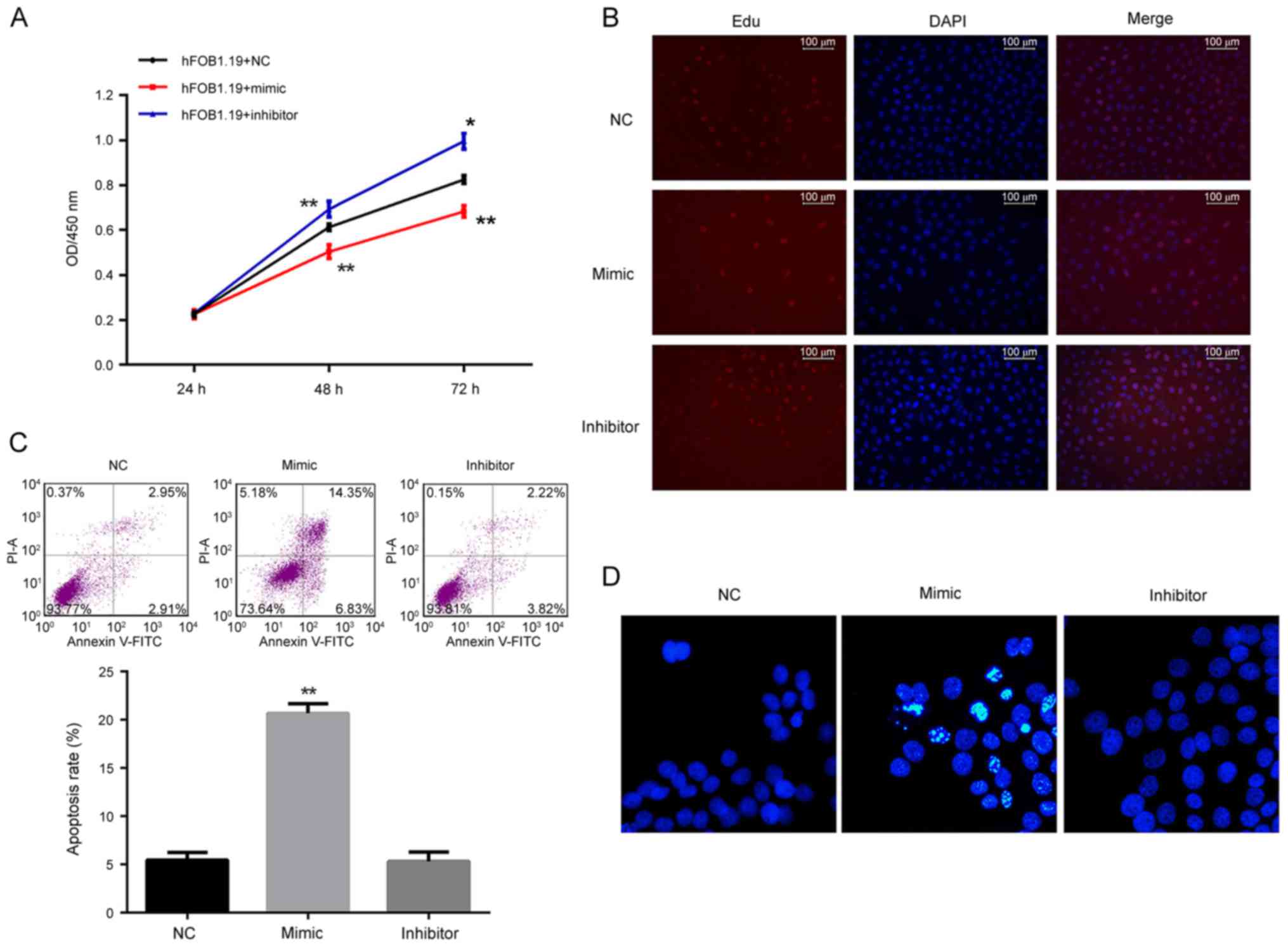 | Figure 3.Analysis of the effect of regulation
of miR-206 on cell viability, proliferation and apoptosis in
hFOB1.19 cells. (A) Cell viability as detected by the Cell Counting
kit-8 assay was decreased by the miR-206 mimic and increased by the
miR-206 inhibitor. (B) Cell proliferation as detected by EdU assay
demonstrated that proliferation of hFOB1.19 cells was inhibited by
the miR-206 mimic and induced by the miR-206 inhibitor.
Proliferating cells were stained red by EdU. (C) Cell apoptosis
detected by flow cytometry and (D) Hoechst staining demonstrated
that the miR-206 mimic induced apoptosis and while miR-206
inhibitor had no influence on apoptosis in hFOB1.19 cells.
*P<0.05 vs. NC. **P<0.01 vs. NC. miR, microRNA; EdU,
5-ethynyl-2′-deoxyuridine; NC, negative control; FITC, fluorescein
isothiocyanate; PI, propidium iodide; OD, optical density; DAPI,
DAPI, 4,6-diamino-2-phenylindole. |
miR-206 impairs the osteogenic process
by suppressing PDCD4-associated signaling
At the molecular level, transfection of an miR-206
mimic downregulated the expression of PCDCD4 (Fig. 4A-C). Additionally, ALP was also
suppressed by transfection with an miR-206 mimic (Fig. 4A and B), indicating the impaired
proliferation and differentiation potential of hFOB1.19 cells as a
result of miR-206 overexpression. The level of the pro-apoptotic
factor Bax was increased while the level of anti-apoptosis factor
Bcl-2 was suppressed in miR-206 overexpressing cells (Fig. 4B). For cells transfected with the
miR-206 inhibitor, the expression of Bax was decreased and the
expression of PCDCD4 and Bcl-2 was increased (Fig. 4B and C), representing an inhibited
apoptotic process in hFOB1.19 cells, which explained the augmented
cell viability by miR-206 knockdown.
 | Figure 4.Regulation of miR-206 influences the
expression of the different signaling molecules associated with
differentiation and apoptosis in hFOB1.19 cells. (A) Relative
expression of miR-206, PDCD4 and ALP in hFOB1.19 cells transfected
with an miR-206 inhibitor, mimic or NC. (B) Western blotting
demonstrating the expression of PDCD4, ALP, Bcl-2, Bax and GAPDH in
hFOB1.19 cells transfected with an miR-206 inhibitor, mimic or NC.
(C) Transfection of miR-206 mimic suppressed the expression of
PDCD4. (D) Transfection of miR-206 inhibitor induced the expression
of PDCD4. *P<0.05, **P<0.01 vs. NC. ##P<0.01 vs. mimic.
PDCD4, programmed cell death 4; ALP, alkaline phosphatase; Bcl-2,
B-cell lymphoma 2; Bax, Bcl-2-X-associated protein; miR, microRNA;
NC, negative control; DAPI, 4,6-diamino-2-phenylindole. |
The involvement of PDCD4 in osteoblastogenesis had
been previously reported (22),
but the exact function of the factor on bone metabolism was
neglected. The effect of PDCD4 and miR-206 on the differentiation
potential, and apoptosis was investigated (Fig. 5). In the present study, the
expression of PDCD4 was inhibited in miR-206 knockdown hFOB1.19
cells, which resulted in significantly decreased expression of ALP
(P<0.001; Fig. 5A). It was
demonstrated that the knockdown of PDCD4 significantly induced
cellular apoptosis in a similar pattern to that of miR-206
overexpression (P<0.01; Fig. 5B and
D). In addition, knockdown of PDCD4 induced the expression of
Bax while the expression of ALP and Bcl-2 was suppressed (Fig. 5C), which relieved the hFOB1.19
cells of the effect of miR-206 inhibition. The above results
indicated that the pro-SANFH function of miR-206 depended on the
suppression of PDCD4 in osteoblasts.
 | Figure 5.Analysis of the effect of miR-206 on
the differentiation potential and apoptosis in hFOB1.19 cells
depended on the suppression of PDCD4 associated signaling. Analysis
of the relative expression of (A) PDCD4 and ALP in hFOB1.19 cells
transfected with the miR-206 inhibitor and an siRNA against PDCD4.
(B) Cell apoptosis and quantification, detected by flow cytometry
of cells transfected with miR-206 inhibitor and an siRNA against
PDCD4. (C) Western blotting and quantification demonstrating the
expression of PDCD4, ALP, Bcl-2 and Bax, in hFOB1.19 cells
transfected with miR-206 inhibitor and an siRNA against PDCD4. (D)
Hoechest staining of hFOB1.19 cells transfected with the miR-206
inhibitor and an siRNA against PDCD4. Data are expressed as the
mean ± standard deviation. *P<0.05, **P<0.01, ***P<0.001
vs. miR-206 inhibited hFOB1.19 cells. NC, negative control; PDCD4,
programmed cell death 4; ALP, alkaline phosphatase; Bcl-2, B-cell
lymphoma 2; Bax, Bcl-2-X-associated protein; miR, microRNA; si,
small interfering; FITC, fluorescein isothiocyanate; PI, propidium
iodide. |
Discussion
The incidence of SANFH has continued to increase due
to the excessive use of exogenous steroids in recent years
(17). Although numerous studies
have attempted to investigate the molecular alterations associated
with the disease and develop novel treatment or prevention
modalities, the onset and progression mechanism underlying SANFH
remain partially understood. Based on clinical statistics, the
prognosis of SANFH children is significantly better than that of
adults (23–25). Given that children possess a
stronger osteogenic ability than adults, the viability and
differentiation potential of osteoblasts is potentially serving a
key role in the onset of SANFH. Therefore, the identification of
biological markers indicating the proliferation/apoptosis of
osteoblasts is a reasonable target for analysis to promote
improvement in the treatment and prevention of SANFH. In the
present study, the results demonstrated that the apoptotic process
of human osteoblast lineage hFOB1.19 was governed by miR-206 in a
PDCD4-inhibition-dependent manner.
The integrity and function of the skeletal system is
maintained by a balance between osteoclasts and osteoblasts,
interruption of this during any stage of life will lead to bone
disease, such as osteonecrosis (26). Based on several previous studies,
the apoptosis of osteoblasts is a critical event in the onset of
SANFH as the process disrupts the balance between osteoclasts and
osteoblasts (7,8,27).
Multiple factors are reported to be associated with the viability
and survival of osteoblasts. Among these, miRs have emerged as
novel regulatory molecules closely associated with bone diseases
(17). A number of miRs exhibit a
tissue or developmental stage-specific expression pattern, and have
been depicted as signaling network nodes modulating osteoblastic
differentiation processes. For example, miR-21, miR-26a and miR-196
are miR members that serve central roles in the osteogenic
differentiation of mesenchymal stem cells (28–30).
Additionally, miR-29, miR-let-7 and miR-26 attenuate extracellular
matrix protein synthesis and mineralization in osteoblasts as well
as support the maturation of osteoblasts (10). In the present study, the expression
of miR-206 was first investigated in clinical SANFH specimens.
Consistent with a previous study (17), it was demonstrated that the
expression of miR-206 was associated with the onset of SANFH. A
previous study demonstrated that the overexpression of miR-206
inhibits the differentiation of osteoblasts via the Wnt/β-catenin
signaling pathway (17). With a
series of in vitro assays, the present study confirmed the
negative effect miR-206 on viability and differentiation potential
on osteoblasts. Induced expression of miR-206 not only increased
the apoptotic rate but also inhibited the cell viability of
hFOB1.19 cells. In addition, the expression of the osteoblastic
differentiation marker ALP was also inhibited by the miR-206 mimic.
To investigate the downstream signaling involved in the effect of
miR-206 on osteoblasts, the function of PDCD4 was also investigated
in the present study. PDCD4 is originally characterized as an
inhibitor of cellular transformation (31). In several cancer types, the
expression of PDCD4 is inhibited (32,33).
In the study of Sugatani et al (22) the authors confirm a novel molecular
mechanism regulating osteoclastogenesis: enforced expression of
miR-21 inhibited osteoclast development via down-regulation of
PDCD4 expression. The conclusion infers a pro-osteoblastogenic role
for PDCD4, which was validated by the results of the present study.
Similar to the effect of increased miR-206 expression, knockdown of
PDCD4 induced apoptosis and impaired the differentiation potential
of hFOB1.19 cells, which inhibited the pro-osteoblastogenesis
effect of miR-206 inhibition.
In conclusion, the results of the present study
supplement the knowledge about the function of miR-206 in the onset
and progression of SANFH. miR-206 induced the apoptosis and
suppressed proliferation and differentiation of osteoblasts in a
PDCD4-dependent manner. Modulation of miR-206 signaling will be an
innovative method for alleviating the deleterious effect of SANFH.
Our results support the potential of miR-206 to be used in
therapies for the treatment of SANFH but further studies need to be
conducted.
Acknowledgements
This work is supported by Science and Technology
Department of Guangdong Province (2014A020212041) and Shenzhen
Science and Technology Innovation Committee
(JCYJ20140411091151442).
References
|
1
|
Herring JA: Management of Perthes'
disease. J Pediatr Orthop. 16:1–2. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Beckmann R, Shaheen H, Kweider N, Ghassemi
A, Fragoulis A, Hermanns-Sachweh B, Pufe T, Kadyrov M and Drescher
W: Enoxaparin prevents steroid-related avascular necrosis of the
femoral head. ScientificWorldJournal. 2014:3478132014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nishida K, Yamamoto T, Motomura G,
Jingushi S and Iwamoto Y: Pitavastatin may reduce risk of
steroid-induced osteonecrosis in rabbits: A preliminary
histological study. Clin Orthop Relat Res. 466:1054–1058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takano-Murakami R, Tokunaga K, Kondo N,
Ito T, Kitahara H, Ito M and Endo N: Glucocorticoid inhibits bone
regeneration after osteonecrosis of the femoral head in aged female
rats. Tohoku J Exp Med. 217:51–58. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Samara S, Dailiana Z, Chassanidis C,
Koromila T, Papatheodorou L, Malizos KN and Kollia P: Expression
profile of osteoprotegerin, RANK and RANKL genes in the femoral
head of patients with avascular necrosis. Exp Mol Pathol. 96:9–14.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weinstein RS, Jilka RL, Parfitt AM and
Manolagas SC: Inhibition of osteoblastogenesis and promotion of
apoptosis of osteoblasts and osteocytes by glucocorticoids.
Potential mechanisms of their deleterious effects on bone. J Clin
Invest. 102:274–282. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kerachian MA, Séguin C and Harvey EJ:
Glucocorticoids in osteonecrosis of the femoral head: A new
understanding of the mechanisms of action. J Steroid Biochem Mol
Biol. 114:121–128. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yun SI, Yoon HY, Jeong SY and Chung YS:
Glucocorticoid induces apoptosis of osteoblast cells through the
activation of glycogen synthase kinase 3beta. J Bone Miner Metab.
27:140–148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim SJ, Bahk WJ, Chang CH, Jang JD and
Suhl KH: Treatment of osteonecrosis of the femoral head using
autologous cultured osteoblasts: A case report. J Med Case Rep.
2:582008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Wijnen AJ, van de Peppel J, van
Leeuwen JP, Lian JB, Stein GS, Westendorf JJ, Oursler MJ, Im HJ,
Taipaleenmäki H, Hesse E, et al: MicroRNA functions in osteogenesis
and dysfunctions in osteoporosis. Curr Osteoporos Rep. 11:72–82.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lian JB, Stein GS, van Wijnen AJ, Stein
JL, Hassan MQ, Gaur T and Zhang Y: MicroRNA control of bone
formation and homeostasis. Nat Rev Endocrinol. 8:212–227. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Miyaki S and Asahara H: Macro view of
microRNA function in osteoarthritis. Nat Rev Rheumatol. 8:543–552.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xia Z, Chen C, Chen P, Xie H and Luo X:
MicroRNAs and their roles in osteoclast differentiation. Front Med.
5:414–419. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goldring MB and Marcu KB: Epigenomic and
microRNA-mediated regulation in cartilage development, homeostasis,
and osteoarthritis. Trends Mol Med. 18:109–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Taipaleenmäki H, Hokland Bjerre L, Chen L,
Kauppinen S and Kassem M: Mechanisms in endocrinology: micro-RNAs:
Targets for enhancing osteoblast differentiation and bone
formation. Eur J Endocrinol. 166:359–371. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Inose H, Ochi H, Kimura A, Fujita K, Xu R,
Sato S, Iwasaki M, Sunamura S, Takeuchi Y, Fukumoto S, et al: A
MicroRNA regulatory mechanism of osteoblast differentiation. Proc
Natl Acad Sci USA. 106:20794–20799. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu G, Luo G, Bo Z, Liang X, Huang J and
Li D: Impaired osteogenic differentiation associated with
connexin43/microRNA-206 in steroid-induced avascular necrosis of
the femoral head. Exp Mol Pathol. 101:89–99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Krichevsky AM and Galina G: miR-21: A
small multi-faceted RNA. J Cell Mol Med. 13:39–53. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Loh PG, Yang HS, Walsh MA, Wang Q, Wang X,
Cheng Z, Liu D and Song H: Structural basis for translational
inhibition by the tumour suppressor Pdcd4. EMBO J. 28:274–285.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu H, Shi L, Qi G, Zhao S, Gao Y and Li Y:
Gypenoside protects cardiomyocytes against ischemia-reperfusion
injury via the inhibition of mitogen-activated protein kinase
mediated nuclear factor kappa B pathway in vitro and in vivo. Front
Pharmacol. 7:1482016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sugatani T, Vacher J and Hruska KA: A
microRNA expression signature of osteoclastogenesis. Blood.
117:3648–3657. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cheng JC, Lam TP and Ng BK: Prognosis and
prognostic factors of Legg-Calve-Perthes disease. J Pediatr Orthop.
31 2 Suppl:S147–S151. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ito H, Matsuno T and Kaneda K: Prognosis
of early stage avascular necrosis of the femoral head. Clin Orthop
Relat Res. 149–157. 1999.PubMed/NCBI
|
|
25
|
Ohzono K, Saito M, Takaoka K, Ono K, Saito
S, Nishina T and Kadowaki T: Natural history of nontraumatic
avascular necrosis of the femoral head. J Bone Joint Surg Br.
73:68–72. 1991.PubMed/NCBI
|
|
26
|
Hao C, Yang S, Xu W, Shen JK, Ye S, Liu X,
Dong Z, Xiao B and Feng Y: MiR-708 promotes steroid-induced
osteonecrosis of femoral head, suppresses osteogenic
differentiation by targeting SMAD3. Sci Rep. 6:225992016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bai R, Feng W, Liu WL, Zhao ZH, Zhao AQ,
Wang Y, Wang WX, Sun L, Wu LS and Cui SH: Roles of osteocyte
apoptosis in steroid-induced avascular necrosis of the femoral
head. Genet Mol Res. 15:2016. View Article : Google Scholar
|
|
28
|
Meng YB, Li X, Li ZY, Zhao J, Yuan XB, Ren
Y, Cui ZD, Liu YD and Yang XJ: microRNA-21 promotes osteogenic
differentiation of mesenchymal stem cells by the PI3K/β-catenin
pathway. J Orthop Res. 33:957–964. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Su X, Liao L, Shuai Y, Jing H, Liu S, Zhou
H, Liu Y and Jin Y: MiR-26a functions oppositely in osteogenic
differentiation of BMSCs and ADSCs depending on distinct activation
and roles of Wnt and BMP signaling pathway. Cell Death Dis.
6:e18512015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Candini O, Spano C, Murgia A, Grisendi G,
Veronesi E, Piccinno MS, Ferracin M, Negrini M, Giacobbi F, Bambi
F, et al: Mesenchymal progenitors aging highlights a miR-196 switch
targeting HOXB7 as master regulator of proliferation and
osteogenesis. Stem Cells. 33:939–950. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jansen AP: A novel transformation
suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB
or ODC transactivation. Oncogene. 20:669–676. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jansen AP, Camalier CE, Stark C and
Colburn NH: Characterization of programmed cell death 4 in multiple
human cancers reveals a novel enhancer of drug sensitivity. Mol
Cancer Ther. 3:103–110. 2004.PubMed/NCBI
|
|
33
|
Göke R, Barth P, Schmidt A, Samans B and
Lankat-Buttgereit B: Programmed cell death protein 4 suppresses
CDK1/cdc2 via induction of p21 (Waf1/Cip1). Am J Physiol Cell
Physiol. 287:C1541–C1546. 2004. View Article : Google Scholar : PubMed/NCBI
|