Introduction
Microgravity has a profound effect on the physiology
of humans, leading to increased susceptibility to many diseases
(1). Both spaceflight and
ground-based human and animal studies have reported many
physiological adaptations to microgravity in immune and endocrine
systems (2). The major endocrine
changes associated with exposure to hypogravity include disrupted
reproductive cycles. For example, microgravity simulated using a
hindlimb suspension (HLS) model in rats demonstrated lengthened
estrous cycles and prolonged diestrus due to hypoestrogenism
(3). It has in recent years been
established that the gut microbiota play a crucial role in the
maturation of the immune and endocrine systems. Mechanisms through
which microbiota mediate these functions include the cometabolism
of steroid hormones and low-molecular weight dietary compounds with
hormone-like activities that regulate the immune system (4). This microbial/hormonal interplay is
bidirectional, stress-induced neuroendocrine hormones have been
observed to increase bacterial adhesion to host tissues and alter
the growth and virulence of bacteria through the regulation
bacterial gene expression (4).
Intestinal microbiota and intestinal mucous barrier
both respond to simulated microgravity and contribute to the
susceptibility to inflammation in the gut microenvironment
(5). Simulated microgravity has
been reported to result in a transient increase in circulating LPS
and a stimulation of the innate immune system (6). Simulated microgravity disrupts
intestinal microflora and the innate immune system, leading to a
proinflammatory shift in the gut microenvironment and an increase
in colitis susceptibility (7).
Estrogen, the most prominent affected hormone under simulated
microgravity, regulates the permeability of the colonic mucosa
barrier (3,8). Estrogen gene knock-out mice have been
shown to present with many prepathogenic phenotypes, including
abnormal colonic histology and disrupted cellular tight junctions
(9). These architectural
abnormalities facilitate the invasion of intestinal bacteria,
resulting in localized infection and enhanced levels of colonic
inflammation (10). However, there
remains little knowledge regarding the effects of estrogen on the
composition of the gut microbiota and their downstream influences
on intestine barrier dysfunction.
Studies have found that the alteration of even a
single host gene can significantly change host-driven selective
pressures that lead to changes in the structure and function of the
commensal gastrointestinal microbiota (9,11,12).
We thus sought to characterize the possible impact of simulated
hypogravity on gut commensal microbiota and to evaluate the
protective effect of estrogen on the homeostasis of intestinal
microbiota in rats under simulated hypogravity. The predominant
fecal microbiota was analyzed using two universal primers targeting
the V3 regions of the 16S rRNA gene. Specific subpopulations were
quantified with genus- and group-specific primers to complement the
analysis and interpretation of the results obtained with the
universal primers. The study was helpful to more thoroughly
evaluate the potential role of hypogravity in the pathogenesis of
gastrointestinal diseases in humans.
Materials and methods
Animals and treatment
The animals were treated as previously described
(13). Briefly, Male Wistar rats
(Eight-week-old, approximately 250 g) were purchased from the
Experimental Animal Center, Academy of Military Medical Sciencs.
The mice were kept in a specific pathogen-free animal facility at
the State Key Laboratory of Space Medicine Fundamentals. All
experiments were performed in accordance with the ‘Guide for the
Care and Use of Laboratory Animals’ published by the US NIH
(National Institutes of Health Publication No. 85-23, revised 1996)
and were approved by the Committee on the Ethics of Animal
Experiments of the 306th Hospital of the PLA. The rats were caged
separately in a room maintained at 21°C and controlled light/dark
cycles (12/12 h). The rats were randomly assigned to four groups of
10 rats each as follows: A control group (without any treatment), a
simulated microgravity group (tail suspension for 8 weeks group)
and simulated microgravity combined with estrogen for 4 weeks and 8
weeks groups (estrogen was administered via the intramuscular
injection of estradiol benzoate at a dose of 80 µg/Kg daily for
first seven days, then rat were euthanized at the timepoints of 4
weeks and 8 weeks, respectively). To achieve simulated
microgravity, individual rats were subjected to hindlimbs
suspension. The technique of hindlimbs suspension was performed by
the use of a tail harness that partially elevated the hindlimbs
above the floor of the cage according to a previously published
method (14). Briefly, the tail
was firstly fixed with an adhesive tape. Then the tail was
suspended via a tether connecting to a horizontal tube at the top
of the cage. The upper end of the tether included a small pulley,
which allowed the rat rolled freely along the length of the tube.
The animals were maintained in 30 headdown tilt position with the
hindlimbs elevated 0.5 cm above the floor when fully extended. The
animals were euthanized under anesthesia.
Samples and DNA extraction
After the four-week or eight-week tail suspension,
the rats were anesthetized with 1% sodium pentobarbital (45 mg/kg).
Fecal pellets were collected in tubes and weighted. Samples were
quickly frozen in liquid nitrogen and stored at 80°C until nucleic
acid extraction was performed. Fecal bacteria DNA was extracted
using the modified CTAB method as previously described and stored
at 80°C (15). DNA integrity was
analyzed by loading 2 µl DNA on a 1% agarose gel stained with
ethidium bromide. The concentration and purity of the extracted DNA
were analyzed using ultraviolet absorption at 260/280 and 230/280
nm ratios.
PCR amplification and DGGE
analysis
PCR amplification and DGGE analysis were performed
as previously described (16).
Briefly, the variable V3 region of the 16S rRNA gene was amplified
using the universal bacterial primers F357-GC and R518 with the
sequences of 5′-CCGAATTCGTCGACAACAGAGTTTGATCCTGGCTCAG-3′ and
5′-CCCGGGATCCAAGCTTACGGCTACCTTGTTACGACTT-3′, respectively. The PCR
was performed as previously described. The amplified sequences of
16S rRNA were analyzed via DGGE fingerprinting analysis using 35 to
70% denaturing gel. Each lane uploaded 30 µl of PCR product, and
electrophoresis was performed at 70 V for 990 min. Next, the DGGE
gels were stained for 30 min with ethidium bromide (50 µg in 500 ml
1 X TAE buffer), and the band profiles were visualized and analyzed
with a Quantity One Analysis System (Bio-Rad, California, USA). In
short, all fingerprinting profiles were aligned using the reference
lanes and then compared. The numeric value of the relative
intensity per band class was then exported for all profiles and
further analyzed. The Berger-Parker index, which was used to assess
species richness, and α diversity measures, including the Shannon
and Simpson diversity indices, were calculated based on the band
profiles according to previously described method (17).
Cloning of the PCR-amplified products
and sequence analysis
PCR products were extracted from the band using an
Invitrogen PCR product purification kit (Invitrogen Life
Technologies, Shanghai, China) and cloned in E. coli JM109
using the pGEM-T vector system (15). Colonies of ampicillin-resistant
transformants were selected and transferred to Luria broth medium.
After incubation at 37°C overnight, one hundred microliters of
cultures were recovered and dissolved in 10 ml TE buffer. The
solutions were then boiled to lyse the cells. The cell lysates were
used as templates for PCR reactions using the pGEM-T-specific
primers Sp6 and T7 to verify the size of the DNA inserts (15). The plasmids with appropriately
sized inserts were sent for sequencing (Invitrogen Life
Technologies). Homology searches of the GenBank DNA database were
performed with the BLAST Search Tool. A phylogenetic tree was then
determined via MEGA4 software based on the neighbor-joining
algorithm and the Jukes-Cantor model to demonstrate the
evolutionary relationship (18).
Real-time PCR assay
Significant differences of intensity of the
bacterial species after DGGE analysis were confirmed using
real-time PCR. The abundance of specific intestinal bacterial
groups chosen from four representative phyla were measured via qPCR
using group-specific 16S rRNA gene primers. All primers used in
this study are listed in Table I.
The qPCR assay was performed as previously described (19). Briefly, the qPCR assay was
performed with a SYBR Premix Ex Taq (Takara Bio, Inc., Otsu, Japan)
on an Applied Biosystems 7500 fast real-time PCR system (Applied
Biosystems, Ghent, Belgium). For each primer set, a constructed
plasmid was chosen to create a 10-log fold standard curve to
directly quantify all samples. Each qPCR contained 10 µl SYBR
Premix Ex Taq, 0.4 µl of a 10 µmol/l F/R primer mix, and 1 µl of
the respective template DNA. Amplifications were performed under
the following temperature profiles: One cycle at 95°C for 3 min, 40
cycles of denaturation at 95°C for 30 sec, annealing for 40 sec,
and extension for 30 sec. Fluorescence was measured after the
extension phase of each cycle at an appropriate temperature for 10
sec to avoid interference from primer dimers, secondary structure,
or spurious priming. A final extension step was sustained for 5
min.
 | Table I.Bacterial group-specific primers used
in reverse transcription-polymerase chain reaction for microbiota
studies. |
Table I.
Bacterial group-specific primers used
in reverse transcription-polymerase chain reaction for microbiota
studies.
| Group | Primer sequences
(5′-3′) |
|---|
| Eubacteria |
F-ACTCCTACGGGAGGCAGCAGT |
| (all bacteria) |
R-ATTACCGCGGCTGCTGGC |
|
Escherichia |
F-GACCTCGGTTTAGTTCACAGA |
| coli |
R-CACACGCTGACGCTGACCA |
|
Bifidobacterium |
F-GCCGTATCTCTACGACCGTCG |
| longum |
R-TATCGGGGAGCAAGCGAGAG |
|
Bacteroides |
F-ATAGCCTTTCGAAAGRAAGAT |
|
fragilis |
R-CCAGTATCAACTGCAATTTTA |
|
Fusobacterium |
F-GGATTTATTGGGCGTAAAGC |
|
nucleatum |
R-GGCATTCCTACAAATATCTACGAA |
Lipopolysaccharide (LPS) and LPS
binding protein (LBP) detection
Blood was collected via cardiac puncture. LPS and
LBP detection in the serum were based on a previously performed
method (20). Circulating
endotoxin (LPS) was analyzed using a Limulus Amoebocyte Lysate
(LAL) assay QCL-1000 (Lonza AG, Valais Switzerland) in duplicate in
96-well plates according to the manufacturer's instructions;
lipopolysaccharide-binding protein (LBP) levels were detected using
the LBP Elisa kit (Abnova, Taipei, Taiwan). The lower limit of
detection for each assay is 0.1 EU/ml for LPS and 5 ng/ml for
LBP.
Statistical analysis
Differences between the treatment and control groups
were compared regarding band intensity and bacterial count via
RT-PCR between the different groups for significance testing with
an ANOVA program. Statistically significant differences between 2
groups were evaluated using a 2-tailed unpaired Student's t test.
All values are expressed as the means ± SDs of replicates.
Two-sided P-values <0.05 were considered to indicate a
statistically significant difference. The statistical analysis of
the results was performed via SigmaPlot software (Systat Software
Inc., San Jose, California, USA).
Results
PCR-DGGE analysis of changes in fecal
bacterial populations associated with simulated microgravity and
estrogen exposure
DGGE bands from the PCR products of the V3 regions
of 16S rRNA genes from rat fecal samples of control, simulated
microgravity or simulated microgravity combined with estrogen
administration groups are shown in Fig. 1A. Most bands appeared to be
unaffected by simulated microgravity exposure or estrogen
administration. To obtain an objective evaluation of the DGGE
profiles of the various groups, the electrophoretic bands underwent
numerical analysis based on band density. The beta diversity
measures included Shannon and Simpson indices, which were computed
to compare the diversity of the dominant bacterial microbiota in
the rat feces of the control and treatment groups. The
Berger-Parker index, which reflects species richness, was also
calculated to compare the discrepancy between groups. A statistical
analysis showed that the diversity indices did not significantly
differ between the groups (Fig.
1B). However, species richness was slightly lower after
exposure to simulated microgravity.
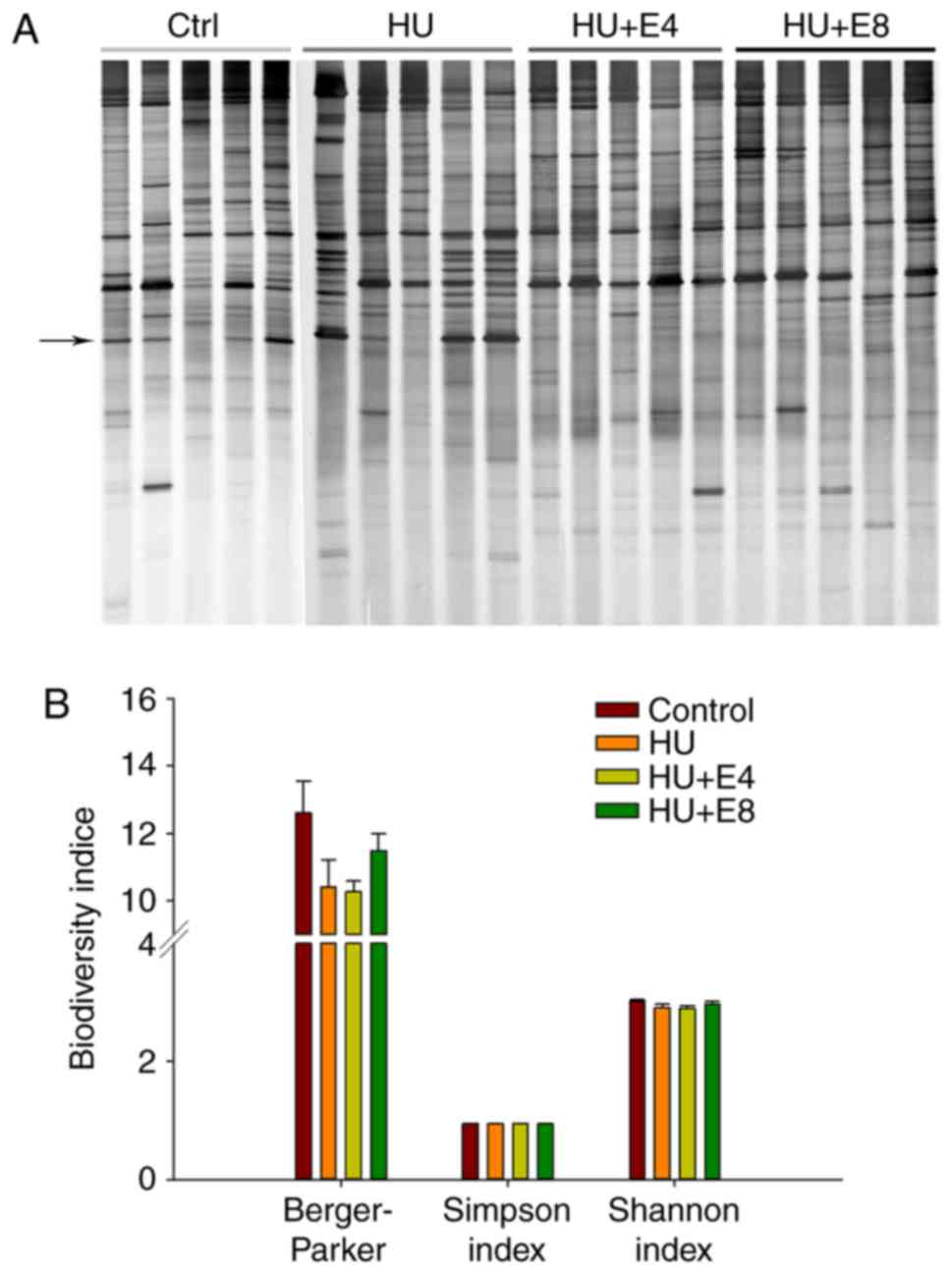 | Figure 1.DGGE profiles showed microbial
diversity in the rat feces of different groups. (A) The figure
shows DGGE gels of the V3 hypervariable 16S rDNA region,
demonstrating the microbiota's composition in the feces of rats
from control (Ctrl), simulated microgravity (HU), simulated
microgravity combined with estrogen for 4 weeks (HU+E4) and
simulated microgravity combined with estrogen for 8 weeks (HU+E8).
The arrow indicates the most significantly changed band. Number
indicated the bands that were cloned and sequenced. (B) Comparison
of biodiversity indices of feces microbiota between the control and
treatment groups. There were no significant difference between
groups (P-value were 0.09, 0.25, 0.17 for Berger-parker, Simpson
index, Shannon index, respectively). Bars indicate the standard
deviation of five samples in the same group. DGGE, denaturing
gradient gel electrophoresis; HU, hindlimb unweighting. |
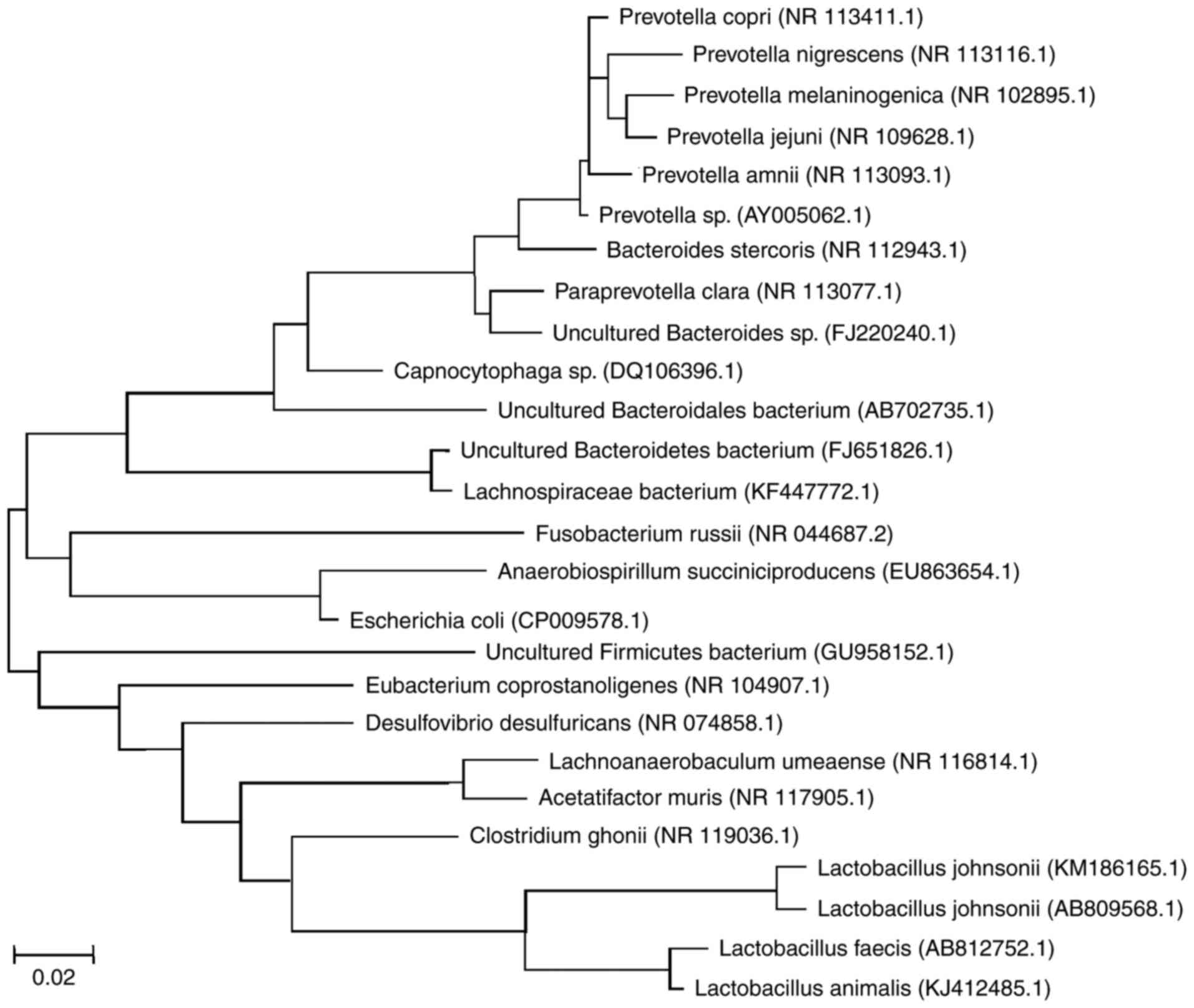
Sequencing of DGGE bands and
predominant gut microflora change
To further identify the predominant populations, 36
bands of the 16S rRNA genes from rat feces samples were extracted
from the gel, and three independent clones were constructed and
subjected to DNA sequencing for each band. The sequences of these
clones were compared with the NCBI database, and representative
aligned bacteria species were demonstrated in the brief
phylogenetic tree as shown in Fig.
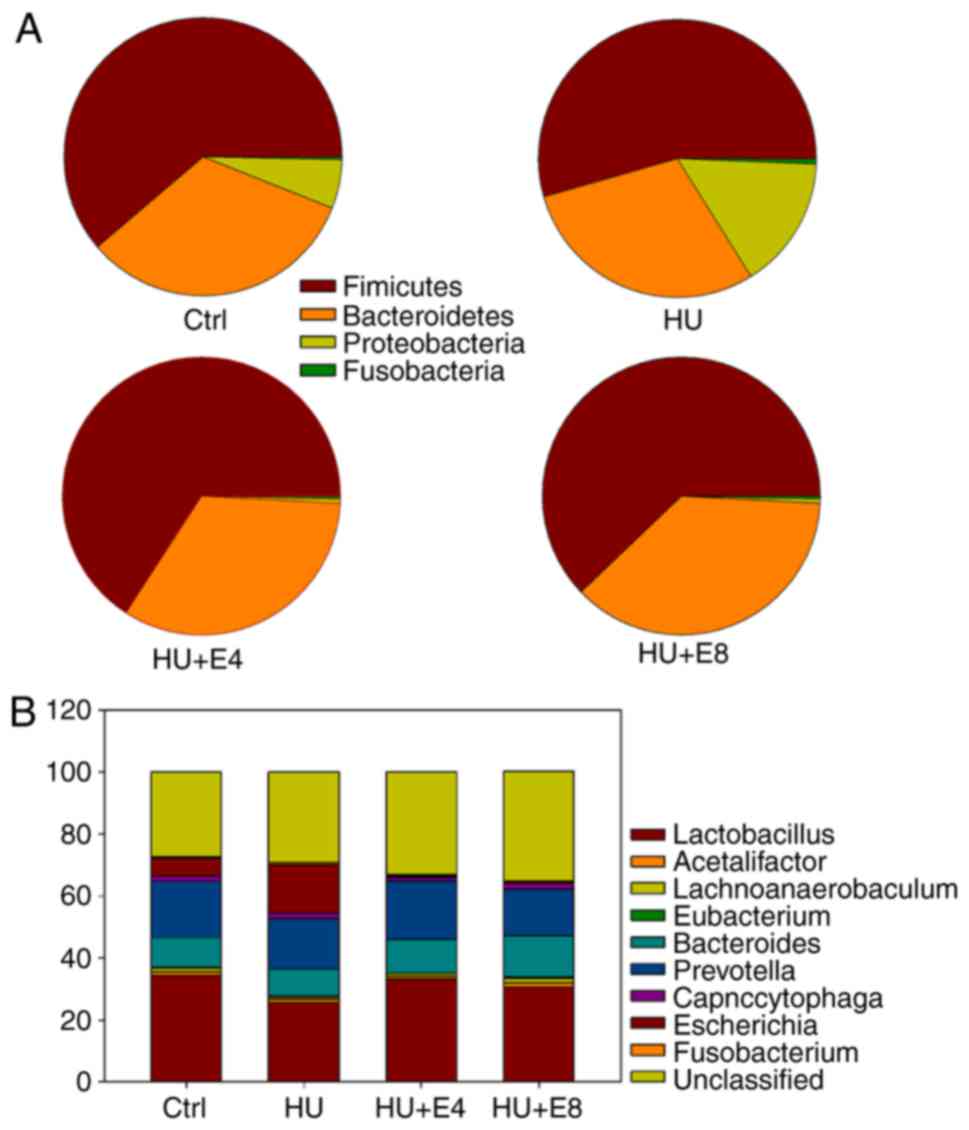
2. Overall, the two major phyla (Firmicutes and
Bacteroidetes) had similar abundance in all four groups,
whereas there were changes in the abundance of the
Proteobacteria and Fusobacteria phyla (Fig. 3A and Table II). Additionally, treatment with
microgravity and estrogen changed the abundance of the colonic
microbiota at the genus level (Fig.
3B and Table II).
Escherichia coli, which belongs to the Gram-negative
Enterobacteriaceae family, showed the most remarkable band
change during exposure to simulated microgravity and estrogen. As
shown in Fig. 1, the
Escherichia coli band was significantly intensified after
exposure to microgravity, indicating the overgrowth of
Escherichia coli under simulated microgravity. However,
estrogen greatly protected the gut microbiota from changes under
microgravity in rats.
 | Table II.Relative distributions of bacterial
phylotypes and genera in the rat feces of different groups. |
Table II.
Relative distributions of bacterial
phylotypes and genera in the rat feces of different groups.
| Groups | Control | HU | HU+E4 | HU+E8 |
|---|
| Phylum |
|
Firmicutes | 61.2 | 54.4 | 65.8 | 62.1 |
|
(range) | (51.2–69.1) | (36.3–61.5) | (51.5–70.1) | (43.1–69.4) |
|
SEM | 4.9 | 5.9 | 4.2 | 6.3 |
|
P-value |
| 0.2134 | 0.2514 | 0.1972 |
|
Bacteroidetes | 32.8 | 29.5 | 33.4 | 37.1 |
|
(range) | (20.9–38.3) | (22.4–37.4) | (27.2–45.7) | (28.6–55.2) |
|
SEM | 4.5 | 3.4 | 5.7 | 6.2 |
|
P-value |
| 0.3131 | 0.4512 | 0.2578 |
|
Proteobacteria | 5.7 | 15.5 | 0.5 | 0.5 |
|
(range) | (0.0–13.4) | (0.2–24.7) | (0.0–1.8) | (0.0–1.6) |
|
SEM | 1.8 | 2.9 | 0.21 | 0.17 |
|
P-value |
| 0.0035 | 0.0075 | 0.0067 |
|
Fusobacteria | 0.3 | 0.6 | 0.3 | 0.3 |
|
(range) | (0.0–1.2) | (0.0–2.4) | (0.0–1.4) | (0.0–1.1) |
|
SEM | 0.028 | 0.044 | 0.038 | 0.041 |
|
P-value |
| 0.15 | 0.47 | 0.43 |
| Genus |
|
Lactobacillus | 34.4 | 25.7 | 33.3 | 30.5 |
|
SEM | 4.5 | 3.4 | 3.5 | 2.7 |
|
P-value |
| 0.2109 | 0.2732 | 0.3578 |
|
Acetatifactor | 1.1 | 1.1 | 0.9 | 1.4 |
|
SEM | 0.15 | 0.14 | 0.11 | 0.21 |
|
P-value |
| 0.1354 | 0.2036 | 0.2677 |
|
Lachnoanaerobaculum | 1.2 | 0.6 | 0.8 | 1.6 |
|
SEM | 0.15 | 0.07 | 0.12 | 0.23 |
|
P-value |
| 0.0284 | 0.0439 | 0.2421 |
|
Eubacterium | 0.2 | 0.3 | 0.2 | 0.4 |
|
SEM | 0.035 | 0.023 | 0.021 | 0.047 |
|
P-value |
| 0.0522 | 0.4189 | 0.159 |
|
Bacteroides | 9.7 | 8.7 | 10.8 | 13.5 |
|
SEM | 0.75 | 1.08 | 1.23 | 1.42 |
|
P-value |
| 0.16 | 0.21 | 0.11 |
|
Prevotella | 18.1 | 16.3 | 19 | 14.9 |
|
SEM | 1.34 | 1.54 | 1.26 | 0.86 |
|
P-value |
| 0.127 | 0.138 | 0.058 |
|
Capnocytophaga | 1.8 | 1.9 | 1.1 | 1.8 |
|
SEM | 0.24 | 0.18 | 0.16 | 0.21 |
|
P-value |
| 0.38 | 0.26 | 0.31 |
|
Escherichia | 5.7 | 15.5 | 0.5 | 0.5 |
|
SEM | 0.61 | 3.2 | 0.06 | 0.07 |
|
P-value |
| 0.0035 | 0.0067 | 0.0075 |
|
Fusobacterium | 0.3 | 0.6 | 0.3 | 0.3 |
|
SEM | 0.028 | 0.044 | 0.038 | 0.041 |
|
P-value |
| 0.15 | 0.47 | 0.43 |
|
Unclassified | 27.5 | 29.3 | 33.1 | 35.2 |
|
SEM | 1.88 | 2.54 | 2.06 | 2.26 |
|
P-value |
| 0.258 | 0.138 | 0.085 |
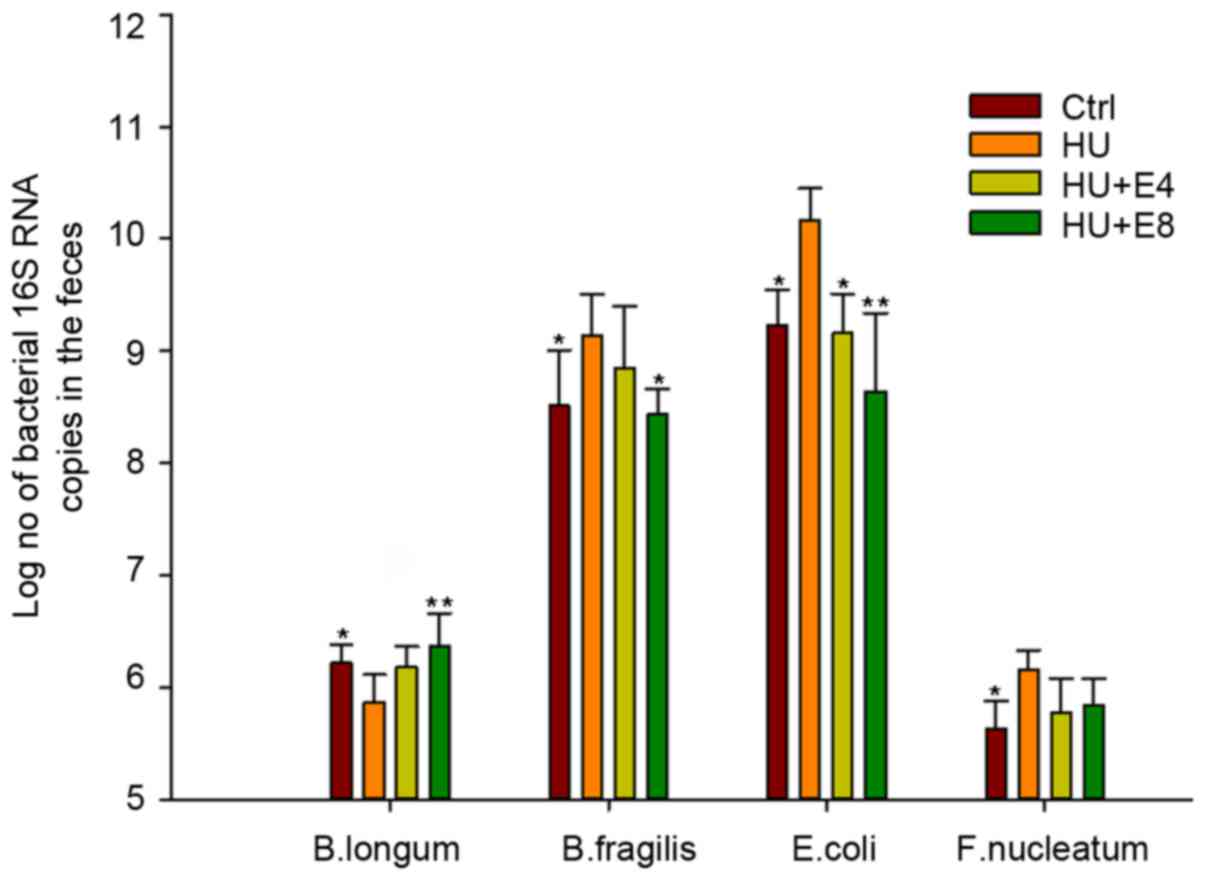
qPCR to analyze specific bacterial
population change
A qPCR assay was used to specifically analyze the
abundance of representative bacterial species. The four bacterial
species of Bifidobacteria longum (B. longum),
Bacteroides fragilis (B. fragilis), Escherichia
coli (E. coli) and Fusobacterium nucleatum (F.
nucleatum) were selected from Firmicutes,
Bacteriodetes, Proteobacteria and
Fusobacteria, respectively, based on the findings of
specific primers and for being with known functions within the four
major phyla. The results were expressed as the estimated average
number of 16S rRNA genes of the target bacteria species. Because
qPCR measures the number of 16S rRNA gene copies per sample and not
actual bacterial numbers, these values are directly related and
strongly correlated (21). The
subgroups of Bacteroides fragilis (B. fragilis),
Escherichia coli (E. coli) and Fusobacterium
nucleatum (F. nucleatum) increased significantly in
number under simulated microgravity, whereas the species of
Bifidobacteria longum (B. longum) significantly
decreased under this circumstance (P<0.05 for all bacterial
groups, Fig. 4). However, the
administration of estrogen resulted in a reversion of the
microbiota change to that of the control group at intervals of 4
and 8 weeks.
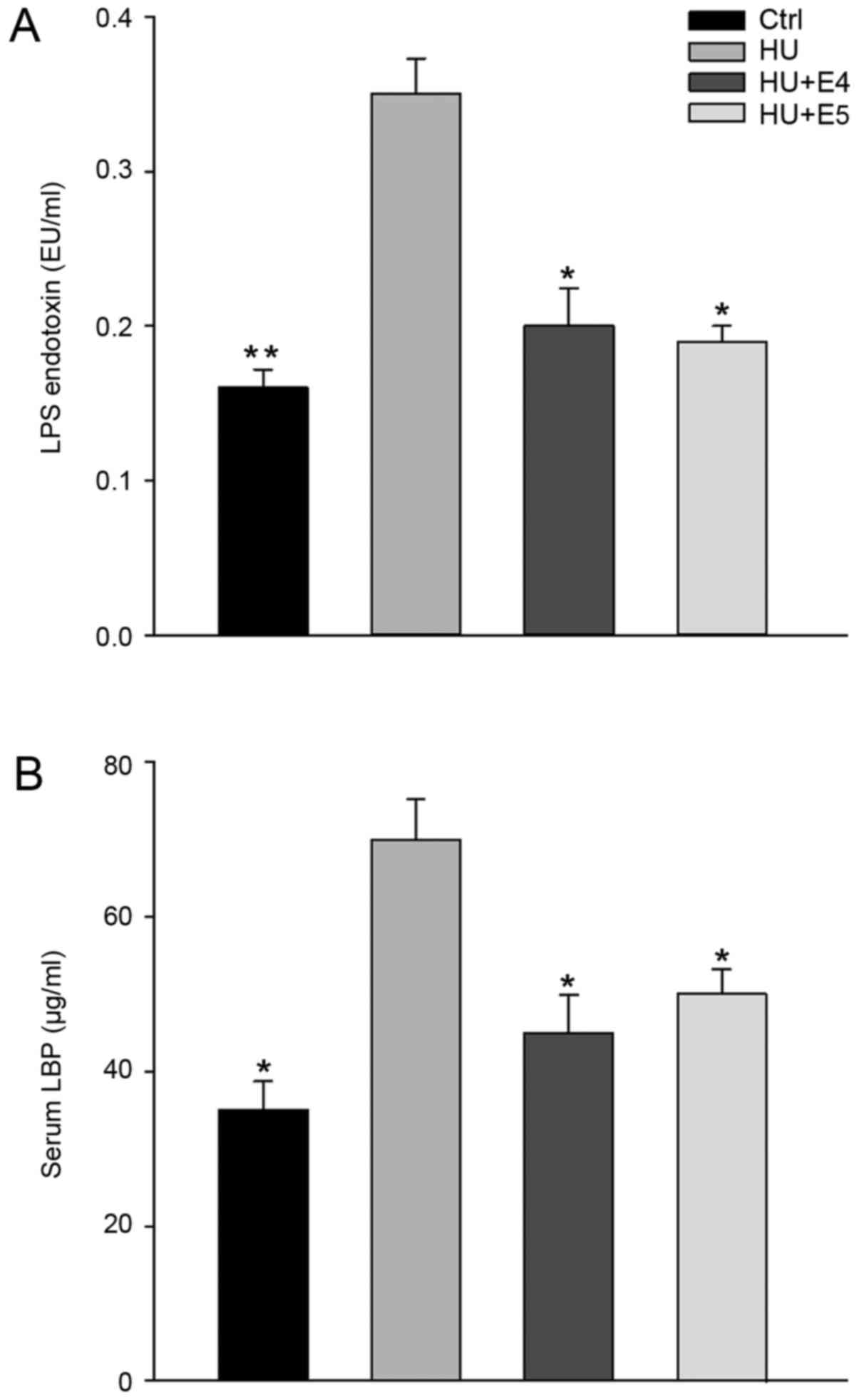
Simulated microgravity and estrogen
affected the serum level of LPS and LBP
To evaluate whether simulated microgravity and the
administration of estrogen under simulated microgravity affect
bacterial translocation, we detected serum LPS and LBP levels as
surrogate markers. It is rational that bacterial translocation
enhances circulating levels of bacterial components including LPS
(7). LPS is released from
Gram-negative bacteria in an aggregate form due to its amphiphilic
nature. LPS aggregates are converted into monomers by LBP. LBP then
transfers LPS monomers to CD14 by binding to the amphipathic lipid
A of bacterial LPS, which in turn transfers it to Toll-like
receptor (TLR) 4, leading to signaling (7). As shown in Fig. 5, both LPS and LPB levels were
significantly increased in rats under microgravity; however, a
significant reduction was observed when estrogen was administered
at 4 and 8 weeks (P<0.05).
Discussion
The long-duration of exposure to the space travel
environment exerts huge impacts on human health. Many immune
functions have been reported to be compromised during space travel.
Spaceflight-associated stresses were shown to compromise the
important capability of neutrophils and monocytes to engulf E.
coli and also reduced their ability to degranulate and elicit
an oxidative burst (1).
Spaceflight represents a unique environment characterized by
decreased gravity. Although immunological and physiological aspects
during spaceflight have been extensively studied, little effort has
been focused on the intestinal microbiota under microgravity. The
present study demonstrated that the DGGE profiles of 16s RNA PCR
amplicons changed after exposure to simulated microgravity,
suggesting a corresponding change in the rat fecal bacteria flora,
although the overall microbiota alpha diversity indices (that is,
diversity within a given community), including Shannon's and
Simpson's index from the microgravity group, were not statistically
significant compared with the control groups. The most significant
change at the phyla level occurred for Proteobacteria, which
is dominated by an E. coli population. Although the
abundance of dominant intestinal bacteria phyla (such as
Firmicutes and Bacteroides) did not significantly
change overall, the specific sub-phylum composition of microbiota,
such as B. fragilis and F. nucleatum, were variable,
as demonstrated by qPCR with a specific probe.
It has been reported that microgravity-induced
decrease in stress on the surface of microorganisms may influence
the growth and physiology of bacteria (22). So it would be rational to speculate
that the microgravity-induced decrease in tension on the surface of
microorganisms may lead to the overgrowth of some species of
bacteria, although the exact mechanism need further elucidate. On
the other side, the microbiota within the body responds and adapts
to changes in its host, such as those induced by stress. There has
been a concept that space traveling as a source of stress can alter
the composition of the intestinal microbiome, which may result in
the transient or permanent overgrowth of pathogenic gut bacteria
(23). Consistently, as shown in
Fig. 4, we demonstrated that gene
expression of E. coli increased in HU group, which provide
evidence that simulated microgravity lead to overgrowth of
potential pathogenic gut bacteria E. coli. Bacteria (such as
S. typhimurium) grown in spaceflight exhibited global
changes in gene expression characterized with increased virulence
expression (24). Besides,
microgravity rendered E. coli more adherent to an intestinal
epithelial-like Caco-2 cell line and markedly increased the
production of the heat-labile enterotoxin (25). Whether the pathogenic bacteria grew
under simulated microgravity with increased virulence to host
requires further research.
Adaptations to new selective pressures may lead to
microbiota at a tissue site breaking through the traditional host
and microbe boundaries. Usually, intestinal microbiota and its
human host coexist in good health and with mutual benefit. These
commensal microbiota seldom cause disease except in rare cases such
as in immunocompromised hosts or where the normal gastrointestinal
barriers are breached (26). To
explore the possibility that the overgrowth of some bacteria under
microgravity circumstances can overcome epithelial barriers to
initiate inflammation, we detected the endotoxin level in the
blood. LPS, the major component of endotoxin, can bind to Toll-like
receptor 4 (TLR4), initiating a potent cytokine cascade and
inflammatory reaction. Unsurprisingly, we observed an increase in
both LPS and LBP in the microgravity rat group. This was consistent
with previous evidence indicating that changes in the dominant
communities of microbiota compromise the host resistance to
infection challenge by intestinal pathogens and predisposes the
invasion of bacteria from the lumen to the interior body (23). Although E. coli is an
important member of the normal intestinal microflora of humans and
other mammals, this bacteria cause diseases in immunocompromised
hosts or where the normal gastrointestinal barriers are breached
(27). It have been reported that
the expressions of hundred genes of E. coli were
significantly altered in simulated microgravity conditions compared
to that of normal gravity conditions (28). Together with the great changes in
number of E. coli as shown in this study, we speculated that
E. coli largely contributed to the increased level of
endotoxin in the blood. The great changes of E. coli as
observed under simulated microgravity may have huge influence on
the health.
Changes in the diversity and number of gut
microflora have been linked to immunological dysregulation, which
is associated with many human noninfectious diseases such as
autoimmunity, allergies and cancer (26). Indeed, simulated microgravity has
recently been shown to increases susceptibility to colitis in mice
(5). Sex hormones influence the
development of autoimmune diseases (29). As we known, females suffer a higher
incidence of many major autoimmune diseases and undergo changes in
disease severity during pregnancy (29). Studies have recently linked this
sexual dimorphism with the gut microbiota (30). For example, the most prominent case
is microbiota in the development of autoimmune disease of Type 1
diabetes (T1D) in non-obesity diabetes (NOD) mice (30). Germ-free NOD males and females have
been observed to develop T1D with a similar incidence. Moreover,
male and female NOD mice suffer different T1D incidence after
puberty with different intestine microbiota profiles (30). This indicates that sex
hormone-mediated microbiota differences at least partially lead to
variations in susceptibility to T1D. Based on all those results, we
speculated that estrogen, the most well-known sex hormone, might
counteract microgravity-induced gut flora changes and maintain
intestinal mucosa homeostasis. As demonstrated in Fig. 4 that (HU+E4) and (HU+E8) groups
with lower E. coli gene expresssion than HU group, it was
agreed with our speculation that estrogen reversed
microgravity-induced gut microbiota changes, especially inhibited
the overgrowth of E. coli in the intestine of male rat under
microgravity. Both the biodiversity index and level of serum LPS
recovered to approximately normal when estrogen was administered to
the blood, which also suggested a protective role for estrogen in
maintaining the homeostasis of intestine mucosa. Also, we observed
estrogen inhibited the overgrowth of B. fragilis and F.
nucleatumis under microgravity. It have been reported that
invasion of those bacteria are involved in the pathogenesis of
colitis (31). So estrogen may
decrease the susceptibility of autoimmune diseases such as colitis
through regulation gut microbiota under microgravity.
Changes in the diversity and number of gut
microflora, including reductions in the defense group of
microorganisms and the overgrowth of opportunistic pathogens, such
as E. coli, B. fragilis and F. nucleatumis, may
profoundly influence health over the short and long term. The
adapted bacteria clones under microgravity acquire specific
virulence attributes, which confers an increased ability to evade
the host defense and increase risk of many infectious and
noninfectious diseases (32–34).
Another concern is antibiotic resistance in bacteria under
microgravity (35). It has
reported that the minimum inhibitory concentration (MIC) of both
colistin and kanamycin sharply increased in E. coli grown on
a spaceflight module compared with that on the ground (35). This presents great challenges in
preventing and controlling infection under microgravity. A better
understanding of estrogen-microbiome crosstalk not only help in
determining additional mechanisms involved in the pathogenesis of
hormone-related diseases and may provide further evidences that
administration of estrogen as a countermeasure to prevent
intestinal microbiota-related disease during space flight
missions.
Acknowledgements
The present study was supported by the China
Scholarship Council (201400930009) and Beijing Natural Science
Foundation (7162207).
References
|
1
|
Saei AA and Barzegari A: The microbiome:
The forgotten organ of the astronaut's body-probiotics beyond
terrestrial limits. Future Microbiol. 7:1037–1046. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cervantes JL and Hong BY: Dysbiosis and
immune dysregulation in outer space. Int Rev Immunol. 35:67–82.
2016.PubMed/NCBI
|
|
3
|
Tou JC, Grindeland RE and Wade CE: Effects
of diet and exposure to hindlimb suspension on estrous cycling in
Sprague-Dawley rats. Am J Physiol Endocrinol Metab. 286:E425–E433.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Neuman H, Debelius JW, Knight R and Koren
O: Microbial endocrinology: The interplay between the microbiota
and the endocrine system. FEMS Microbiol Rev. 39:509–521. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li P, Shi J, Zhang P, Wang K, Li J, Liu H,
Zhou Y, Xu X, Hao J, Sun X, et al: Simulated microgravity disrupts
intestinal homeostasis and increases colitis susceptibility. FASEB
J. 29:3263–3273. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ritchie LE, Taddeo SS, Weeks BR, Lima F,
Bloomfield SA, Azcarate-Peril MA, Zwart SR, Smith SM and Turner ND:
Space environmental factor impacts upon murine colon microbiota and
mucosal homeostasis. PLoS One. 10:e01257922015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou Y, Ni H, Li M, Sanzari JK,
Diffenderfer ES, Lin L, Kennedy AR and Weissman D: Effect of solar
particle event radiation and hindlimb suspension on
gastrointestinal tract bacterial translocation and immune
activation. PLoS One. 7:e443292012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Looijer-van Langen M, Hotte N, Dieleman
LA, Albert E, Mulder C and Madsen KL: Estrogen receptor-β signaling
modulates epithelial barrier function. Am J Physiol Gastrointest
Liver Physiol. 300:G621–G626. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Menon R, Watson SE, Thomas LN, Allred CD,
Dabney A, Azcarate-Peril MA and Sturino JM: Diet complexity and
estrogen receptor β status affect the composition of the murine
intestinal microbiota. Appl Environ Microbiol. 79:5763–5773. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang MH and Achkar JP: Gene-environment
interactions in inflammatory bowel disease pathogenesis. Curr Opin
Gastroenterol. 31:277–282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Walter J and Ley R: The human gut
microbiome: Ecology and recent evolutionary changes. Annu Rev
Microbiol. 65:411–429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Benson AK, Kelly SA, Legge R, Ma F, Low
SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, et al:
Individuality in gut microbiota composition is a complex polygenic
trait shaped by multiple environmental and host genetic factors.
Proc Natl Acad Sci USA. 107:pp. 18933–18938. 2010; View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du F, Ding Y, Zou J, Li Z, Tian J, She R,
Wang D, Wang H, Lv D and Chang L: Morphology and molecular
mechanisms of hepatic injury in rats under simulated weightlessness
and the protective effects of resistance training. PLoS One.
10:e01270472015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang R, Ran HH, Cai LL, Zhu L, Sun JF,
Peng L, Liu XJ, Zhang LN, Fang Z, Fan YY and Cui G: Simulated
microgravity-induced mitochondrial dysfunction in rat cerebral
arteries. FASEB J. 28:2715–2724. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Su Y, Yao W, Perez-Gutierrez ON, Smidt H
and Zhu WY: 16S ribosomal RNA-based methods to monitor changes in
the hindgut bacterial community of piglets after oral
administration of Lactobacillus sobrius S1. Anaerobe. 14:78–86.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Breton J, Massart S, Vandamme P, De Brandt
E, Pot B and Foligné B: Ecotoxicology inside the gut: Tmpact of
heavy metals on the mouse microbiome. BMC Pharmacol Toxicol.
14:622013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lozupone CA and Knight R: Species
divergence and the measurement of microbial diversity. FEMS
Microbiol Rev. 32:557–578. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu M, Wang B, Fu Y, Chen Y, Yang F, Lu H,
Chen Y, Xu J and Li L: Changes of fecal Bifidobacterium species in
adult patients with hepatitis B virus-induced chronic liver
disease. Microb Ecol. 63:304–313. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen Y, Yang F, Lu H, Wang B, Chen Y, Lei
D, Wang Y, Zhu B and Li L: Characterization of fecal microbial
communities in patients with liver cirrhosis. Hepatology.
54:562–572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Du Plessis J, Vanheel H, Janssen CE, Roos
L, Slavik T, Stivaktas PI, Nieuwoudt M, van Wyk SG, Vieira W,
Pretorius E, et al: Activated intestinal macrophages in patients
with cirrhosis release NO and IL-6 that may disrupt intestinal
barrier function. J Hepatol. 58:1125–1132. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barman M, Unold D, Shifley K, Amir E, Hung
K, Bos N and Salzman N: Enteric salmonellosis disrupts the
microbial ecology of the murine gastrointestinal tract. Infect
Immun. 76:907–915. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lynch SV, Brodie EL and Matin A: Role and
regulation of sigma S in general resistance conferred by low-shear
simulated microgravity in Escherichia coli. J Bacteriol.
186:8207–8212. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bailey MT, Dowd SE, Galley JD, Hufnagle
AR, Allen RG and Lyte M: Exposure to a social stressor alters the
structure of the intestinal microbiota: Implications for
stressor-induced immunomodulation. Brain Behav Immun. 25:397–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rosenzweig JA, Abogunde O, Thomas K, Lawal
A, Nguyen YU, Sodipe A and Jejelowo O: Spaceflight and modeled
microgravity effects on microbial growth and virulence. Appl
Microbiol Biotechnol. 85:885–891. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Allen CA, Niesel DW and Torres AG: The
effects of low-shear stress on Adherent-invasive Escherichia coli.
Environ Microbiol. 10:1512–1525. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang YJ, Li S, Gan RY, Zhou T, Xu DP and
Li HB: Impacts of gut bacteria on human health and diseases. Int J
Mol Sci. 16:7493–7519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gombošová L, Lazúrová I, Zakuciová M,
Curová K, Kmeťová M, Petrášová D and Siegfried L: Genes of
intestinal Escherichia coli and their relation to the inflammatory
activity in patients with ulcerative colitis and Crohn's disease.
Folia Microbiol (Praha). 56:367–372. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arunasri K, Adil M, Venu Charan K, Suvro
C, Himabindu Reddy S and Shivaji S: Effect of simulated
microgravity on E. Coli K12 MG1655 growth and gene expression. PLoS
One. 8:e578602013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rubtsova K, Marrack P and Rubtsov AV:
Sexual dimorphism in autoimmunity. J Clin Invest. 125:2187–2193.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yurkovetskiy L, Burrows M, Khan AA, Graham
L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y and Chervonsky
AV: Gender bias in autoimmunity is influenced by microbiota.
Immunity. 39:400–412. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xavier RJ and Podolsky DK: Unravelling the
pathogenesis of inflammatory bowel disease. Nature. 448:427–434.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bashir A, Miskeen AY, Bhat A, Fazili KM
and Ganai BA: Fusobacterium nucleatum: An emerging bug in
colorectal tumorigenesis. Eur J Cancer Prev. 24:373–385. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sears CL, Geis AL and Housseau F:
Bacteroides fragilis subverts mucosal biology: From symbiont to
colon carcinogenesis. J Clin Invest. 124:4166–4172. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Castellarin M, Warren RL, Freeman JD,
Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P,
Allen-Vercoe E, Moore RA and Holt RA: Fusobacterium nucleatum
infection is prevalent in human colorectal carcinoma. Genome Res.
22:299–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mermel LA: Infection prevention and
control during prolonged human space travel. Clin Infect Dis.
56:123–130. 2013. View Article : Google Scholar : PubMed/NCBI
|