Introduction
Antioxidant of bamboo leaves (AOB) consists mainly
of several bioactive flavonoids (orientin, isoorientin, vitexin and
isovitexin), and has been approved by the Ministry of Health of the
People's Republic of China as a natural food additive since 2007
(1,2). AOB has anti-cancer and cardiovascular
protective functions; however, its effects on early embryo vascular
development is not well known (3).
Platelet-derived growth factor receptor alpha (PDGFRA) is
identified as a functional gene that is restrictedly expressed
during in vitro differentiation and mouse embryogenesis
(4). PDGFRA expression is
evident in undifferentiated mouse embryonic stem cells (ESCs), at
sites of blood island formation and vasculogenesis during early
embryogenesis, as well as during angiogenesis in the adult mice
(5). MicroRNAs (miRNAs/miRs) are
non-coding single-stranded RNAs that can bind to the
3′-untranslated regions of target mRNAs, leading to their
translation or degradation (6,7).
miRNAs participate in various biological processes (8) and it has been highlighted that miRNAs
serve an important role in early embryo vascular development
through promoting or inhibiting angiogenesis (9).
To address whether miRNAs have an important role in
early embryo vascular development, the miRNA profile in embryonic
bodies (EBs) in response to AOB treatment was investigated. In the
current study, subtle alterations in the miRNA profile in EBs
following AOB treatment were identified. Through functional
enrichment analysis and target prediction, few candidate pathways
as putative targets of miR-146a were identified. In the ‘Wnt
signaling pathway’ which was among the top ranked predictions,
PDGFRA was verified as a functional target in angiogenesis.
Additionally, knockdown of PDGFRA could significantly
inhibit the proliferation and migration of EBs.
Materials and methods
ESC culture
Wild type mouse ESCs (WT ESCs; strain 129; ESD3
cells from the American Type Culture Collection, Manassas, VA, USA)
were plated in a flask at a density of 35,000 cells/cm2
and incubated at 37°C with 5% CO2 for 24 h. ESCs were
firstly expanded on a mitotically inactivated feeder layer followed
by at least one passage on gelatin-coated plastic in Dulbecco's
modified Eagle's medium (DMEM) containing 15% ESC-qualified fetal
bovine serum (FBS; Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 2 mM L-glutamine, 0.1 mM non-essential amino
acids, 0.1 mM beta-mercaptoethanol, 1,000 U/ml Leukemia Inhibitory
Factor (LIF; EMD Millipore, Billerica, MA, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). The study was approved by the Ethics Committee
of Linzi District People's Hospital (Linzi, China).
EB differentiation
WT ESCs were spontaneously differentiated as EBs in
3-D suspension culture. ESCs were dissociated and cultured at a
density of 800 cells/well for 24 h at 37°C. Subsequently, EBs were
transferred to culture dishes (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) with coated agar and without the feeder layer
and LIF, and cultured for up to 10 days. From day 4 onwards, medium
and dishes were changed every 24 h.
Morphological assessment
The morphology of pluripotent cell colonies and EBs
were determined by using phase contrast microscopy. The size of EBs
was evaluated using Image J software (version 3.4; National
Institutes of Health, Bethesda, MD, USA) after taking cross
sectional areas and phase contrast images for EBs.
Transfection of miR-146a antagomir and
mimic, and infection of lentiviral PDGFRA small interfering
(si)RNA
miR-146a mimic (cat. no. 4464066;
5′-UGAGAACUGAAUUCCAUGGGUU-3′), antagomir (cat. no. 4464084;
5′-AACCCAUGGAAUUCAGUUCUCA-3′) and scramble control (cat. no.
4464058) were purchased from Ambion; Thermo Fisher Scientific, Inc.
10 ng miRNA mimic, antagomir and scramble control was transfected
into EBs using Lipofectamine® RNAi Max (Thermo Fisher
Scientific, Inc.) for 6 h at room temperature. Subsequently, cells
were harvested and stored at 4°C for further assays. Pooled
lentiviral siRNA targeting PDGFRA (cat. no. sc-29444-V) and
scramble siRNA as a negative control (cat. no. sc-108080) were
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Infection with the lentivirus expressing PDGFRA or scramble
siRNA was performed as described previously (10).
miRNA microarray analysis
RNA quantity and quality was assessed by using an
Agilent 2100 Bioanalyzer. The complementary (c)RNA was generated
from 100 ng of total RNA using the FlashTag™ HSR
labeling kit (Genisphere LLC, Hatfield, PA, USA) following the
manufacturer's protocol. The cRNA was biotinylated to hybridize the
GeneChip miRNA Array (Affymetrix; Thermo Fisher Scientific, Inc.).
An Affymetrix Model 3000 scanner was used for scanning arrays.
miRNA QC tool software (version 5.1; Affymetrix; Thermo Fisher
Scientific, Inc.) was used to generate expression values, which
were normalized and log transformed. Differential expression,
defined as log2 fold change ≥1.5 or ≤-1.5 (P<0.05), was detected
by the limma tool, in R (version 3.4.2; www.r-project.org).
Target and pathway common
analysis
TargetScan Release 7.0 (www.targetscan.org) was employed to predict targets of
miR-146a. A total of 2,898 predictions were ranked by the
‘cumulative weighted context ++ score’, and the top 200 were
selected as putative targets of miR-146a for pathway common
analysis to identify functional clusters. Pathway enrichment
analysis was performed using an online webserver, Webgestalt
(www.webgestalt.org) for functional
enrichment analysis (11). Pathway
analysis was initially used to identify significant linkage of
differentially expressed genes. Minimum genes contained in each
pathway were set to ‘2’ and significance ‘P=0.05’ corrected by
Benjamini-Hochberg method.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from EBs by TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), followed by quality
assessment. A total of 10 ng total RNA was reverse-transcribed to
cDNA using Taqman microRNA Reverse Transcription kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Quantification of the
expression of PDGFRA mRNA was performed using a 7500 Fast
Real-Time PCR System. The primers were as follows: PDGFRA forward,
5′-GATCCGGGCTAAGGAAGAAG-3′ and reverse, 5′-CCAAAATGGATGCAGGAACT-3′;
GAPDH forward, 5′-CACAATTTCCATCCCAGACC-3′ and reverse,
5′-GTGGGTGCAGCGAACTTTAT-3′. GAPDH was used as the internal control.
To quantify miR-146a, cDNA was synthesized using mir-XTM miRNA
first-strand synthesis and a SYBR qRT-PCR kit (Clontech
Laboratories, Inc., Mountainview, CA, USA) according to the
manufacturer's protocol. Hsa-miR-146a (forward,
5′-GGGGAGAACTAGGTGCCAAA-3′ and reverse, 5′-GCCAGAAAGGAACTTGAACT-3′)
was used as a primer for RT-qPCR. U6 small nuclear RNA (forward,
5′-GAGAAGGGCTATCCAGGAAG-3′ and reverse, 5′-CCGAAAGGAATTGAAGCACT-3′)
was used as the internal standard. Initial denaturation was at 94°C
for 15 min followed by amplification for 45 cycles (95°C for 35 sec
and 55°C for 50 sec). The relative expression was calculated using
the 2−ΔΔCq method (12).
Cell viability assay
Cell viability was detected by
methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). A total of
6×103 cells/well were seeded in plates and incubated for
6 h at 37°C. Following washing with PBS to remove the non-adherent
cells, 10 µl MTT reagent was added to each well, and cells were
incubated for 3 h at 37°C until purple precipitate was visible.
Subsequently, 100 µl detergent reagent (Gibco; Thermo Fisher
Scientific, Inc.) was added to each well and the plates were kept
in the dark for 2 h. The absorbance was measured at a wavelength of
570 nm on day 1, 2 and 3 using an ELx800 Absorbance Reader (BioTek
Instruments, Inc., Winooski, VA, USA).
Wound-healing assay
At 24 h following transfection, 6×103
cells were digested with 0.25% trypsin (Bio-Rad Laboratories,
Inc.), centrifuged at 800 × g for 10 min at 4°C and resuspended in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS. Cells were seeded as single-cell
suspensions of 1×105/ml, 1 ml into 6-well plates or as
whole-cell monolayers cultured in serum-containing medium (1–2 ml).
Adherent cells in the medium were aspirated with a
micropipette-like tip to create a 1-mm cell-free zone that was
washed with serum-free medium or PBS to remove any remaining cells.
Cells were observed under a light microscope 24 h after
transfection and images were captured.
Transwell assay
At 24 h following transfection, cells were digested
with 0.25% trypsin, centrifuged at 800 × g for 10 min at 4°C and
resuspended in RPMI-1640 medium supplemented with 10% FBS. Cells
were seeded as single-cell suspensions of 1×105
cells/well/200 µl in Transwell chambers with 700 µl RPMI-1640
medium supplemented with 10% FBS. Cells from the two groups
(control and experimental group) were incubated for 48 h at 37°C
with 5% CO2. Cells were fixed in a neutral formalin
solution and examined on an inverted microscope at low
magnification; three random fields were chosen, and the number of
cells was counted under high magnification to determine the mean
number of cells per field.
Western blot analysis
Cells (6×103) were lysed in NP-40 buffer
containing 1 mM phenylmethyl sulfonylfluoride, 150 mM NaCl, 1.0%
NP-40, 50 mM Tris-HCl and protease inhibitors (Roche Diagnostics,
Basel, Switzerland) for 30 min on ice. Cell lysates were
centrifuged at 12,000 × g for 20 min at 4°C following sonication on
ice and supernatants were separated prior to boiling for 10 min in
the presence of 2-mercaptoethanol. Protein concentration was
determined using the bicinchoninic assay and 40 µg/lane was
separated on 10% SDS-PAGE and subsequently transferred onto
nitrocellulose membranes. Membranes were blocked in 10% dry
milk-TBST [20 mM Tris-HCl (PH 7.5), 0.1% Tween 20] for 1 h at 37°C.
Membranes were then washed three times and incubated with
monoclonal antibodies against PDGFRA (1:1,000; cat. no. 3164; Cell
Signaling Technology, Inc., Danvers, MA, USA) and GAPDH (1:1,000;
cat. no. 2118; Cell Signaling Technology, Inc.) overnight at 4°C.
The resultant membranes were washed and incubated with a
horseradish peroxidase-conjugated goat anti-rabbit secondary
antibody (1:3,000; cat. no. sc-2004; Santa Cruz Biotechnology,
Inc.) in TBST at 4°C overnight. Bands were visualized with an
enhanced chemiluminescence kit (GE Healthcare, Chicago, IL,
USA).
Luciferase reporter gene assay
The 3′-UTR of PDGFRA was amplified by PCR and
inserted into the pGL3-controlvector (Promega Corporation, Madison,
WI, USA). To generate the PDGFRA3′-UTR mutant reporter, the seed
region of PDGFRA 3′-UTR was mutated to remove all complementarity
to nucleotides 17–22 of miR-146a, using the QuikChangeII XL
mutagenesis kit (Stratagene; Agilent technologies, Inc., Santa
Clara, CA, USA). The PDGFRA3′-UTR mutant was further verified by
sequencing. Cells were seeded in a 24-well plate at a density of
1×105 cells/well. Following a 24-h culture, cells were
co-transfected with miR-146a and firefly luciferase reporter
plasmids containing WT or PDGFRA 3′-UTR mutant, using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 36 h
following transfection, firefly and Renilla luciferase
activities were measured by Dual Luciferase Reporter assay (Promega
Corporation). The assay was performed in triplicate.
Spheroid sprouting assay
Multicellular spheroids were generated by seeding
1×104 cells/well in 96-well plates filled with DMEM
(Gibco; Thermo Fisher Scientific, Inc.) containing 5% FBS and 0.24%
high-viscosity methylcellulose (Tecan Group, Ltd., Männedorf,
Switzerland). Spheroids were collected following culture at 37°C
for 24 h, and embedded in collagen gels and kept in DMEM containing
2% FBS at 37°C for another 24 h. AOBs were prepared from the leaves
of Phyllostachys nigra var. henonis (Zhejiang University
Innoessen Bio-Technology Co., Ltd). AOB was added at a final
concentration of 100 µg/ml from a stock solution of 10 mg/ml.
Following treatment for 24 h at 4°C, cells were observed under a
Zeiss Axiovert 25 microscope (Carl Zeiss AG, Oberkochen, Germany)
and analyzed using the KS400 Kontron image analysis software
(version 1.2, Kontron Elektronik GmbH, Munich, Germany).
Statistical analysis
GraphPad Prism 5 software (version 5; GraphPad
Software, Inc., La Jolla, CA, USA) was used to perform the
statistical analysis. Data is presented as the mean ± standard
error and was analyzed using the one-way analysis of variance.
Multiple comparisons between the groups were performed using the
Student-Newman-Keuls method. P<0.05 was considered to indicate a
statistically significant difference.
Results
AOB modulates microRNA expression in
EBs
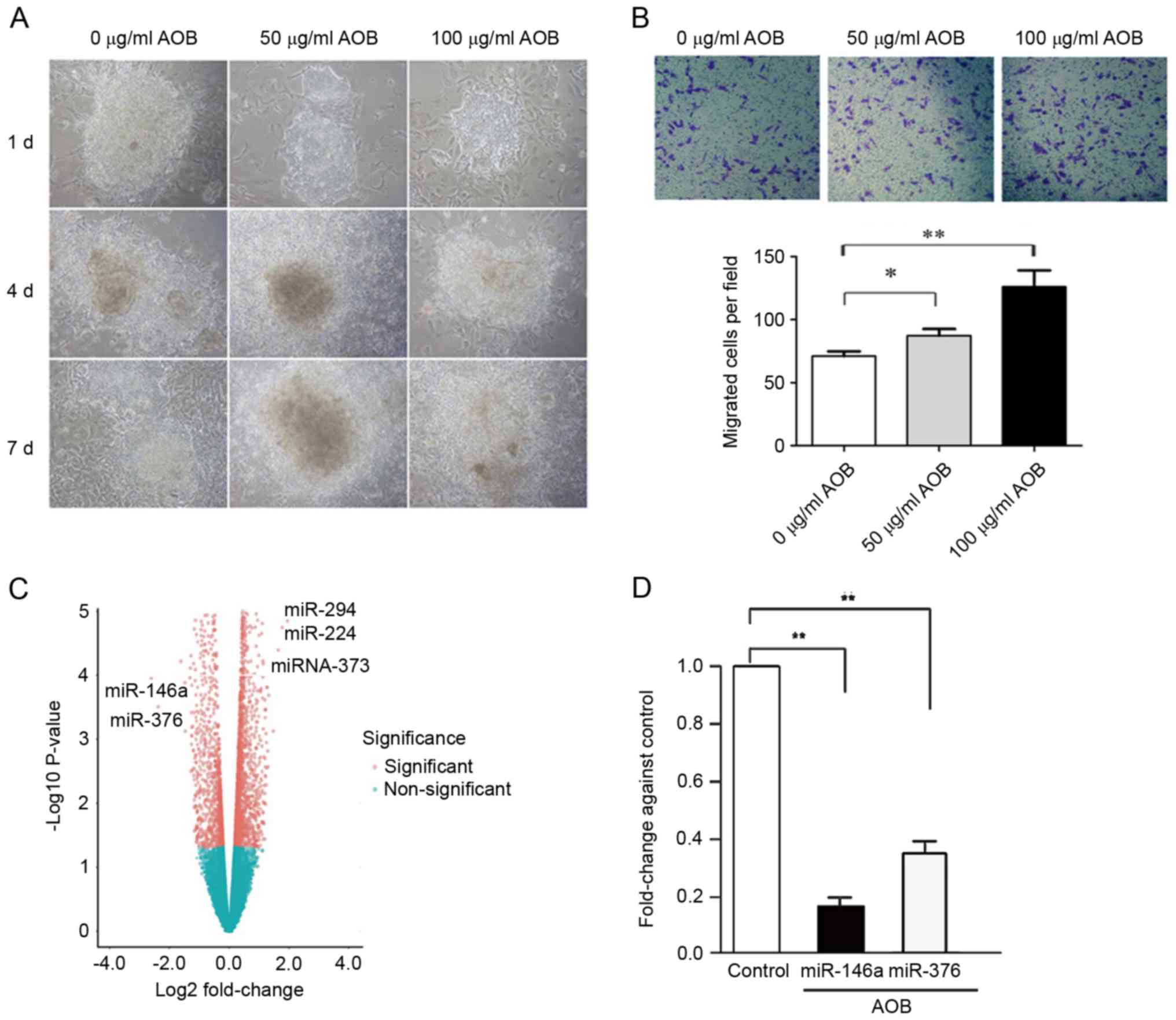
In this study, treatment with AOB increased the
proliferation and migration of EBs (Fig. 1A and B, respectively). To further
detect the microRNA expression profile involved in AOB-treated EBs,
microarray analysis was conducted. As depicted in Fig. 1, expression differences were
compared between AOB-treated and the control cells. In all, 15
miRNAs were downregulated, among which miR-146a was the most
downregulated one, followed by miR-376. Significant differences
were also identified in miRNA-294, miRNA-224 and miR-373 (Fig. 1C). RT-qPCR results demonstrated
that the expression levels of miR-146a (log FC=−1.97, P=2.03E-11)
and miR-376 (log FC=−1.23, P=6.49E-13) were both markedly
downregulated following AOB treatment compared with the control
(Fig. 1D).
miR-146a regulates EB
proliferation
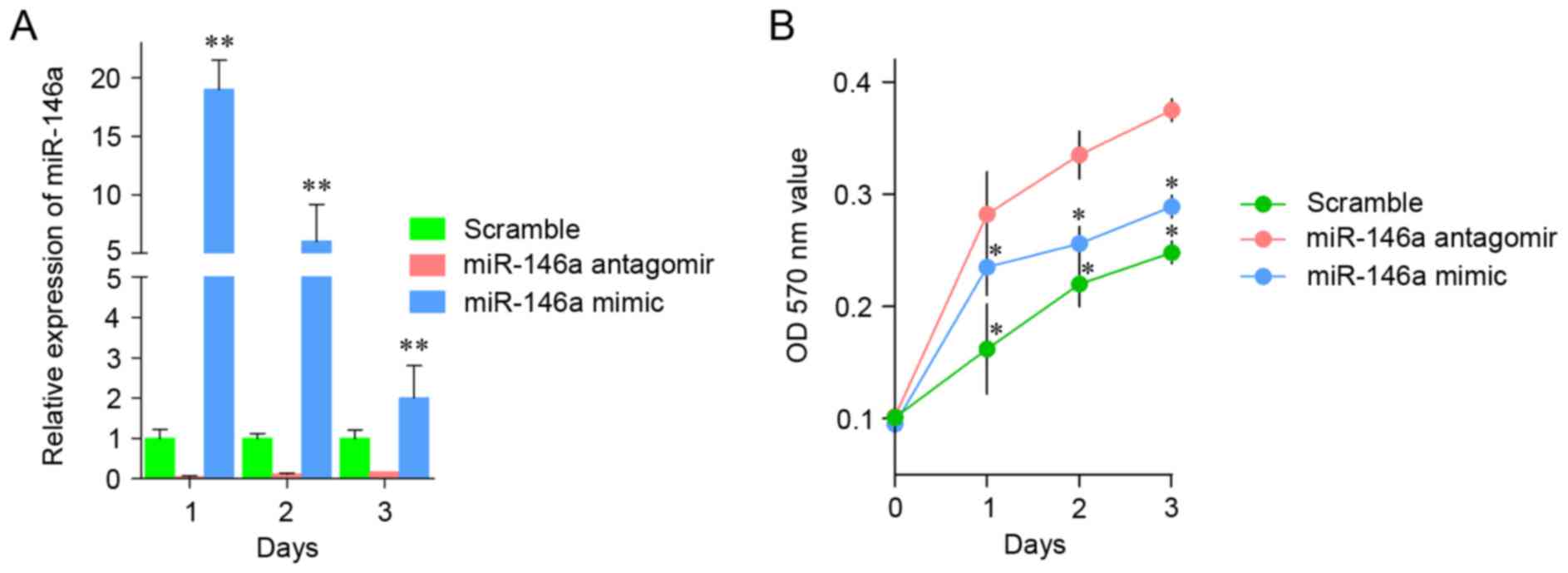
To examine whether miR-146a is essential in the
angiogenesis process following AOB treatment, miR-146a antagomir or
mimic was transfected into EBs, and the proliferation was
determined. As illustrated in Fig.
2A, transfection of miR-146a antagomir significantly reduced
the expression of miR-146a, whereas transfection of miR-146a mimic
markedly increased the level of miR-146a, compared with
transfection with the scramble. The cell viability of EBs was
continually measured for 3 days following transfection of miR-146a
antagomir or mimic. The results demonstrated that the growth rate
of EBs transfected with miR-146a antagomir was higher than that of
EBs transfected with the scramble sequence. By contrast, cells
transfected with miR-146a mimics had an increased growth rate
compared with the control cells transfected with the scramble
sequence (Fig. 2B).
Putative targets of miR-146a are
abundant in the phosphatidylinositol 3-kinase/protein kinase B
(PI3K/AKT) signaling pathway
By Targetscan software, a total of 1,327 transcripts
were predicted as putative targets of miR-146a, namely the
‘cumulative weighted context ++ score’. Genes were chosen for the
pathway common analysis. These predicted genes were enriched in
various pathways, including the ‘wingless-type MMTV integration
site (Wnt) signaling pathway’, the ‘vascular endothelial growth
factor (VEGF) and VEGF receptor (VEGFR) signaling network’, the
‘PI3K/AKT pathway’, ‘Endothelins’, and the ‘mitogen-activated
protein kinase (MAPK) signaling network’ (Table I). In particular, the ‘PI3K/AKT
pathway’ and the ‘VEGF/VEGFR signaling network pathway’ were
involved in angiogenesis, indicating that miR-146a might regulate
angiogenesis through them.
 | Table I.Over represented pathways of top 200
predicted target genes of microRNA-146a. |
Table I.
Over represented pathways of top 200
predicted target genes of microRNA-146a.
| ID | Pathway | Observed | Expected | adjP |
|---|
| 1585 | Arf6 downstream
pathway | 22 | 5.65 |
3.2×107 |
| 1575 | VEGF and VEGFR
signaling network | 22 | 5.64 |
3.3×107 |
| 1556 | Plasma membrane
estrogen receptor signaling | 22 | 5.71 |
3.5×105 |
| 1472 | Nectin adhesion
pathway | 22 | 5.73 |
3.7×105 |
| 1461 | GMCSF-mediated
signaling events | 22 | 5.65 |
3.7×107 |
| 1615 | Arf6 trafficking
events | 22 | 5.65 |
3.7×107 |
| 15744 | Signaling events
mediated by focal adhesion kinase | 22 | 5.64 |
3.4×107 |
| 1602 | ErbB1 downstream
signaling | 22 | 5.64 |
3.4×105 |
| 1571 | mTOR signaling
pathway | 22 | 5.64 |
3.4×105 |
| 1619 | Endothelins | 22 | 5.71 |
3.4×107 |
| 1517 | Beta1 integrin cell
surface interactions | 22 | 5.90 |
5.6×107 |
| 1499 | Integrin family
cell surface interactions | 22 | 6.01 |
7.66×107 |
| 1546 | Integrin-linked
kinase signaling | 13 | 2.19 |
2.63×105 |
| 1488 | CDC42 signaling
events | 13 | 3.36 | 0.0001 |
| 1544 | E-cadherin
signaling in the nascent adherens junction | 7 | 1.23 | 0.0006 |
| 1494 | N-cadherin
signaling events | 6 | 1.12 | 0.0023 |
| 1611 | Regulation of
nuclear SMAD2/3 signaling | 6 | 1.35 | 0.0055 |
| 936 | Cell junction
organization | 3 | 0.02 | 0.0074 |
| 935 | Cell-Cell
communication | 3 | 0.58 | 0.0245 |
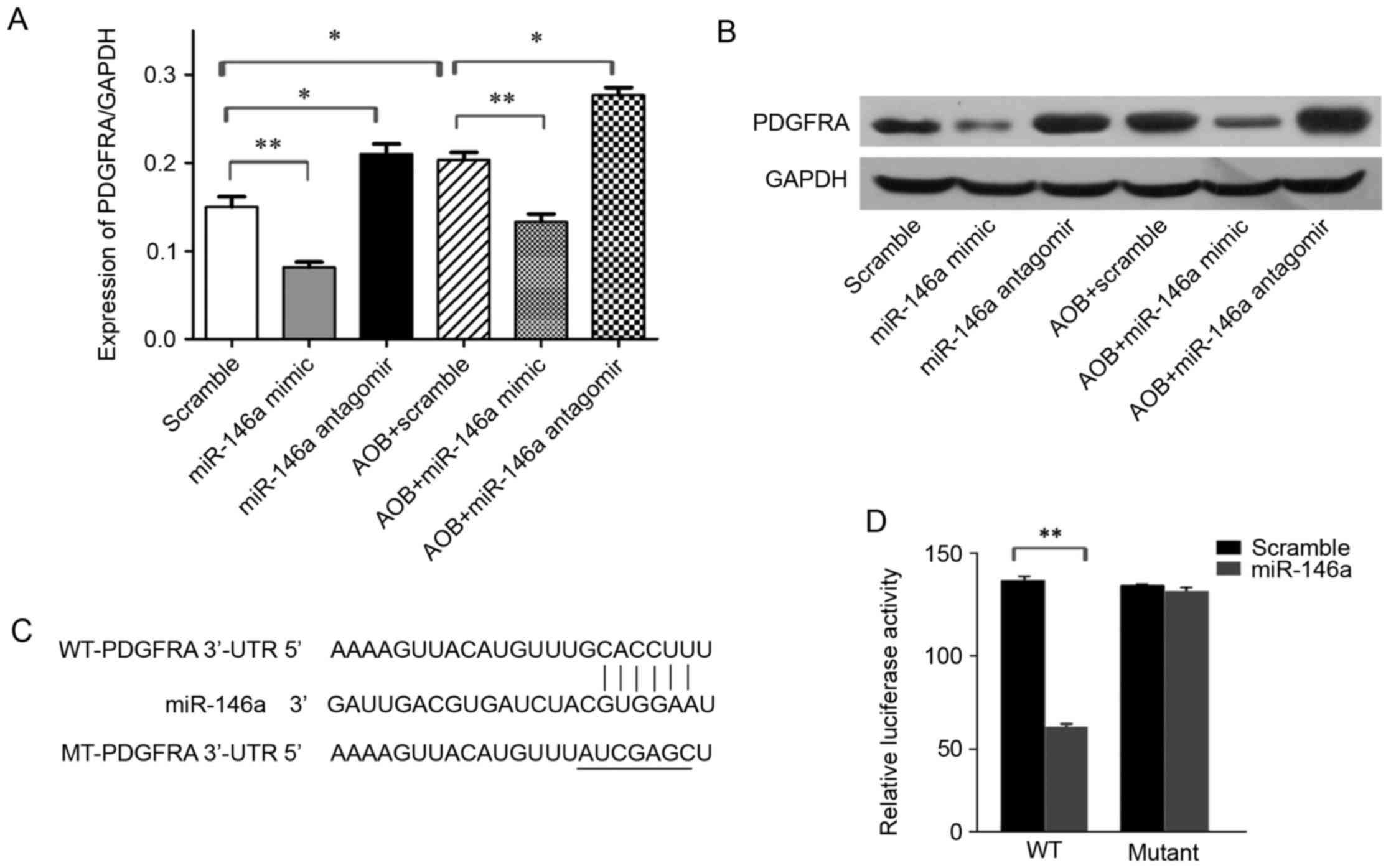
PDGFRA is a target of miR-146a
PDGFRA has been reported to be associated with
angiogenesis and cell migration (13). Therefore, it was postulated that
miR-146a might function via targeting PDGFRA. To access this
assumption, the mRNA and protein expression levels of PDGFRA in EBs
was determined following transfection with miR-146a mimic or
antagomir, using RT-qPCR and western blotting, respectively. The
results demonstrated that transfection with miR-146a mimics
significantly decreased both mRNA and protein expression levels of
PDGFRA, whereas transfection with miR-146a antagomir led to a
remarkable increase of PDGFRA expression. In addition, AOB
treatment markedly elevated the level of PDGFRA (Fig. 3A and B). Notably, transfection with
miR-146a mimic obstructed AOB-mediation PDGFRA upregulation
(Fig. 3A and B), while
transfection of miR-146a antagomir led to a further increase in
PDGFRA expression following AOB treatment (Fig. 3A and B). To further confirm that
PDGFRA is a target of miR-146a, a dual luciferase reporter assay
was performed (Fig. 3C). PDGFRA
3′-UTR or PDGFRA 3′-UTR mutant was co-transfected with miR-146a
mimics into EBs, and 36 h following this, the luciferase activity
was detected. The result illustrated that miR-146a mimic decreased
the luciferase activity of PDGFRA 3′-UTR but had a minimal effect
on that of PDGFRA 3′-UTR mutant (Fig.
3D), suggesting that PDGFRA is a target of miR-146a.
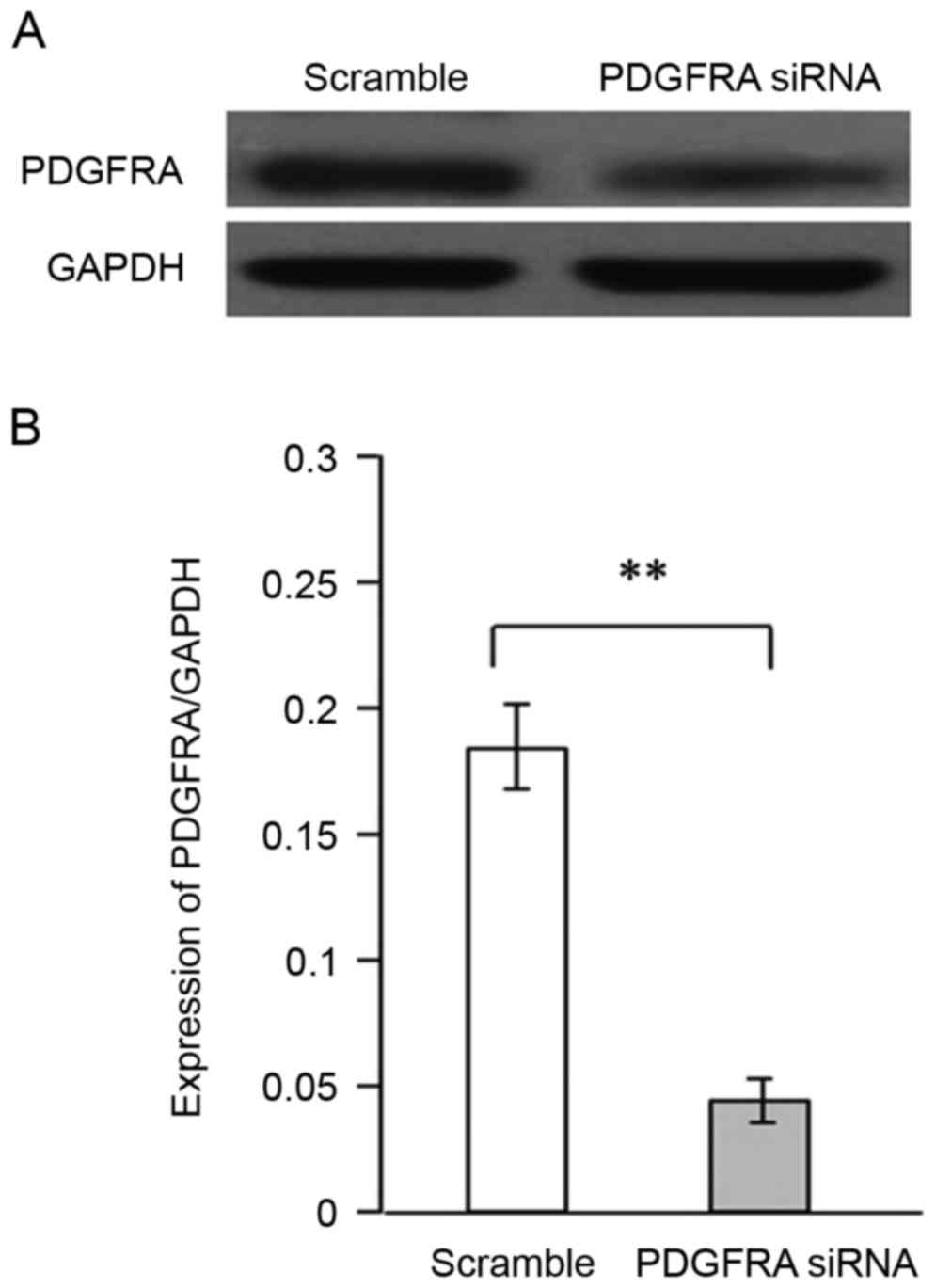
Silencing of PDGFRA inhibits the
proliferation and migration induced by AOB
Whether PDGFRA is involved in AOB-mediated
proliferation and migration of EBs was also investigated. The cell
viability and angiogenic sprouting of EBs were measured following
PDGFRA knockdown and AOB treatment. Knockdown efficiency of
PDGFRA was confirmed at the protein and mRNA levels
(Fig. 4A and B, respectively).
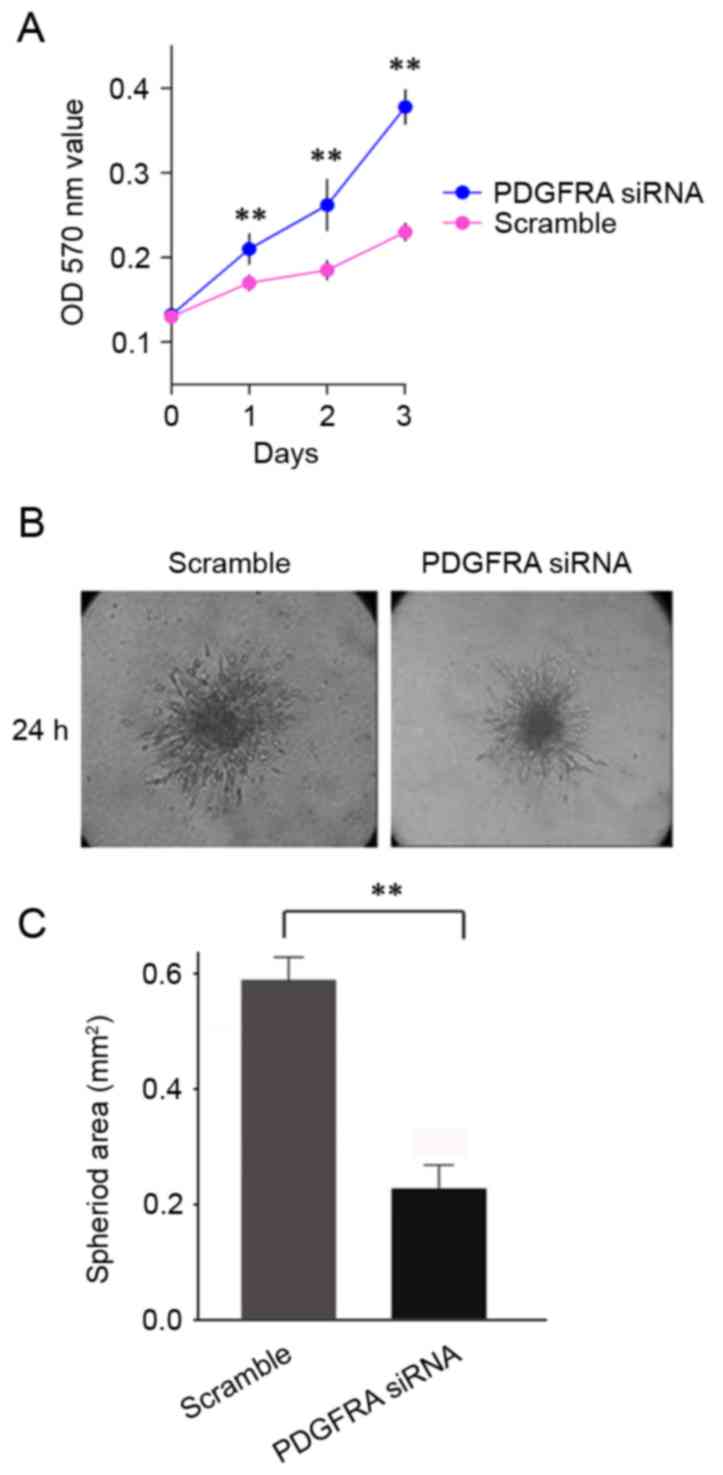
Cell viability analysis demonstrated that silencing of
PDGFRA significantly increased the growth rate of EBs
(Fig. 5A). PDGFRA-ablated EBs
demonstrated decreased angiogenic sprouting in comparison with the
control cells (Fig. 5B and C). In
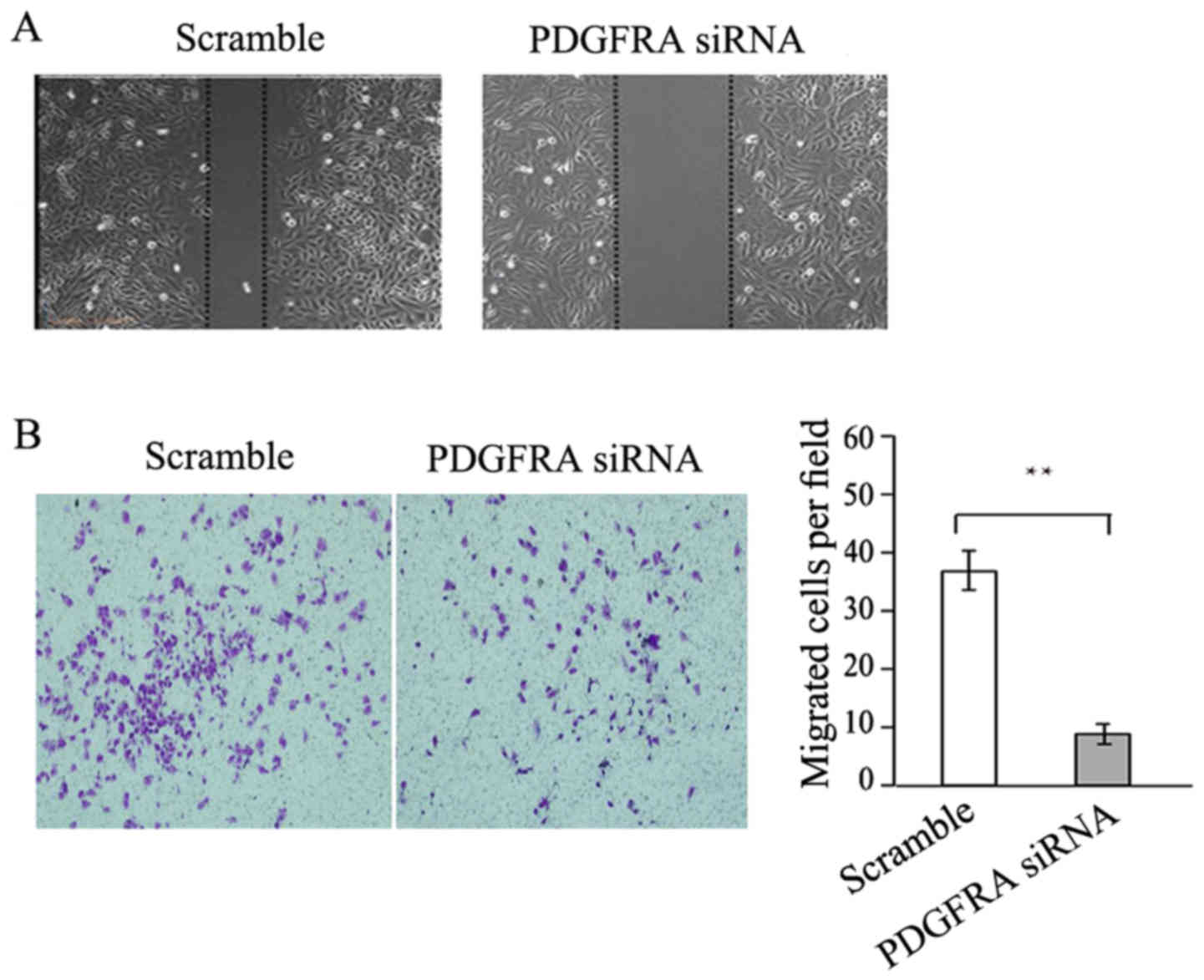
addition, PDGFRA knockdown significantly reduced the
migration ability of EBs as demonstrated by the wound-healing assay
and Transwell assay (Fig. 6A and
B, respectively). Together, PDGFRA deficiency inhibited EB
proliferation and migration, demonstrating a vital role in EB
angiogenesis.
Discussion
ESCs can be induced under certain conditions to form
ectoderm, mesoderm and endoderm cell types, including EBs (14). EBs are basically formed during
early embryonic development of embryos following gastrulation, and
are easy to handle and to carry out various operations, avoiding
experiments with embryos and ethical controversy (15,16).
Therefore, EB is a preferred experimental model for studying early
embryonic development, genetic, mesoderm mutual induction, the
impact of environmental factors on the development of the early
embryos, exogenous substances embryonic teratogenic, cytotoxic and
tire-borne diseases and other diseases (17). In recent years, increasing research
on miRNA has emphasized the importance of miRNAs in gene regulation
(18). miRNAs are a class of
non-coding single-stranded small RNA nucleotides from pri-miRNA
processing by Drosha and Dicer (19). Typically, miRNAs can recognize
specific mRNAs, and bind to their 3′-UTR by inexact match,
affecting mRNA stability, or inhibit their translation, regulating
the expression levels of target genes (20–22).
Studies on miRNA functions are mainly achieved with the usage of
in vitro miRNA mimics.
The Wnt signaling pathway is recognized as the most
associative pathway to epithelialization. In the Wnt signaling
pathway, PDGFRA which has been reported to function in
angiogenesis, was further verified as a target of miR-146a
(23,24). Zhu et al (25) have reported that miR-146a can
target PDGFRA 3′-UTR, and its aberrant expression markedly
decreases PDGFRA expression, which is associated with the activity
of EC angiogenesis in hepatic cell carcinoma. This report indicated
that the processes of angiogenesis and cancer metastasis to some
extent share common biological signaling pathways. Based on the
above statements, PDGFRA is identified as a vital element in
angiogenesis.
In conclusion, changes in the miRNA profile in
AOB-treated EBs were explored, among which miR-146a was
demonstrated to be the most susceptible one. Although target
prediction yielded a large number of candidates, the list was
narrowed by functional enrichment analysis. Among all putative
pathways, five were highly implicated in angiogenesis, suggesting a
target pool of miR-146a, and PDGFRA was verified as one of its
targets. Although the underlying mechanism remains to be
elucidated, the present study implies the potential therapeutic
value of AOB on EB angiogenesis via downregulation of miR-146a.
Furthermore, the functional clusters identified based on the
putative targets of miR-146a may offer candidates for further
exploring their role in angiogenesis and associated diseases.
References
|
1
|
Ma X, Wang E, Lu Y, Wang Y, Ou S and Yan
R: Acylation of antioxidant of bamboo leaves with fatty acids by
lipase and the acylated derivatives' efficiency in the inhibition
of acrylamide formation in fried potato crisps. PLoS One.
10:e01306802015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu L, Xia B, Jin C and Zhang Y and Zhang
Y: Chemical acylation of water-soluble antioxidant of bamboo leaves
(AOB-w) and functional evaluation of oil-soluble AOB (cAOB-o). J
Food Sci. 79:C1886–C1894. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y, Luo Z, Shao Z, Yu C and Wang S:
Effects of antioxidants of bamboo leaves and flavonoids on
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) formation in
chemical model systems. J Agric Food Chem. 62:4798–4802. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hirota S and Ogawa M: Activin A in
combination with OP9 cells facilitates development of Flk-1(+)
PDGFRα(−) and Flk-1(+) PDGFRα(+) hematopoietic mesodermal cells
from murine embryonic stem cells. Biochem Biophys Res Commun.
467:583–588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moriya J and Ferrara N: Inhibition of
protein kinase C enhances angiogenesis induced by platelet-derived
growth factor C in hyperglycemic endothelial cells. Cardiovasc
Diabetol. 14:192015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ro S, Park C, Young D, Sanders KM and Yan
W: Tissue-dependent paired expression of miRNAs. Nucleic Acids Res.
35:5944–5953. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mallory AC and Vaucheret H: MicroRNAs:
Something important between the genes. Curr Opin Plant Biol.
7:120–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hayes J, Peruzzi PP and Lawler S:
MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol
Med. 20:460–469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang TY, Huang TS, Wang HW, Chang SJ, Lo
HH, Chiu YL, Wang YL, Hsiao CD, Tsai CH, Chan CH, et al: miRNome
traits analysis on endothelial lineage cells discloses biomarker
potential circulating microRNAs which affect progenitor activities.
BMC Genomics. 15:8022014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sundaravinayagam D, Kim HR, Wu T, Kim HH,
Lee HS, Jun S, Cha JH, Kee Y, You HJ and Lee JH: miR146a-mediated
targeting of FANCM during inflammation compromises genome
integrity. Oncotarget. 7:45976–45994. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
An integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33(Web Server Issue): W741–W748. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Scmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McCarthy N, Liu JS, Richarte AM, Eskiocak
B, Lovely CB, Tallquist MD and Eberhart JK: Pdgfra and Pdgfrb
genetically interact during craniofacial development. Dev Dyn.
245:641–652. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dziedzicka D, Markouli C, Barbé L, Spits
C, Sermon K and Geens M: A high proliferation rate is critical for
reproducible and standardized embryoid body formation from
laminin-521-based human pluripotent stem cell cultures. Stem Cell
Rev. 12:721–730. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Boxman J, Sagy N, Achanta S, Vadigepalli R
and Nachman I: Integrated live imaging and molecular profiling of
embryoid bodies reveals a synchronized progression of early
differentiation. Sci Rep. 6:316232016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Moon SH, Ju J, Park SJ, Bae D, Chung HM
and Lee SH: Optimizing human embryonic stem cells differentiation
efficiency by screening size-tunable homogenous embryoid bodies.
Biomaterials. 35:5987–5997. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim JE, Lee JM and Chung BG: Microwell
arrays for uniform-sized embryoid body-mediated endothelial cell
differentiation. Biomed Microdevices. 16:559–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Crabbé MA, Gijbels K, Visser A, Craeye D,
Walbers S, Pinxteren J, Deans RJ, Annaert W and Vaes BL: Using
miRNA-mRNA interaction analysis to link biologically relevant
miRNAs to stem cell identity testing for next-generation culturing
development. Stem Cells Transl Med. 5:709–722. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang Y, Xing Y, Liang C, Hu L, Xu F and
Chen Y: Crucial microRNAs and genes of human primary breast cancer
explored by microRNA-mRNA integrated analysis. Tumour Biol.
36:5571–5579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tutar L, Tutar E, Özgür A and Tutar Y:
Therapeutic targeting of microRNAs in cancer: Future perspectives.
Drug Dev Res. 76:382–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Breda J, Rzepiela AJ, Gumienny R, van
Nimwegen E and Zavolan M: Quantifying the strength of miRNA-target
interactions. Methods. 85:90–99. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miao CG, Xiong YY, Yu H, Zhang XL, Qin MS,
Song TW and Du CL: Critical roles of microRNAs in the pathogenesis
of systemic sclerosis: New advances, challenges and potential
directions. Int Immunopharmacol. 28:626–633. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rudat C, Norden J, Taketo MM and Kispert
A: Epicardial function of canonical Wnt-, Hedgehog-, Fgfr1/2- and
Pdgfra-signalling. Cardiovasc Res. 100:411–421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li F, He Z, Li Y, Liu P, Chen F, Wang M,
Zhu H, Ding X, Wangensteen KJ, Hu Y and Wang X: Combined activin
A/LiCl/Noggin treatment improves production of mouse embryonic stem
cell-derived definitive endoderm cells. J Cell Biochem.
112:1022–1034. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu K, Pan Q, Zhang X, Kong LQ, Fan J, Dai
Z, Wang L, Yang XR, Hu J, Wan JL, et al: MiR-146a enhances
angiogenic activity of endothelial cells in hepatocellular
carcinoma by promoting PDGFRA expression. Carcinogenesis.
34:2071–2079. 2013. View Article : Google Scholar : PubMed/NCBI
|