Introduction
Gastric cancer (GC) is one of the most common solid
malignant tumors (1), and is
typically associated with a poor prognosis due to the high
frequency of metastases and relapse. In addition, the limitations
of chemotherapy and surgery have contributed to the low survival
rates of patients with GC. The five-year survival rate of patients
with GC is 30–50% (2). Therefore,
a comprehensive understanding of the mechanisms involved in the
development and progression of GC is essential for improving the
diagnosis, prevention and treatment of this disease.
Long non-coding RNA (lncRNA) is a type of
non-protein encoding endogenous RNA that is ~200 nucleotides in
length. This feature permits the formation of secondary structures.
Some lncRNAs possess the same sequences corresponding to
protein-coding genes (3). Previous
studies have revealed that lncRNAs and mRNAs may compete for shared
microRNA (miRNA) response elements (4–6). It
has been reported that lncRNAs might function as competing
endogenous RNAs (ceRNAs) to sequester miRNAs, thereby modulating
the expression of miRNA target genes (4–7). The
H19 lncRNA is a paternally imprinted gene located close to the
telomeric region of chromosome 11p15.5 (8,9),
which is a region that is frequently associated with tumor
development (10,11). It has been demonstrated that H19
lncRNA binds and sequesters let-7, which inhibits its function
(12). In addition, Gao et
al (13) identified that
let-7c is negatively associated with human epidermal growth factor
receptor 2 (HER2) expression. The results of these studies suggest
that a correlation and potential crosstalk between H19, let-7c and
HER2 may exist. Therefore, the aim of the present study was to
investigate this hypothesis.
Materials and methods
Tissue samples
A total of 24 GC and adjacent benign tissues
(located 5 cm from the tumor margin) were collected during the
surgery from patients admitted to the General Hospital of Daqing
Oil Field (Daqing, China) between February 2016 and August 2016.
None of the patients had received radiotherapy and chemotherapy
before surgery. The differentiation and tumor, node and metastasis
(TNM) stage of the tumor tissues were determined by histopathology.
The sex ratio between female and male was 1:2. The average age of
the patients was 63±2.6. The clinicopathological features of all
patients are listed in Table I.
The present study was approved by the Ethics Committee of the
General Hospital of Daqing Oil Field, and written informed consent
was obtained from all patients. Tumor diameters were measured with
a Vernier caliper and were stored in liquid nitrogen.
 | Table I.Correlation between H19 expression and
clinicopathologic features of patients with gastric cancer. |
Table I.
Correlation between H19 expression and
clinicopathologic features of patients with gastric cancer.
| Clinical pathologic
features | Number of patients
(%) | Relative expression
of H19 (95% CI) | P-value |
|---|
| Sex |
|
| 0.660 |
| Male | 16 (66.7) | 3.21 (0.75–4.53) |
|
|
Female | 8
(33.3) | 4.01 (1.34
−4.85) |
|
| Tumor size (cm) |
|
| 0.038 |
|
>5 | 14 (58.3) | 3.20 (1.34–4.85) |
|
|
<5 | 10 (41.7) | 1.90 (0.75–3.12) |
|
| Differentiation |
|
| 0.019 |
| Poor | 13 (54.2) | 3.60 (1.25–4.36) |
|
|
High/moderate | 11 (45.8) | 1.10
(0.75–3.14) |
|
| Lymph node
metastasis |
|
| 0.015 |
| N0 | 8
(33.3) | 1.14
(0.75–1.42) |
|
|
N1-3 | 16 (66.7) | 3.44
(1.15–4.85) |
|
| Metastatic disease
stage |
|
| 0.383 |
| M0 | 7
(29.2) | 2.35
(1.53–3.64) |
|
| M1 | 17 (70.8) | 3.82
(0.75–4.85) |
|
Cell culture
Human GC cell lines, BGC823 and SGC7901, and the
normal gastric epithelial GES-1 cell line, were purchased from the
China Academy of Chinese Medical Sciences (Beijing, China). Cells
were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(Sanofi Genzyme, Cambridge MA, USA) at 37°C in an atmosphere
containing 5% CO2.
Immunohistochemistry (IHC)
Tissues were fixed in 4% paraformaldehyde and
subjected to standard (4 mm) paraffin sectioning. For
immunohistological staining, the sections were treated with
hydrogen peroxide to black the endogenous peroxidase activity,
followed by heating to 96°C for 10 min for antigen retrieval.
Following blocking with 1% goat serum (G9023; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) at room temperature for 1 h, the sections
were then incubated with anti-HER2 primary antibody (1:800, cat.
no. 4290; Cell Signaling Technology, Inc., Danvers, MA, USA) for 2
h at room temperature. The anti-PCNA antibody (1:50, BZ00678;
Bioworld Technology Inc., St. Louis Park, MN, USA) was also used to
stain the nucleus. Immunoperoxidase staining was performed with the
3,3′ diaminobenzidine chromogen (K3647, Dako; Agilent Technologies,
Inc., Santa Clara, CA, USA) for 5 min at room temperature. The
images were captured by light microscopy (Leica Microsystems Ltd.,
Milton Keynes, UK) under high magnification (×200). IHC-staining
was confirmed independently by three pathologists. The IHC scoring
system was used to determine the scores of HER2 expression
(14–19). Scores ≥2+ were defined as
HER2-positive, and IHC scores of 0 and 1+ were defined as
HER2-negative, indicating tumor differentiation stage.
Transfection of GC cells
All plasmid vectors for transfection were extracted
from DH5α competent cells (Thermo Fisher Scientific, Inc.) using a
DNA Midiprep kit (Qiagen GmbH, Hilden, Germany) according to the
manufacturer's instructions. Three individual H19 small interfering
(si)RNAs (si-H19) and a scrambled negative control siRNA (si-NC)
were purchased from Invitrogen; Thermo Fisher Scientific, Inc.
Target sequences for H19 siRNAs were listed as H19-siRNA1,
5′-UAAGUCAUUUGCACUGGUUdTdT-3′; H19-siRNA2,
5′-GCAGGACAUGACAUGGUCCdTdT-3′; and H19-siRNA3
5′-CCAACAUCAAAGACACCAUdTdT-3′. BGC-823 and SGC7901 cells were
transfected with si-NC and the three siRNAs of H19 using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
RT-qPCR
Total RNA was extracted from frozen GC tissue
samples or BGC823 and SGC7901 cells using TRIzol reagent (Thermo
Fisher Scientific, Inc.), and the reverse transcription reactions
were performed using the SuperScript™ IV First-Strand
Synthesis System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. H19 and let-7c
expression levels were quantified relative to GAPDH expression.
The forward and reverse primers were as follows:
GAPDH, forward, 5′-CATGAGAAGTATGACAACAGCCT-3′ and reverse,
5′-AGTCCTTCCACGATACCAAAGT-3′; H19 forward,
5′-GGGTCTGTTTCTTTACTTCCTCCAC-3′ and reverse,
5′-GATGTTGGGCTGATGAGGTCTGG-3′; let-7c forward,
5′-UGAGGUAGUAGGUUGUAUGGUU-3′ and reverse,
5′-UGAGGUAGUAGGUUGUAUGGUU-3′. qPCR was performed using the
SYBR-Green PCR kit with the ABI Prism 7900 HT Sequence Detection
System (both from Applied Biosystems; Thermo Fisher Scientific,
Inc.) The PCR reaction conditions were: Forty-two cycles at 95°C
for 30 sec, 1 cycle at 60°C for 30 sec and 1 cycle at 72°C for 30
sec. The relative expression levels were determined with the
2−ΔΔCq method (20).
Agarose gel
The amplified cDNA was separated by 2% agarose gel
electrophoresis. The bands were visualized with the ethidium
bromide staining [Tiangen Biotech (Beijing) Co., Ltd., Beijing,
China]. The DNA fragments were visualized with a long wave UV light
monitor at 254 nm.
Western blot analysis
Total protein was extracted from the tissues and GC
cells with the Protein Extraction kit (Nanjing KeyGen Biotech Co.,
Ltd., Nanjing, China). The protein concentration was determined
with the bicinchoninic protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Cell protein lysates (20 µg/lane) were separated
by 10% SDS-PAGE, transferred to 0.22 µm nitrocellulose membranes
(Sigma-Aldrich; Merck KGaA). The membrane was firstly blocked with
5% non-fat milk at room temperature for 1 h. The membrane was then
incubated with primary antibodies, including anti-GAPDH (1:2,000,
ab37168; Abcam, Cambridge, MA, USA) and anti-HER2 (1:1,000, cat.
no. 4290; Cell Signaling Technology, Inc., Danvers, MA, USA) for 2
h at room temperature. GAPDH served as the control. Subsequently,
the membrane was incubated with horseradish peroxidase-linked goat
anti-rabbit or anti-mouse immunoglobulin G (1:5,000, sc-2007 or
sc-2005, respectively; Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA). The bands were visualized with the Pierce™ ECL
Western Blotting Substrate (Pierce; Thermo Fisher Scientific,
Inc.). Autoradiograms were quantified by densitometry analysis
using Quantity One software (version 4.62; Bio-Rad Laboratories,
Inc., Hercules, CA, USA). GAPDH was used as the control of protein
expression level.
Statistical analysis
The results are presented as the mean ± standard
error of the mean of five independent experiments. Statistical
analyses were performed using GraphPad Prism 5.0 software (GraphPad
Software, Inc., La Jolla, CA, USA). The Student's t-test was used
to analyze differences between groups. The association between H19
expression and pathological characteristics were analyzed by
one-way analysis of variance followed by a Tukey's test and binary
logistic regression analysis. P<0.05 was considered to indicate
a statistically significant difference.
Results
H19 is highly expressed in GC tissues
and cell lines
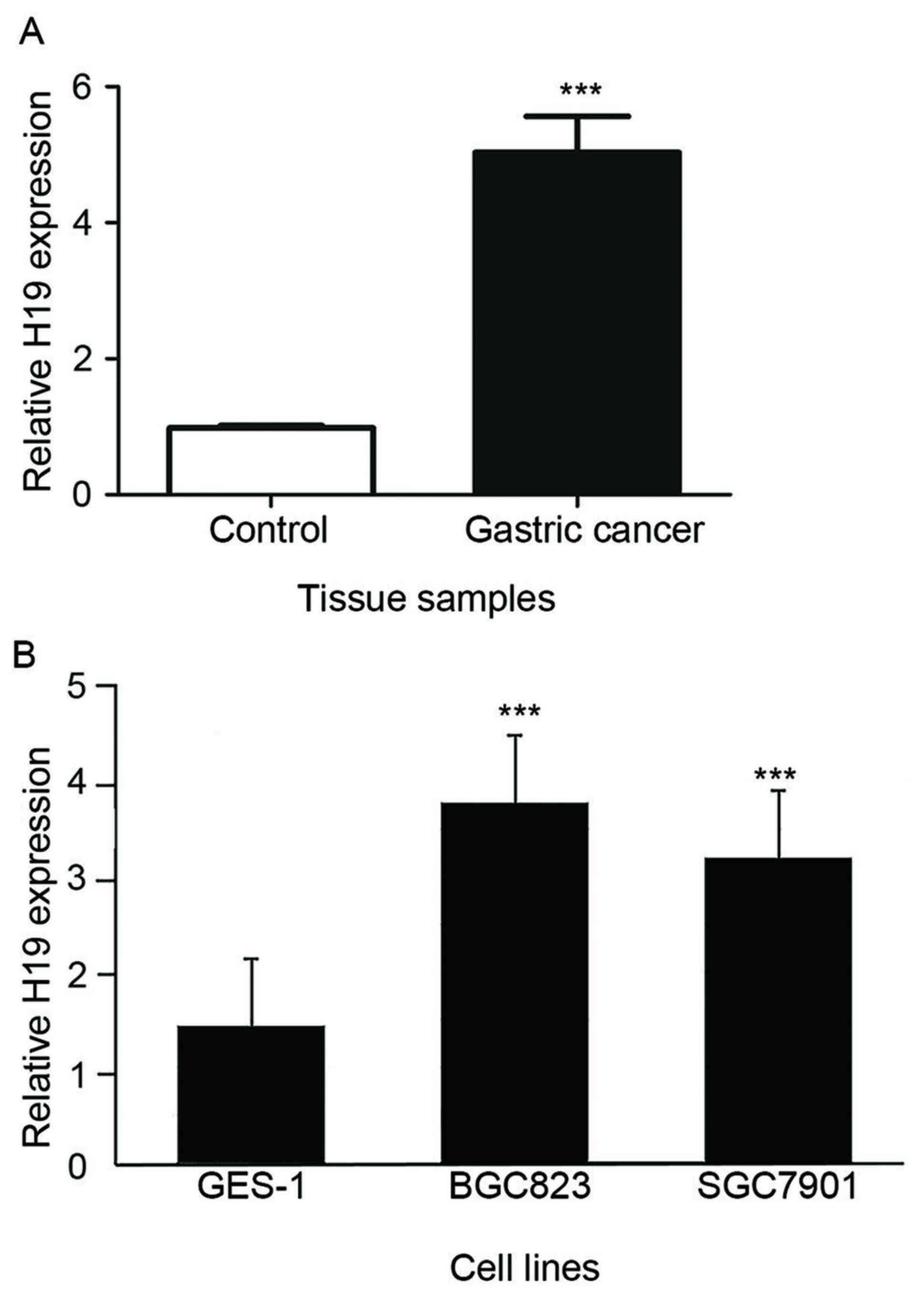
RT-qPCR analysis indicated that H19 expression was
significantly higher in 24 GC tissues when compared the normal
adjacent control tissue samples (5.23±0.34, P<0.001; Fig. 1A). In addition, the expression
levels of H19 in GC cell lines, BGC823 and SGC7901 (3.68±0.23 and
3.14±0.24, respectively), were significantly increased when
compared with normal control GES-1 cells (P<0.001; Fig. 1B).
GC tissues that express high levels of
H19 expression are HER2-positive
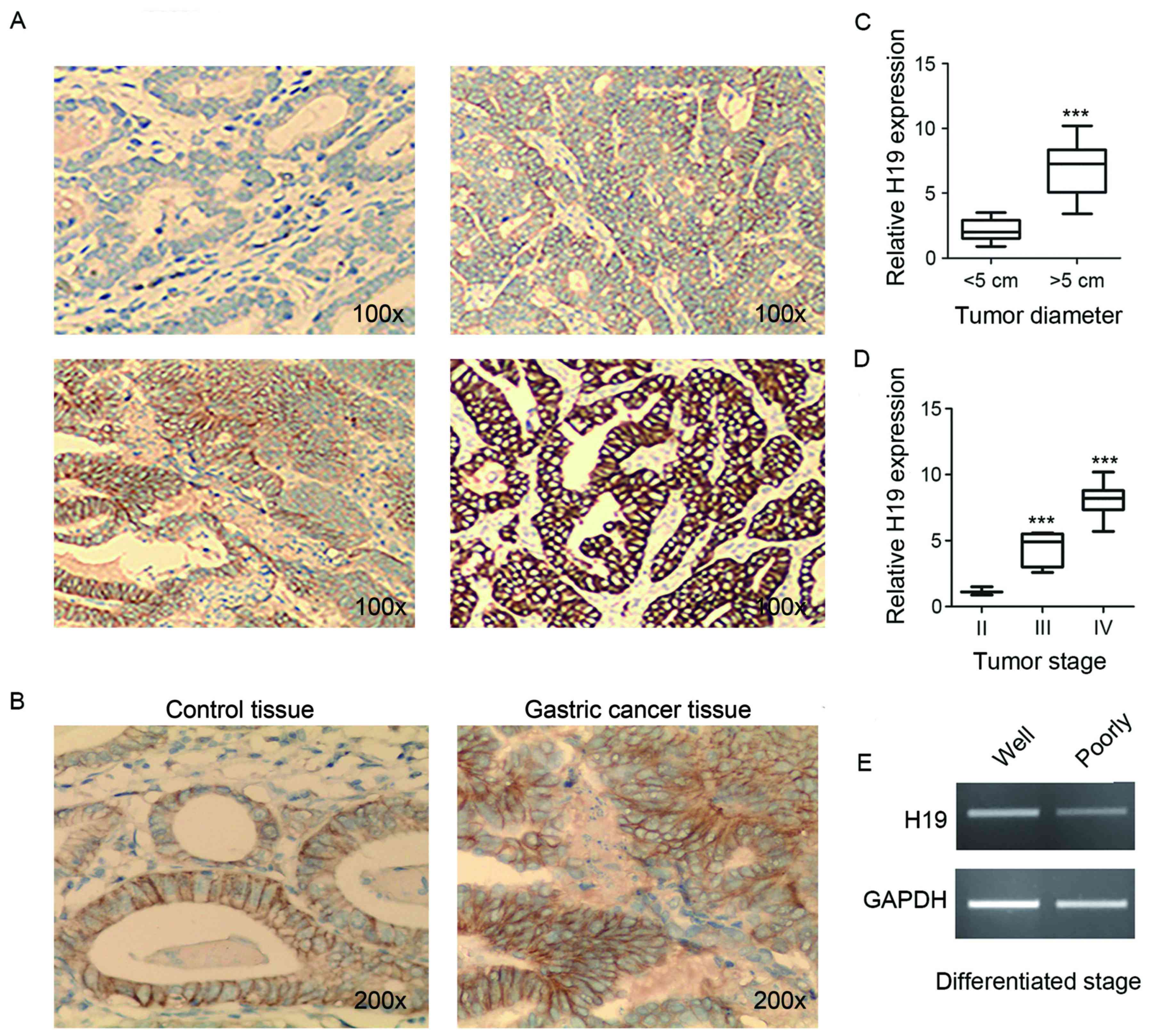
Immunostaining analysis of the 24 GC tissue samples
and paired normal controls indicated that 50.0% of GC tissue
samples that highly expressed H19 were HER2-positive, and lower H19
expression samples were observed to exhibit a lower HER2-positive
rate (8.3%; Table II). As
presented in Fig. 2A, the standard
score of HER2: Top left panel, 0; top right panel, 1; bottom left
panel, 2; and bottom right panel, 3. The standard score indicated
that the score of HER2 was significantly higher in gastric tissue
compared with in the control. As presented in Fig. 2B, the score of HER2 was nearly 0
within the control and nearly 2 with in the gastric cancer tissue.
In addition, H19 expression levels were significantly higher in the
group with tumor diameters >5 cm compared with in the group with
diameters <5 cm (Fig. 2C). The
more advance the TNM stage, the grater the average expression
levels of H19 (Fig. 2D).
Furthermore, H19 expression levels were higher in poorly
differentiated tissues compared with in well-differentiated tissues
(Fig. 2E).
 | Table II.Concordance of HER2 status with H19
expression levels in gastric cancer and adjacent normal gastric
tissue samples. |
Table II.
Concordance of HER2 status with H19
expression levels in gastric cancer and adjacent normal gastric
tissue samples.
| A, Gastric
cancer |
|---|
|
|---|
| HER2 score | H19 high | H19 low |
|---|
| 0/1+ | 4 | 6 |
| 2+/3+ | 12 | 2 |
| Percentage of HER2
positive (%) | 50.0 (12/24) | 8.3 (2/24) |
|
| B, Adjacent
benign tissues |
|
| HER2
score | H19
high | H19 low |
|
| 0/1+ | 14 | 7 |
| 2+/3+ | 2 | 1 |
| Percentage of HER2
positive (%) | 8.3 (2/24) | 4.2 (1/24) |
H19 silencing increases let-7c
expression in GC cells
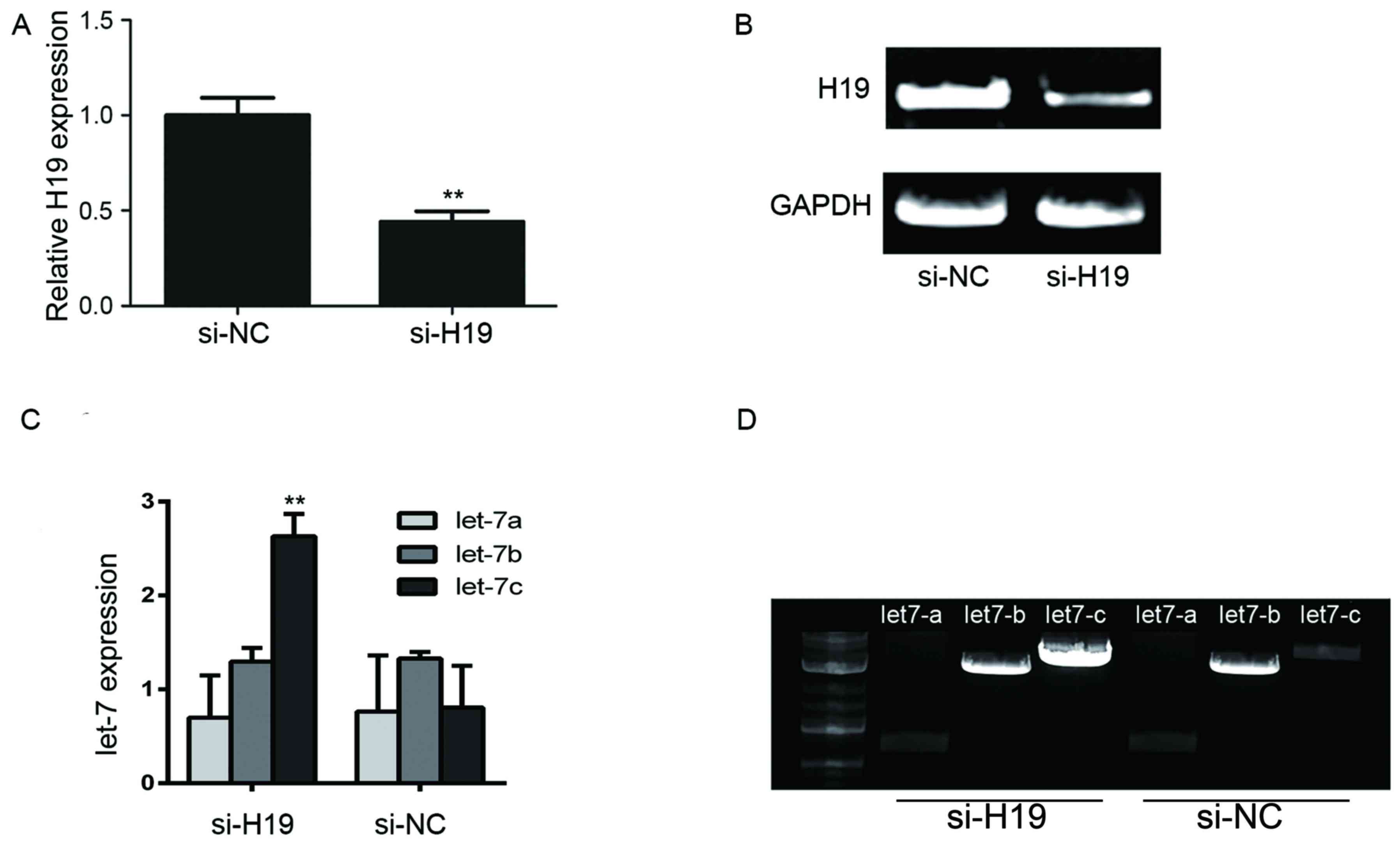
To assess the effect of H19 silencing on the
expression levels of let-7a, let-7b and let-7c miRNAs in GC,
BGC-823 cells were transfected with H19 siRNA sequences, and
RT-qPCR was used to analyze the expression of let-7 miRNAs. BGC-823
cells transfected with H19 siRNA expressed significantly lower
levels of H19 expression when compared with scrambled controls
(P<0.01; Fig. 3A and B). In
addition, the expression of let-7a/b/c with or without si-H19
transfection was detected; the expression of let-7c was
significantly increased in the H19 siRNA-transfected BGC-823 cells
compared with in si-NC transfected cells (P<0.01; Fig. 3C and D). As presented in Fig. 3C, let-7a expression levels were and
let-7b exhibited high expression levels within BGC-823 cell
expressing si-NC. The results of the present study revealed that
the expression of let-7a/b may not significantly change with or
without H19 siRNA transfection.
H19 silencing decreases HER2
expression in GC cells
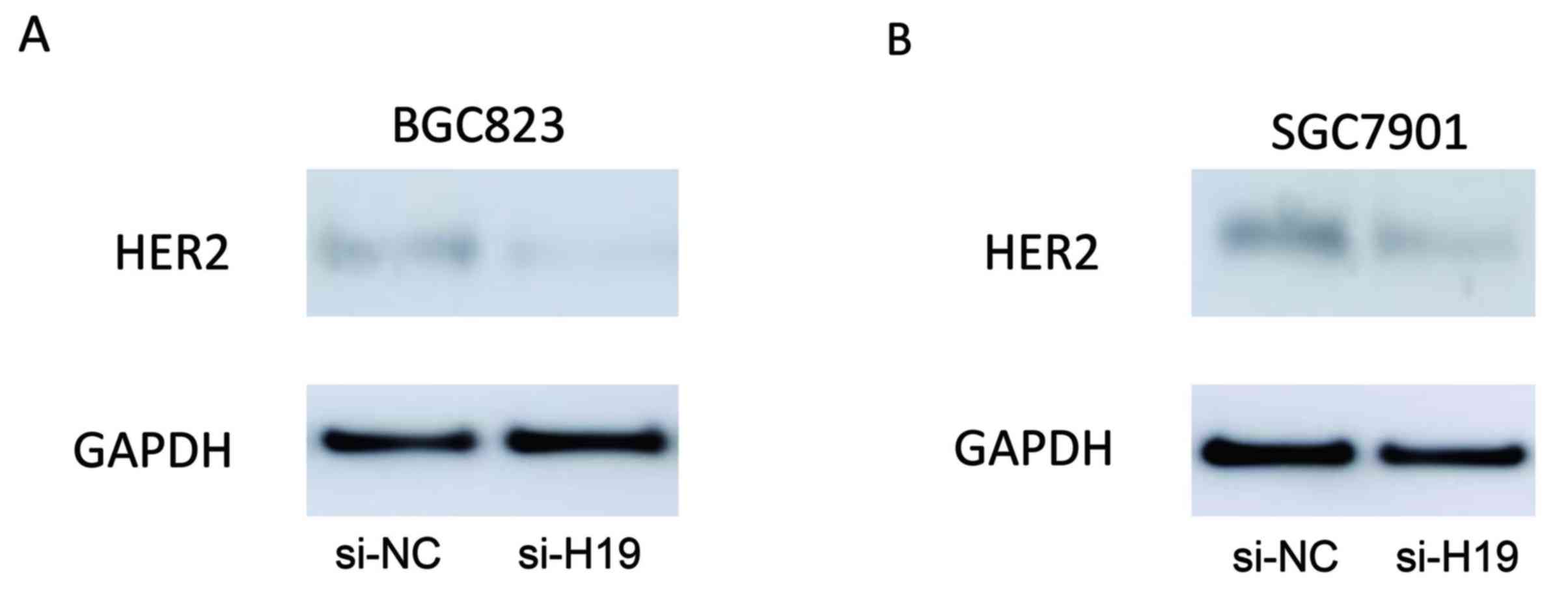
The expression levels of HER2 in BGC823 and SGC7901
cells transfected with H19 siRNA were assessed using western blot
analysis. As demonstrated in Fig.
4, H19-silenced BGC823 and SGC7901 cells expressed markedly
reduced levels of HER2 protein expression when compared with their
respective scrambled controls.
Discussion
lncRNAs are RNA transcripts consisting of ~200
nucleotides with no protein-encoding functions (21). An increasing number of studies have
suggested that the molecular mechanisms of carcinogenesis are
relevant not only to protein-encoding genes, but also to non-coding
RNAs (22–27). Previous findings have indicated
that functional alterations of specific lncRNAs promote tumor
formation, progression and metastasis in various human malignancies
(28).
The H19 lncRNA was first identified to be expressed
in developing embryos and adult muscles (29). H19 binds to and sequesters let-7
miRNA family members, thus inhibiting their function (30). The let-7-binding sites on H19 have
been demonstrated to sequester let-7 miRNAs in a variety of cell
types and among various species.
The majority of studies have indicated that
decreased miRNA expression is associated with cancer progression
(31–35). These studies suggested the tumor
suppressor or oncogenic function of miRNAs in tumors. A previous
study revealed that the let-7 family may serve estrogen-dependent
and estrogen-independent roles in estrogen receptor-positive cancer
types (36).
The present study examined the expression of HER2 in
GC tissues and cell lines. The results indicated a positive
correlation between H19 and HER2 expression in GC tissue samples.
The results of the current study suggest that high levels of H19
expression in GC tumors may be associated with increased tumor size
and a more advanced tumor stage compared with tumors expressing
lower levels of H19. Similarly, previous studies have reported that
GC patients with increased H19 or HER2 expression demonstrated a
poorer outcome and response to endocrine therapy (37–39).
Notably, Peng et al (39)
identified that HER2 expression may correlate with the expression
of Lin28 and its homolog Lin28b. These proteins bind to the
stem-loop of let-7 miRNA precursors to directly inhibit the Drosha-
and Dicer-mediated processing of their primary-miRNA precursors
into mature let-7 miRNAs (40).
Furthermore, Lin28 expression has been demonstrated to regulate the
expression of let-7 miRNA family members in tumors and cell lines
(41). Lin28 is transcriptionally
regulated by Myc, which is an estrogen receptor-regulated gene that
is associated with H19 expression (42). In addition, Lin28 is targeted by
let-7, which suggests that H19, Lin28, let-7 and Myc may function
as part of a regulatory loop. These findings suggest a novel
endogenous gene target as a therapeutic strategy for GC.
In conclusion, the results of the present study
demonstrated that H19 may function as a ceRNA to regulate HER2
expression by sequestering let-7c in GC cells. In addition, high
expression levels of H19 may be associated with poorer prognosis
for patients with GC. Further studies may be encouraged to
investigate the outcome of GC patients with high or low expression
abundance of H19 and determine the correlation between the
expression of H19 and the overall survival of GC patients.
References
|
1
|
Bailey ST, Westerling T and Brown M: Loss
of estrogen-regulated microRNA expression increases HER2 signaling
and is prognostic of poor outcome in luminal breast cancer. Cancer
Res. 75:436–445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen R, Zhou X, Liu J and Huang G:
Relationship between 18F-FDG PET/CT findings and HER2 expression in
gastric cancer. J Nucl Med. 57:1040–1044. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Darb-Esfahani S, Denkert C, Stenzinger A,
Salat C, Sinn B, Schem C, Endris V, Klare P, Schmitt W, Blohmer JU,
et al: Role of TP53 mutations in triple negative and HER2-positive
breast cancer treated with neoadjuvant anthracycline/taxane-based
chemotherapy. Oncotarget. 7:67686–67698. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudogene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
De Martino M, Forzati F, Marfella M,
Pellecchia S, Arra C, Terracciano L, Fusco A and Esposito F:
HMGA1P7-pseudogene regulates H19 and Igf2 expression by a
competitive endogenous RNA mechanism. Sci Rep. 6:376222016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Degrauwe N, Suvà ML, Janiszewska M, Riggi
N and Stamenkovic I: IMPs: An RNA-binding protein family that
provides a link between stem cell maintenance in normal development
and cancer. Genes Dev. 30:2459–2474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao Y, Wu F, Zhou J, Yan L, Jurczak MJ,
Lee HY, Yang L, Mueller M, Zhou XB, Dandolo L, et al: The H19/let-7
double-negative feedback loop contributes to glucose metabolism in
muscle cells. Nucleic Acids Res. 42:13799–13811. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Wang H, Yao B, Xu W, Chen J and Zhou
X: lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by
targeting VDAC1 in diabetic cardiomyopathy. Sci Rep. 6:363402016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liang WC, Fu WM, Wang YB, Sun YX, Xu LL,
Wong CW, Chan KM, Li G, Waye MM and Zhang JF: H19 activates Wnt
signaling and promotes osteoblast differentiation by functioning as
a competing endogenous RNA. Sci Rep. 6:201212016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gao P, Zhang C, Bian X, Guo Y, Wei Y,
Zhang L, Liu Z, Wang X and Huang S: The increasingly anti-tumor
effect of a colonic carcinoma DNA vaccine carrying HER2 by the
adjuvanticity of IL-12. Immunopharmacol Immunotoxicol. 1–6.
2016.(Epub ahead of print). PubMed/NCBI
|
|
14
|
Hanna MG, Bleiweiss IJ, Nayak A and Jaffer
S: Correlation of Oncotype DX recurrence score with histomorphology
and immunohistochemistry in over 500 patients. Int J Breast Cancer.
2017:12570782017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lambein K, Van Bockstal M, Vandemaele L,
Geenen S, Rottiers I, Nuyts A, Matthys B, Praet M, Denys H and
Libbrecht L: Distinguishing score 0 from score 1+ in HER2
immunohistochemistry-negative breast cancer: Clinical and
pathobiological relevance. Am J Clin Pathol. 140:561–566. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meller S, Meyer HA, Bethan B, Dietrich D,
Maldonado SG, Lein M, Montani M, Reszka R, Schatz P, Peter E, et
al: Integration of tissue metabolomics, transcriptomics and
immunohistochemistry reveals ERG- and gleason score-specific
metabolomic alterations in prostate cancer. Oncotarget.
7:1421–1438. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Seyed Jafari SM and Hunger RE: IHC optical
density score: A new practical method for quantitative
immunohistochemistry image analysis. Appl Immunohistochem Mol
Morphol. 25:e12–e13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Viúdez A, Carvalho FL, Maleki Z, Zahurak
M, Laheru D, Stark A, Azad NS, Wolfgang CL, Baylin S, Herman JG and
De Jesus-Acosta A: A new immunohistochemistry prognostic score
(IPS) for recurrence and survival in resected pancreatic
neuroendocrine tumors (PanNET). Oncotarget. 7:24950–24961. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Viudez A, Carvalho FL, Maleki Z, Zahurak
M, Laheru D, Stark A, Azad NS, Wolfgang CL, Baylin S, Herman JG and
De Jesus-Acosta A: Correction: A new immunohistochemistry
prognostic score (IPS) for recurrence and survival in resected
pancreatic neuroendocrine tumors (PanNET). Oncotarget.
8:186172017.PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu C, Chen Z, Fang J, Xu A, Zhang W and
Wang Z: H19-derived miR-675 contributes to bladder cancer cell
proliferation by regulating p53 activation. Tumour Biol.
37:263–270. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herrera-Marcos LV, Lou-Bonafonte JM, Arnal
C, Navarro MA and Osada J: Transcriptomics and the mediterranean
diet: A systematic review. Nutrients. 9:E4722017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu C, Zhang YH, Deng Q, Li Y, Huang T,
Zhou S and Cai YD: Cancer-Related Triplets of mRNA-lncRNA-miRNA
revealed by integrative network in uterine corpus endometrial
carcinoma. Biomed Res Int. 2017:38595822017.PubMed/NCBI
|
|
24
|
Mao Y, Liu R, Zhou H, Yin S, Zhao Q, Ding
X and Wang H: Transcriptome analysis of miRNA-lncRNA-mRNA
interactions in the malignant transformation process of gastric
cancer initiation. Cancer Gene Ther. 24:267–275. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu Q, Guo L, Jiang F, Li L, Li Z and Chen
F: Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER-breast
cancer cell lines. J Cell Mol Med. 19:2874–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yao K, Wang Q, Jia J and Zhao H: A
competing endogenous RNA network identifies novel mRNA, miRNA and
lncRNA markers for the prognosis of diabetic pancreatic cancer.
Tumour Biol. 39:10104283177078822017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye S, Yang L, Zhao X, Song W, Wang W and
Zheng S: Bioinformatics method to predict two regulation mechanism:
TF-miRNA-mRNA and lncRNA-miRNA-mRNA in pancreatic cancer. Cell
Biochem Biophys. 70:1849–1858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cai H, Yao J, An Y, Chen X, Chen W, Wu D,
Luo B, Yang Y, Jiang Y, Sun D and He X: lncRNA HOTAIR acts a
competing endogenous RNA to control the expression of notch3 via
sponging miR-613 in pancreatic cancer. Oncotarget. 8:32905–32917.
2017.PubMed/NCBI
|
|
29
|
Keniry A, Oxley D, Monnier P, Kyba M,
Dandolo L, Smits G and Reik W: The H19 lincRNA is a developmental
reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell
Biol. 14:659–665. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin LT, Chang CY, Chang CH, Wang HE, Chiou
SH, Liu RS, Lee TW and Lee YJ: Involvement of let-7 microRNA for
the therapeutic effects of Rhenium-188-embedded liposomal
nanoparticles on orthotopic human head and neck cancer model.
Oncotarget. 7:65782–65796. 2016.PubMed/NCBI
|
|
31
|
Lodewijk L, Prins AM, Kist JW, Valk GD,
Kranenburg O, Rinkes IH and Vriens MR: The value of miRNA in
diagnosing thyroid cancer: A systematic review. Cancer Biomark.
11:229–238. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Srivastava K and Srivastava A:
Comprehensive review of genetic association studies and
meta-analyses on miRNA polymorphisms and cancer risk. PLoS One.
7:e509662012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tilghman SL, Rhodes LV, Bratton MR,
Carriere P, Preyan LC, Boue SM, Vasaitis TS, McLachlan JA and Burow
ME: Phytoalexins, miRNAs and breast cancer: A review of
phytochemical-mediated miRNA regulation in breast cancer. J Health
Care Poor Underserved. 24 Suppl 1:S36–S46. 2013. View Article : Google Scholar
|
|
34
|
Wang QX, Zhu YQ, Zhang H and Xiao J:
Altered miRNA expression in gastric cancer: A systematic review and
meta-analysis. Cell Physiol Biochem. 35:933–944. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu X, Yang X, Xing C, Zhang S and Cao J:
miRNA: The nemesis of gastric cancer (Review). Oncol Lett.
6:631–641. 2013.PubMed/NCBI
|
|
36
|
Zhao Y, Deng C, Wang J, Xiao J, Gatalica
Z, Recker RR and Xiao GG: Let-7 family miRNAs regulate estrogen
receptor alpha signaling in estrogen receptor positive breast
cancer. Breast Cancer Res Treat. 127:69–80. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nonagase Y, Yonesaka K, Kawakami H,
Watanabe S, Haratani K, Takahama T, Takegawa N, Ueda H, Tanizaki J,
Hayashi H, et al: Heregulin-expressing HER2-positive breast and
gastric cancer exhibited heterogeneous susceptibility to the
anti-HER2 agents lapatinib, trastuzumab and T-DM1. Oncotarget.
7:84860–84871. 2016.PubMed/NCBI
|
|
38
|
Ohtsuka M, Ling H, Ivan C, Pichler M,
Matsushita D, Goblirsch M, Stiegelbauer V, Shigeyasu K, Zhang X,
Chen M, et al: H19 noncoding RNA, an independent prognostic factor,
regulates essential Rb-E2F and CDK8-β-catenin signaling in
colorectal cancer. EBioMedicine. 13:113–124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peng F, Li TT, Wang KL, Xiao GQ, Wang JH,
Zhao HD, Kang ZJ, Fan WJ, Zhu LL, Li M, et al: H19/let-7/LIN28
reciprocal negative regulatory circuit promotes breast cancer stem
cell maintenance. Cell Death Dis. 8:e25692017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ravid O, Shoshani O, Sela M, Weinstock A,
Sadan TW, Gur E, Zipori D and Shani N: Relative genomic stability
of adipose tissue derived mesenchymal stem cells: Analysis of
ploidy, H19 long non-coding RNA and p53 activity. Stem Cell Res
Ther. 5:1392014. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Reese KJ, Lin S, Verona RI, Schultz RM and
Bartolomei MS: Maintenance of paternal methylation and repression
of the imprinted H19 gene requires MBD3. PLoS Genet. 3:e1372007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Scrima M, Zito Marino F, Oliveira DM,
Marinaro C, La Mantia E, Rocco G, De Marco C, Malanga D, De Rosa N,
Rizzuto A, et al: Aberrant signaling through the HER2-ERK1/2
pathway is predictive of reduced disease-free and overall survival
in early stage non-small cell lung cancer (NSCLC) patients. J
Cancer. 8:227–239. 2017. View Article : Google Scholar : PubMed/NCBI
|