Introduction
Intervertebral disc degeneration (IDD) has become
the most common cause of low-back pain, which seriously affects the
lives of patients and leads to ~10% of patients becoming
chronically disabled (1,2). Patients with IDD exhibit
characteristics, including the proliferation of intervertebral disc
tissues, which consist of the central gelatinous nucleus pulposus
(NP) and the outer lamellar annulus fibrosus (3). The degeneration of intervertebral
discs is caused by multiple factors, including aging, physical
loading, genetic predisposition and lifestyle (4,5).
Accumulating pathological conditions result in the abnormal
degradation of proteoglycan and type II collagen generated by NP,
leading to the remodeling of disc structures and vertebrae
(3). Increasing studies have
demonstrated that a large number of cellular events are involved in
the degeneration of intervertebral discs, ranging from gene
expression to protein synthesis. Among these events, the complex
progression of gene expression has been found to be vital in IDD
(6). Long non-coding RNAs
(lncRNAs), as an important factor in gene expression in IDD, have
attracted substantial attention in previous years (7–9).
It is known that lncRNAs comprise a variety of
transcriptional byproducts of the mammalian genome without
protein-coding function (2). They
have been identified to be important regulators in differential
biological functions, including genome imprinting, gene expression,
epigenetic regulation and chromatin modification (10–12).
In addition, increasing evidence has shown that the dysregulation
of lncRNAs exerts significant function in several human diseases,
including ovarian cancer, hepatocellular carcinoma, colorectal
cancer and fatty liver disease (13–16).
Fas-associated protein factor-1 (FAF1) is an lncRNA and is a member
of the Fas death-inducing signaling complex, as its name suggests
(17). A previous study indicated
that FAF1 is upregulated in IDD (18). However, to the best of our
knowledge, the role and molecular mechanism underlying the effect
of FAF1 in IDD has not to be investigated.
The present study aimed to investigate the role of
FAF1 in IDD. Initially, the expression of FAF1 and its correlation
with Pfirrmann scores were detected in patients with IDD.
Subsequently, the role of FAF1 on cell growth, proliferation and
apoptosis was examined in patients with IDD. The findings suggested
that the overexpression of FAF1 may act as a suppressor in the
development of IDD by targeting the extracellular signal-regulated
(Erk) signaling pathway.
Materials and methods
Patients and tissue samples
The human central NP specimens were collected from
patients with IDD (n=10; average age 43.79±6.33, range 35–61
years), bulging (n=10; average age 46.47±5.41, range 30–62 years),
hernia (n=10; average age 44.82±3.95, range 37–66 years), and
spinal cord injury (n=10; average age 41.63±2.72, range 31–59
years) between May 2013 and June 2016 from the Department of
Orthopedics and Traumatic Surgery, Nantong Municipal Hospital of
Traditional Chinese Medicine (Nantong, China). Each group comprised
5 males and 5 females. The degree of disc degeneration was
determined by two spinal surgeons from a magnetic resonance imaging
(MRI) scan according to the modified Pfirrmann grading
classification (19). The samples
graded as 1 were classified as a normal control, whereas samples
graded as 3, 4 and 5 were included in bulging, hernia and IDD
groups, respectively. The specimens were first isolated from the
annulus fibrosus and then divided into two, which were frozen in
liquid nitrogen at −80°C or fixed in 4% paraformaldehyde,
respectively. All samples were obtained following the provision of
informed consent. The study was approved by Nantong Municipal
Hospital of Traditional Chinese Medicine.
RNA Extraction and lncRNA expression
assay
The NP tissues were isolated from 10 IDD and 10
spinal cord injury samples, and total RNA was extracted separately
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's protocol.
Following removal of the contaminating genomic DNA, the quality and
quantity of the RNA were detected using a NanoDrop ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Wilmington, DE, USA) and an Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc., Santa Clara, CA, USA). Subsequently, gel
electrophoresis was performed to measure RNA integrity. The total
RNAs obtained were divided equally into IDD and normal control
groups. Reverse transcription was then performed using a Reverse
Transcription kit (Thermo Fisher Scientific, Inc.). The
quantitative polymerase chain reaction (qPCR) analysis was
performed using an Applied Biosystems 7900 Real-time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) using 5 µg RNA
(miRNA cDNA Synthesis kit; Takara Biotechnology Co., Ltd., Dalian,
China), with a reaction volume of 20 µl. Reaction reagents (20 µl)
were: 10 µl SYBR Premix Ex Taq II, 0.8 µl PCR Forward Primer, 0.8
µl PCR Reverse Primer, 0.4 µl ROX Reference Dye, 2.0 µl cDNA
template and 6.0 µl ddH2O. PCR was performed as follows:
10 sec at 95°C, 95 sec at 5°C, and 34 sec at 60°C for 45 cycles.
Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used for
normalization and the relative expression of FAF1was calculated
using the 2−ΔΔCq method (20). The PCR primers were as follows:
FAF1: Forward, AGA GCA AAG AGG GAA G, reverse, AAG AAC TCG CCA CTG;
GAPDH: Forward, GGG AAA CTG TGG CGT G AT; Reverse, GAG TGG GTG TCG
CTG TTG A.
Culture of NP cells
The NP tissues were cut into sections using
ophthalmic scissors, and digested with PBS containing 0.025% type
II collagenase (Invitrogen; Thermo Fisher Scientific, Inc.) for 4 h
at 37°C. Following filtration at 200 µm pore, the well-digested NP
tissues were centrifuged at 500 × g for 10 min at room temperature
and the supernatant was aspirated. The NP cells were seeded
(5×105 cells/well) into a culture flask in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 1%
streptomycin/penicillin, and then placed into an incubator with 5%
CO2, saturated humidity at 37°C for 3 days.
Plasmids and cell transfection
The full-length sequence of FAF1 was PCR-amplified
using PrimeSTAR HS DNA Polymerase (Takara Biotechnology Co., Ltd.)
and cloned into the PcDNA3.1 vector (Invitrogen; Thermo Fisher
Scientific, Inc.). PCR was performed as follows: 15 sec at 95°C, 95
sec at 5°C, and 60 sec at 60°C for 30 cycles. Short hairpin (sh)
RNA-FAF1 (5′-GGCAGCGGTCCCCGAGGAGGCGGCAG-3′) and shRNA-control
(5′-AAGTCCCTGGTCTCTGGAGACTGTTCTG-3′) were synthesized by GenePharma
Co., Ltd. (Shanghai, China). The day prior to transfection,
6×104 cells were inoculated into 6-well dishes with 2 ml
medium in every well. Transfection was performed when the cell
density reached 50–60%. The diluted transfections were mixed
carefully with Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) and cultured for 20 min at room
temperature. The mixture of plasmid oligonucleotide and
transfection reagents was added to each well containing the cells
and culture medium. The cells were incubated under conditions of
37°C and 5% CO2, and then collected following 48–72 h of
transfection.
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was used to determine the
proliferation of the transfected NP cells. Firstly, the cells were
seeded onto 96-well plates (5×103 cells/well) and 10 µl
CCK-8 was added to each well at the indicated time (1, 2, 3 and 4
days) in accordance with the manufacturer's protocol. The cells
were incubated at 37°C for 2 h. According to the absorbance of each
well at 450 nm for each group, the cell viability was assessed
using a Benchmark microplate reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Immunofluorescence staining
Following rinsing with phosphate-buffered saline
three times, the cultured NP cells were fixed with 4% formaldehyde
for 10 min, and blocked with 1% BSA (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) for 45 min. The cells were incubated for 1 h at
room temperature with primary antibody against Ki67 (rabbit
polyclonal antibody; ab15580; 1:200; Abcam, Cambridge, UK). The
cells were then rinsed with PBS and incubated with fluorescein
isothiocyanate (FITC)-conjugated goat anti-rabbit IgG antibody
(goat polyclonal antibody; ab6662; 1:100; Abcam) for 1 h at room
temperature. Subsequently, the cells were analyzed using a
fluorescence microscope (Olympus, Tokyo, Japan).
Analysis of cell cycle and
apoptosis
The effects of FAF1 on cell cycle and apoptosis were
measured using flow cytometry. First, the cells were cultured for
48 h, trypsinized and fixed with 70% ethanol at −20°C for 12 h
according to the manufacturer's protocol. Following washing with
PBS, the cells were treated with 20 µg/ml RNaseA (Sigma-Aldrich;
Merck KGaA) for 2 h at 37°C, then incubated with 50 µg/ml propidium
iodide (Sigma-Aldrich; Merck KGaA) for 30 min. An Annexin-V FITC
Apoptosis kit (Tiangen Biotech Co., Ltd., Beijing, China) was used
to assess cell apoptosis according to the manufacturer's protocol.
Cell cycle and apoptosis were analyzed using FACSDiva 6.1.1 (BD
Biosciences, Franklin Lakes, NJ, USA) and ModFit LT 3.0 (Verity
Software House, Inc., Topsham, ME, USA) software.
Western blot analysis
Total protein was extracted from the transfected NP
cells using radioimmunoprecipitation assay lysis buffer
(Sigma-Aldrich; Merck KGaA) at 4°C for 30 min. Then, the proteins
were extracted by RIPA buffer (Sigma-Aldrich; Merck KGaA) and
separated on a 10% sodium dodecyl sulphate-polyacrylamide
electrophoresis gel by electrophoresis for 120 min and transferred
onto 0.45-mM PVDF membranes (EMD Millipore, Billerica, MA, USA).
The concentration of protein was quantified by a BCA kit (Beyotime
Institute of Biotechnology, Nantong, China). Following blocking in
sealing fluid for 60 min, the membranes were incubated with
anti-phosphorylated (p)-Erk (rabbit polyclonal antibody; 4370;
1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA)
antibodies overnight at 4°C. Following rinsing with TBST three
times, the membranes were incubated with a secondary antibody
(7074; 1:1,000; anti-rabbit polyclonal antibody; Cell Signaling
Technology, Inc.) for 1 h at room temperature. Enhanced
chemiluminescence reagent (Nanjing KeyGen Biotech, Inc., Nanjing,
China) was used to develop the immunoreactivity, and the protein
bands were analyzed using Quantity One software version 4.62
(Bio-Rad Laboratories, Inc.). GAPDH (mouse monoclonal antibody;
97166; 1:2,000; Cell Signaling Technology, Inc.) was used for
normalization.
Statistical analysis
All the data were analyzed using SPSS software
version 18.0 (SPSS, Inc., Chicago, IL, USA) and expressed as the
mean ± standard deviation. Differences between two groups were
evaluated using Student's t-test, and differences among three or
more groups were evaluated using one-way analysis of variance. The
correlation between the expression of FAF1 and Pfirrmann grades was
determined using Spearman's correlation analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of FAF1 is downregulated in
patients with IDD
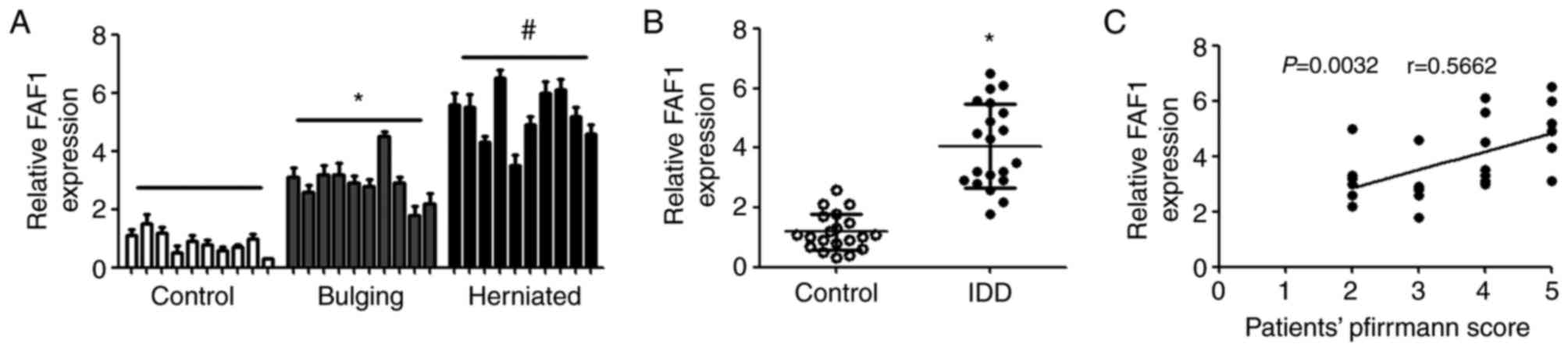
RT-qPCR analysis was performed to detect the
differential expression of FAF1 in patients with spinal cord
injury, bulging, herniated intervertebral disc and IDD. Compared
with the control group, the expression of FAF1 was upregulated in
the patients with bulging and herniation, and the level of FAF1 in
those with hernia was higher, compared with that in patients with
bulging (Fig. 1A). In addition, it
was found that the expression of FAF1 in patients with IDD was
significantly increased, compared with that in patients with spinal
cord injury (Fig. 1B). The
correlations between the expression of FAF1 and the severity of IDD
were also examined. The results showed that there were significant
positive correlations between the expression of FAF1 and patients'
Pfirrmann scores (Fig. 1C).
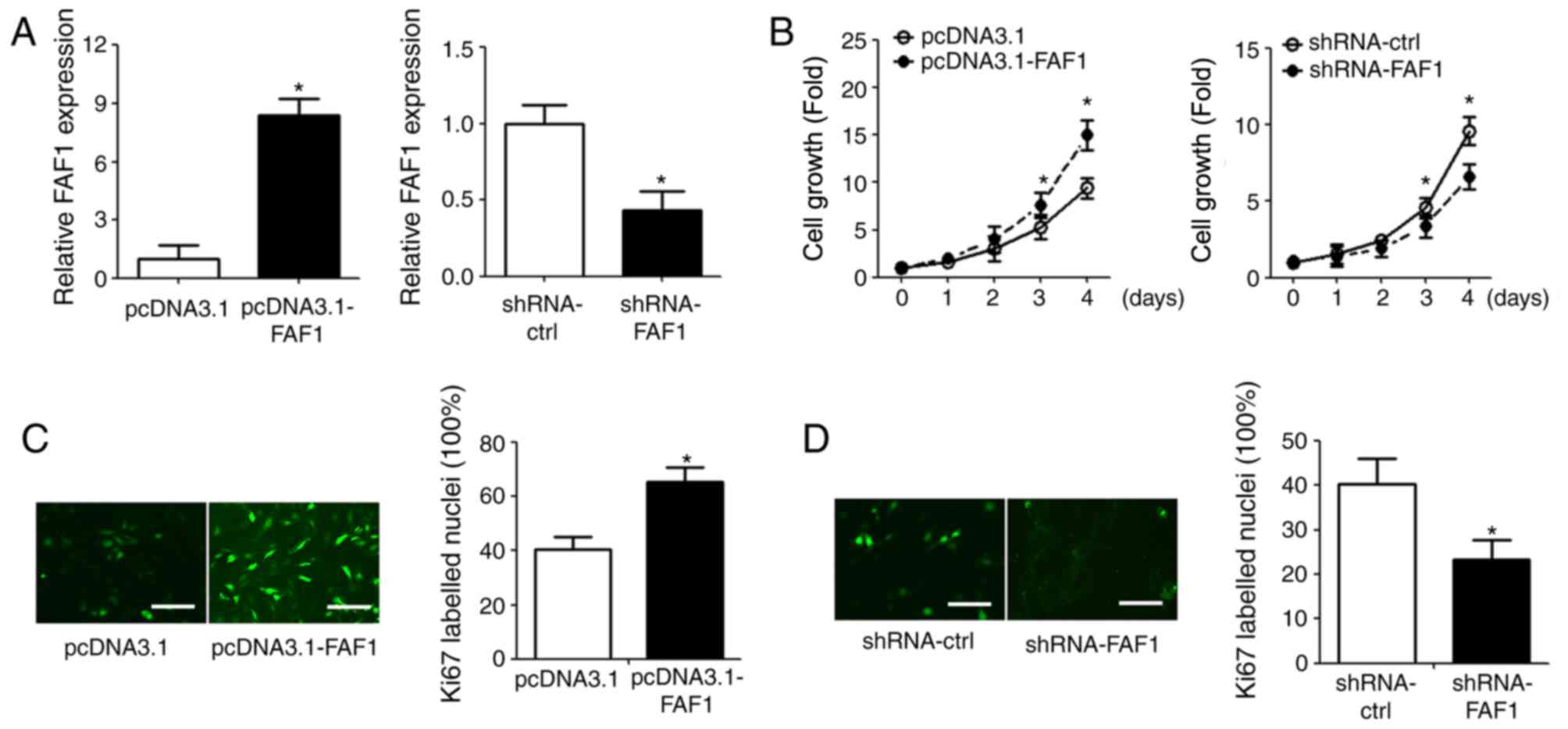
Overexpression of FAF1 promotes NP
cell proliferation
To investigate the role of FAF1 in NP cells, the
FAF1 expression vector and FAF1-specific shRNA were transfected
into NP cells (Fig. 2A). The
effect of FAF1 on NP cell proliferation was measured using a CCK-8
assay. The overexpression of FAF1 in NP cells significantly
promoted cell growth, compared with that in the control group
(P<0.05); the shRNA-FAF1 group exhibited decreased cell growth,
compared with that in the shRNA-ctrl group (Fig. 2B). The results of the
immunofluorescence staining showed similar results; Ki67
fluorescence intensity was upregulated in the FAF1-overexpressing
group and downregulated in the shRNA-FAF1 group, compared with
intensity in the corresponding control groups (Fig. 2C and D).
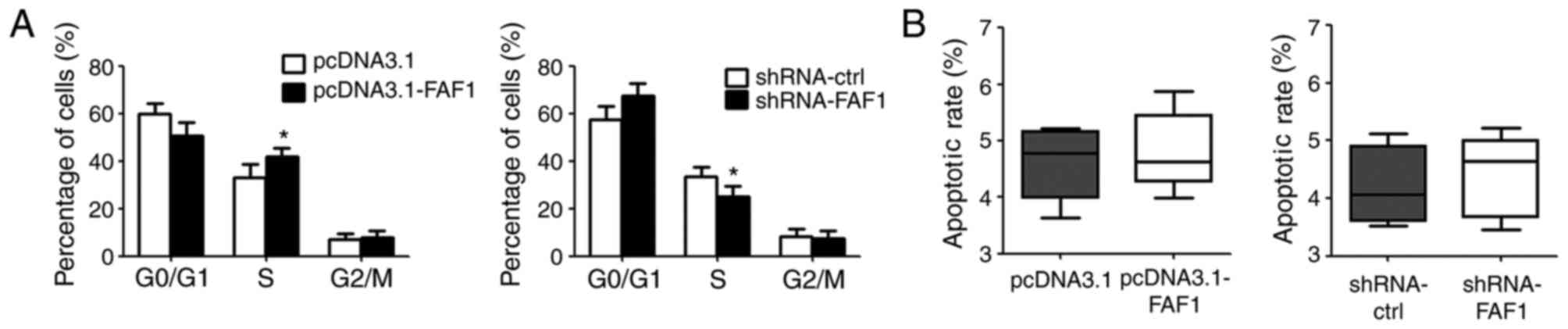
FAF1 increases the percentage of cells
in the S-phase of the cell cycle
To elucidate the pro-growth mechanism of FAF1 in NP
cells, the present study assessed the cell cycle and cell
apoptosis. According to the results of the flow cytometric assay,
the overexpression of FAF1 in NP cells increased the percentage of
cells in the S-phase, compared with that in the control group. By
contrast, the downregulation of FAF1 in NP cells reduced the
percentage of cells in the S-phase (Fig. 3A). However, no significant
differences in apoptotic rate were found between the
FAF1-overexpressing group or shRNA-FAF1 group and their
corresponding control groups (Fig.
3B).
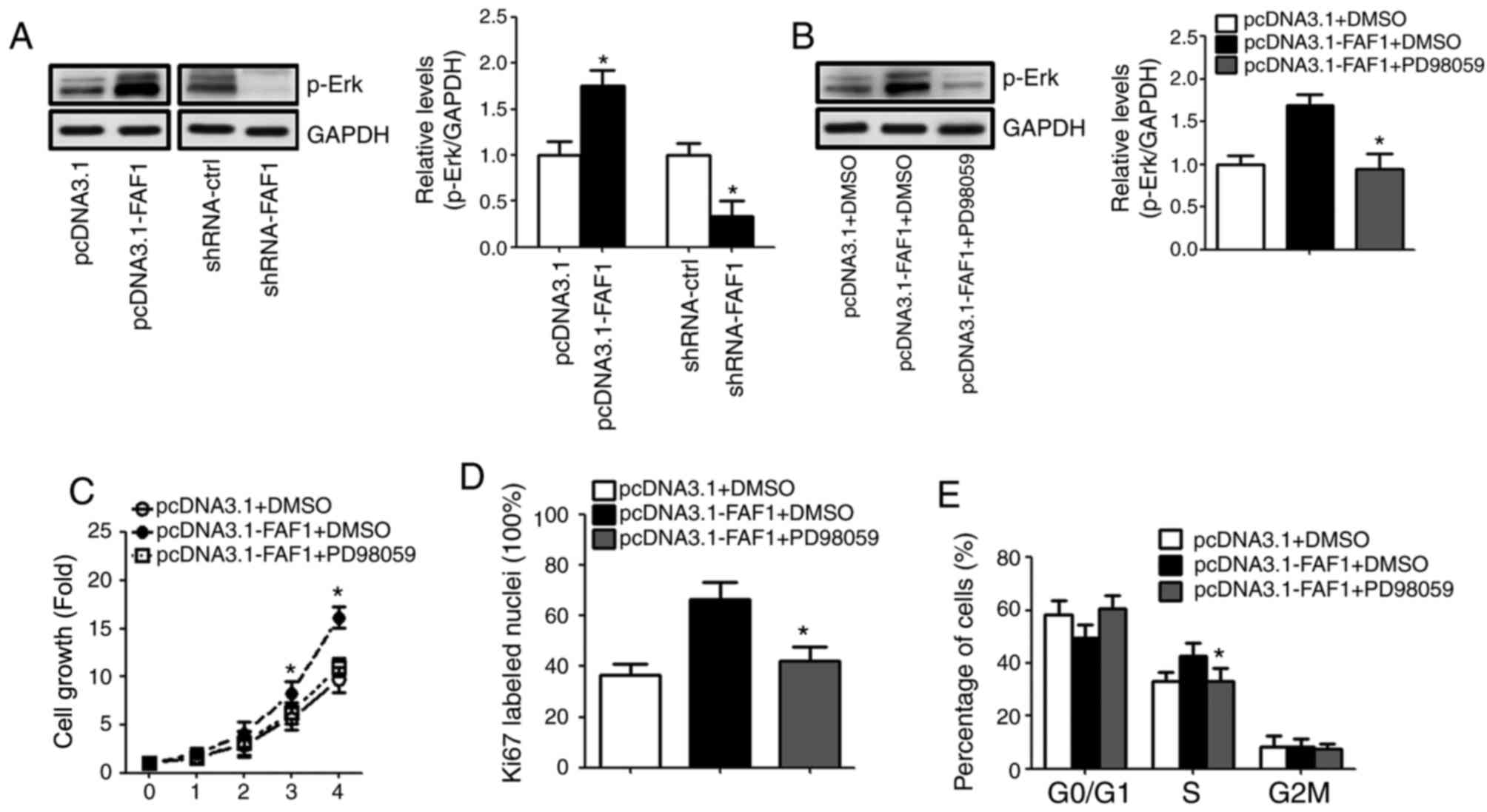
FAF1 promotes NP cell proliferation
directly via targeting the Erk signaling pathway
The results of the western blot analysis showed that
the expression of p-Erk increased with the presence of the FAF1
expression vector. By contrast, the level of p-Erk in the
shRNA-FAF1 group was decreased, compared with that in the control
group (Fig. 4A). To investigate
whether FAF1 promoted NP cell proliferation by modulating Erk
signaling pathway activation, PD98059 was used to inhibit the
activation of the Erk signaling pathway. The results showed that
the expression of p-Erk decreased sharply following treatment with
PD98059 (Fig. 4B). The results
showed that PD98059 eliminated the FAF1-induced cell proliferation
in NP cells (Fig. 4C). Similarly,
the Ki67 immunofluorescence staining and flow cytometry indicated
that PD98059 reduced the percentage of Ki67-labelled nuclei and
S-phase cells in the FAF1-overexpressing NP cells (Fig. 4D and E).
Discussion
Patients with IDD suffer from pain physically and
psychologically. The investigation of novel diagnostic and
therapeutic strategies is required to assist in improving patient
quality of life (6,21–23).
Accumulating evidence has demonstrated that lncRNAs are important
in the development and progression of IDD (24,25).
At present, the diagnosis of IDD is based on advanced imaging
techniques, including MRI (26).
However, the information provided by MRI reveals only intermediate
or late stages of IDD, whereas the degradation of proteoglycans,
particularly aggrecan, and extracellular matrix changes are early
signs of IDD. Therefore, the investigation of more precise
diagnostic methods for the initial stage of IDD is required. It has
been reported that certain small molecules, including lncRNAs, are
associated with IDD, and that the dysregulation of these molecules
is key in the progression of gene transcription (27).
In the present study, the expression of FAF1 was
analyzed in 40 tissue samples from patients with IDD, bulging,
herniation and spinal cord injury. The results demonstrated that
the expression level of FAF1 was upregulated, with the degree of
aggravated degeneration and the expression of FAF1 in IDD being
significantly higher, compared with those of the control group,
suggesting that FAF1 may be closely associated with the development
of IDD. In addition, patients' Pfirrmann scores showed that the
expression of FAF1 was positively correlated with the grade of disc
degeneration, which suggested that FAF1 may act as a promoter in
the development of IDD. To confirm the role of FAF1, FAF was
overexpressed or knocked down in primary NP cells. The CCK-8 assay
and immunofluorescence staining demonstrated that the
overexpression of FAF1 promoted NP cell proliferation and that the
knockdown of FAF1 suppressed NP cell proliferation. Consistently, a
previous study reported that FAF1 was upregulated in degenerative
discs (2). The present study also
found that FAF1 regulated NP cell proliferation via controlling
cell cycle. These results indicated that FAF1 may be a novel
inducer in the progression of IDD by promoting cell
proliferation.
To elucidate the molecular mechanism underlying the
effect of FAF1, p-Erk was confirmed as a target gene of FAF1, and
it was found that the expression of FAF1 was positively associated
with p-Erk. Erk is one of the members of the mitogen-activated
protein kinase pathway, which is important in cell survival, cell
growth and programmed cell death signaling events (28). p-Erk is reported to be closely
associated with cell survival and cell growth in several human
diseases, including ovarian cancer (29) and hepatotoxicity (30). The present study showed that p-Erk
was upregulated with the overexpression of FAF1, but downregulated
when FAF1 was knocked down, compared with that in the control
group, suggesting that FAF1 may be connected with Erk signaling
pathway activation. In addition, PD98059, an inhibitor of the Erk
signaling pathway, eliminated FAF1-induced cell proliferation in NP
cells. These results indicated that FAF1 may promote cell
proliferation by modulating Erk pathway activation, which is
consistent with the results of previous studies (29,30).
In conclusion, the present study found that the
expression of FAF1 was upregulated in IDD and that the expression
level of FAF1 was positively correlated with the grade of disc
degeneration. These results elucidated an important molecular
mechanism, in which FAF1 exerted a positive effect on cell
proliferation in IDD by upregulating p-Erk. Taken together, FAF1
may be a novel marker in the early diagnosis of IDD and a
therapeutic target for patients.
References
|
1
|
Vasiliadis ES, Pneumaticos SG,
Evangelopoulos DS and Papavassiliou AG: Biologic treatment of mild
and moderate intervertebral disc degeneration. Mol Med. 20:400–409.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liu H, Huang X, Liu X, Xiao S, Zhang Y,
Xiang T, Shen X, Wang G and Sheng B: miR-21 promotes human nucleus
pulposus cell proliferation through PTEN/AKT signaling. Int J Mol
Sci. 15:4007–4018. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Jarman JP, Arpinar VE, Baruah D, Klein AP,
Maiman DJ and Muftuler LT: Intervertebral disc height loss
demonstrates the threshold of major pathological changes during
degeneration. Eur Spine J. 24:1944–1950. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Samartzis D, Karppinen J, Mok F, Fong DY,
Luk KD and Cheung KM: A population-based study of juvenile disc
degeneration and its association with overweight and obesity, low
back pain and diminished functional status. J Bone Joint Surg Am.
93:662–670. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bolesta MJ: Commentary on an article by
Dino Samartzis, DSc, et al.: ‘A population-based study of juvenile
disc degeneration and its association with overweight and obesity,
low back pain and diminished functional status’. J Bone Joint Surg
Am. 93:e342011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Freemont AJ: The cellular pathobiology of
the degenerate intervertebral disc and discogenic back pain.
Rheumatology (Oxford). 48:5–10. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Z, Yu X, Shen J, Chan MT and Wu WK:
MicroRNA in intervertebral disc degeneration. Cell Prolif.
48:278–283. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao B, Yu Q, Li H, Guo X and He X:
Characterization of microRNA expression profiles in patients with
intervertebral disc degeneration. Int J Mol Med. 33:43–50. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu P, Feng B, Wang G, Ning B and Jia T:
Microarray based analysis of gene regulation by microRNA in
intervertebral disc degeneration. Mol Med Rep. 12:4925–4930. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mang Y, Li L, Ran J, Zhang S, Liu J, Li L,
Chen Y, Liu J, Gao Y and Ren G: Long noncoding RNA NEAT1 promotes
cell proliferation and invasion by regulating hnRNP A2 expression
in hepatocellular carcinoma cells. Onco Targets Ther. 10:1003–1016.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen T, Wang H, Yang P and He ZY:
Prognostic role of long noncoding RNA NEAT1 in various carcinomas:
A meta-analysis. Onco Targets Ther. 10:993–1000. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang LQ, Yang SQ, Wang Y, Fang Q, Chen
XJ, Lu HS and Zhao LP: Long noncoding RNA MIR4697HG promotes cell
growth and metastasis in human ovarian cancer. Anal Cell Pathol
(Amst). 2017:82678632017.PubMed/NCBI
|
|
16
|
Sookoian S, Rohr C, Salatino A, Dopazo H,
Fernandez Gianotti T, Castaño GO and Pirola CJ: Genetic variation
in long noncoding RNAs and the risk of nonalcoholic fatty liver
disease. Oncotarget. 8:22917–22926. 2017.PubMed/NCBI
|
|
17
|
Song J, Park JK, Lee JJ, Choi YS, Ryu KS,
Kim JH, Kim E, Lee KJ, Jeon YH and Kim EE: Structure and
interaction of ubiquitin-associated domain of human Fas-associated
factor 1. Protein Sci. 18:2265–2276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Urrutia J, Besa P, Campos M, Cikutovic P,
Cabezon M, Molina M and Cruz JP: The Pfirrmann classification of
lumbar intervertebral disc degeneration: An independent inter- and
intra-observer agreement assessment. Eur Spine J. 25:2728–2733.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gore M, Sadosky A, Stacey BR, Tai KS and
Leslie D: The burden of chronic low back pain: Clinical
comorbidities, treatment patterns and health care costs in usual
care settings. Spine (Phila Pa 1976). 37:E668–E677. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Katz JN: Lumbar disc disorders and
low-back pain: Socioeconomic factors and consequences. J Bone Joint
Surg Am. 2 88 Suppl:S21–S24. 2006. View Article : Google Scholar
|
|
23
|
Parthan A, Evans CJ and Le K: Chronic low
back pain: Epidemiology, economic burden and patient-reported
outcomes in the USA. Expert Rev Pharmacoecon Outcomes Res.
6:359–369. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao B, Lu M, Wang D, Li H and He X:
Genome-wide identification of long noncoding RNAs in human
intervertebral disc degeneration by RNA sequencing. Biomed Res Int.
2016:36848752016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen Y, Ni H, Zhao Y, Chen K, Li M, Li C,
Zhu X and Fu Q: Potential role of lncRNAs in contributing to
pathogenesis of intervertebral disc degeneration based on
microarray data. Med Sci Monit. 21:3449–3458. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mehra M, Hill K, Nicholl D and Schadrack
J: The burden of chronic low back pain with and without a
neuropathic component: A healthcare resource use and cost analysis.
J Med Econ. 15:245–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ohrt-Nissen S, Døssing KB, Rossing M,
Lajer C, Vikeså J, Nielsen FC, Friis-Hansen L and Dahl B:
Characterization of miRNA expression in human degenerative lumbar
discs. Connect Tissue Res. 54:197–203. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng J, Son DJ, Lee HL, Lee HP, Kim TH,
Joo JH, Ham YW, Kim WJ, Jung JK, Han SB and Hong JT:
(E)-2-methoxy-4-(3-(4-methoxyphenyl)prop-1-en-1-yl)phenol
suppresses ovarian cancer cell growth via inhibition of ERK and
STAT3. Mol Carcinog. 56:2003–2013. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kandala PK, Wright SE and Srivastava SK:
Blocking epidermal growth factor receptor activation by
3,3′-diindolylmethane suppresses ovarian tumor growth in vitro and
in vivo. J Pharmacol Exp Ther. 341:24–32. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gao XP, Qian DW, Xie Z and Hui H:
Protective role of licochalcone B against ethanol-induced
hepatotoxicity through regulation of Erk signaling. Iran J Basic
Med Sci. 20:131–137. 2017.PubMed/NCBI
|