Introduction
During the development of normal cells, the
proliferation and differentiation remain a dynamic balance, which
is important to the normal physical function of the body (1,2).
Among the pathways that regulate the balance of proliferation and
differentiation, Notch-Hes is one of the key pathways (3–5).
Previous studies have identified that Hes1 protein is extensively
expressed in various cell types in adult mice (6–8). The
Hes1 gene is an important member of the basic Helix-Loop-Helix gene
family, which is abundantly expressed during embryonic development
(9–11). It is located at 3q28-q29 on human
chromosome 3, the mRNA contains 1,471 base pairs and 4 exons. The
Hes1 protein is encoded by the Hes1 gene and contains 280 amino
acids and the relative molecular weight is 29,400 (12,13).
As an important downstream effector molecule of the Notch signaling
pathway, Hes1 serves an important role in the proliferation and
particularly the differentiation of the mammalian cells (14,15).
Previously, in the research of cervical caner, it has been
identified that Hes1 protein is particularly expressed in the
nucleus and cytoplasm of the cervical epithelial cells (16,17).
The Hes1 expression levels in the epithelial cells of cervical
intraepithelial neoplasia and cervical cancer are significantly
increased compared with normal cervical epithelial cells. As the
severity of cervical epithelial neoplasia increases, the Hes1
expression is increased (18–20).
It was hypothesized that Hes1 protein overexpression may be
involved in the carcinogenesis of the cervical epithelium. In the
present study, the Hes1 promoter was cloned and the activity was
analyzed in order to provide a basis for the research on
transcription and regulation of Hes1 during the occurrence and
development of cervical cancer.
Materials and methods
Materials
The human cervical cancer HeLa cell line (Institute
for Regenerative Medicine, ZhuJiang Hospital of Southern Medical
University, Guangzhou, China), GenBank (National Center for
Biotechnology Information, Bethesda, MD USA), Dulbecco's modified
Eagle's medium (DMEM; GE Healthcare Life Sciences, Logan, UT, USA),
fetal bovine serum (Thermo Fisher Scientific, Inc., Waltham, MA,
USA), Ex Taq, T4 DNA ligase, pMD18-T vector, restriction enzymes
SacI and HindIII (all from Takara Bio, Inc., Otsu,
Japan), primer synthesis (Sangon Biotech Co., Ltd., Shanghai,
China), pGL3-Basic vector, PGL3-control vector, PRL-TK vector,
dual-luciferase reporter assay kit (all Promega Corporation,
Madison, WI, USA), Escherichia coli DH5α (Institute for
Regenerative Medicine, ZhuJiang Hospital of Southern Medical
University), genomic DNA extraction kit, agarose gel extraction
kit, plasmid minipreparation medium kit (DP304; Tiangen Biotech
Co., Ltd., Beijing, China), Lipofectamine™ 2000 (Thermo
Fisher Scientific Inc.), and electrophoresis-grade agarose (12%
separation gel and 5% spacer gel) were used.
Cell culture and the extraction of
genome
HeLa cells were cultured in high glucose DMEM
containing 10% fetal bovine serum. The cells were maintained at
37°C, in a saturated humidity and 5% CO2 incubator, and
were passaged every 2–3 days. The cells at the logarithmic phase
were used in all the experiments. The TIANamp Genomic DNA kit
(DP304) was used to extract the genomic DNA of HeLa cells, and all
procedures were conducted according to the manufacturer's protocol
(Tiangen Biotech Co., Ltd.).
Amplification of human Hes1 gene
promoter and the purification of the product
The primer was designed according to the 5′end of
Hes1 gene from GenBanksp16 (ncbi.nlm.nih.gov/nuccore/NT_005612.17?from=100428715&to=100434890&report=genbank).
The Hes1 gene sequence was analyzed using BIO-XM™
(Biomax Informatics AG, Planegg, Germany) and the −747-+66 segment
at the 5 end was amplified. The following primers were used:
Upstream, 5-CGA GCT CAGCGGGAACTTTAGATGTG-3 and downstream, 5-CCCAAG
CTTGTTGACACTGGCTGGGGTA-3. The underlined parts indicate the enzyme
sites-SacI and HindIII. The genomic DNA of HeLa cells
was used as a template, and polymerase chain reaction (PCR) was
used to amplify the Hes1 promoter segment. The Hes1 promoter
sequences are listed in Table
I.
 | Table I.Hes1 promoter sequences and their
associated transcription factors. |
Table I.
Hes1 promoter sequences and their
associated transcription factors.
| Start point | Sequence (5′-3′) |
|---|
| −889 |
gggattcaagaactaccttgctccgaaaaacctgcatttgtgaggtagaaggcaattttt |
| −829 |
cctttttctgcatggaaacaggaaaatttttttggccctt
ttcctttaccatctactttc |
| −769 | accctcctga
atgtaaagtctg*agcgggaactttagatgtgtcggtaactcacattctta |
| −709 | cacccgtccccccctccccccgcccccttttaaccactgctgtttttttctttattgttt |
|
|
MZF1 SP1 SPY |
| −649 |
atacctttaaaaaaaatatgtttcaaatgaacttactacagtcaaagcagctctgttaca |
| −589 |
tatgagagagggcataaagagcaaagaccctggctccaaaagaaatagacaagatcaaga |
|
|
CdxA SRY Evi1 |
| −529 | ccaaagcggaaagaaaaaaaaaatctctaaaccaaagcccagagggagagagcaaagg |
|
|
SRY |
| −469 |
ttaaaatccttttgattgacgttgtagcctccggtgccctgggctcaggcgcgcgccatt |
|
|
SRY |
| −409 |
ggccgccagaccttgtgcctggcggccaatgggggggcgcggtccacgagcggtgccgcg |
|
|
SP1 |
| −349 |
tgtctcctcctcccattggctgaaagttactgtggga*aagaaagtttgggaagtttcaca |
|
|
SPY Lyf1 |
| −289 |
cgagccgttcgcgtgcagtcccagatatatatagaggccgccagggcctagggatcacac |
| −229 |
aggatccggagctggtgctgataacagcggaatcccccgtctacctctctccttggtcct |
|
|
GATA-1 C-REL |
| −169 |
ggaacagcgctactgatcaccaagtagccacaaaatataataaaccctcagcacttgctc |
|
|
CdxA |
| −109 |
agtagttttgtgaaagtctcaagtaaaagagacacaaacaaaaaattctttttcgtgaag |
|
−49 |
aactccaaaaataaaattctctagagataaaaaaaaaaaaaaaaggaaaatgccagctga |
|
|
GATA-1 |
| 12 |
tataatggagaaaaattcctcgtccccggtggctgctaccccagccagtgtcaacacgac |
| 72 |
accggataaaccaaagacagcatctgagcacagaaaggtaagggcggtacctgtatctct |
| 132 |
ttgcagcccctcaaaattaag |
The amplification conditions were pre-denaturation
at 95°C for 5 min, denaturation at 95°C for 30 sec, annealing at
56.2°C for 30 sec, elongation at 72°C for 45 sec, there were 35
cycles in total, followed by elongation at 72°C for 10 min.
Following PCR, 5 µl product was added in 1.5% agarose gel for
electrophoresis. Subsequently, the gel extraction kit was used for
the purification of the PCR product. The procedures were conducted
according to the manufacturer's protocol [DNA amplification and
extraction kit (cat. no. ER103; Tiangen Biotech Co., Ltd.)].
The connection and identification of
purified target segment and cloning vector
The collected PCR product (Hes1 gene) was
transfected by Lipofectamine™ 2000 into the pMD18-T
vector, which was transformed and amplified to extract the
plasmids. Subsequently, restriction enzymes for SacI and
HindIII dual-enzyme digestion were used and the sequences
were detected, which was termed the pMD18-T-Hes1-promoter.
The establishment and identification
of recombinant expression vectors
The recombinant T vector with the correct sequence
and pGL3-Basic plasmid received SacI and HindIII
dual-enzyme digestion and separated by agarose electrophoresis, the
gel extraction kit was used to collect the target segment, and then
T4 DNA ligase was used to transform, amplify and extract the
plasmids. The product received dual-enzyme digestion and sequence
detection, which was termed the pGL3-Hes1-promoter.
Transfection of recombinant expression
vector into the cells and activity detection
The HeLa cells were inoculated in 24-well plate at 4
h before transfection, 0.5 ml complete medium and
0.5×105 cells were added in each well and the
transfection was conducted when the cells reached 70–80%
confluency. The procedures were done according to the
manufacturer's instructions for Lipofectamine 2000. The transfected
plasmids included negative control plasmid pGL3-Basic, positive
control plasmid pGL3-Control containing the SV40 enhancer/promoter,
recombinant plasmid pGL3-Hes1-promoter and internal control PRL-TK
plasmid. A total of 48 h after transfection, the Dual-luciferase
reporter assay kit from Promega was used to detect the activity of
the transfected plasmid, and there were 3 repeated wells for every
group and the experiments were repeated 3 times.
Statistical analysis
The data were analyzed by SPSS software, version
18.0 (SPSS, Inc., Chicago, IL, USA). One-way analysis of variance
and the Q test were used to analyze the data. The data were
presented as mean ± standard deviation. P<0.05 was considered to
indicate a statistically significant difference.
Results
Target segment after PCR product
electrophoresis
The genomic DNA of HeLa cell was used as a template
and the promoter segment of the Hes1 gene was amplified by PCR, and
the electrophoresis results indicated that there was a specific
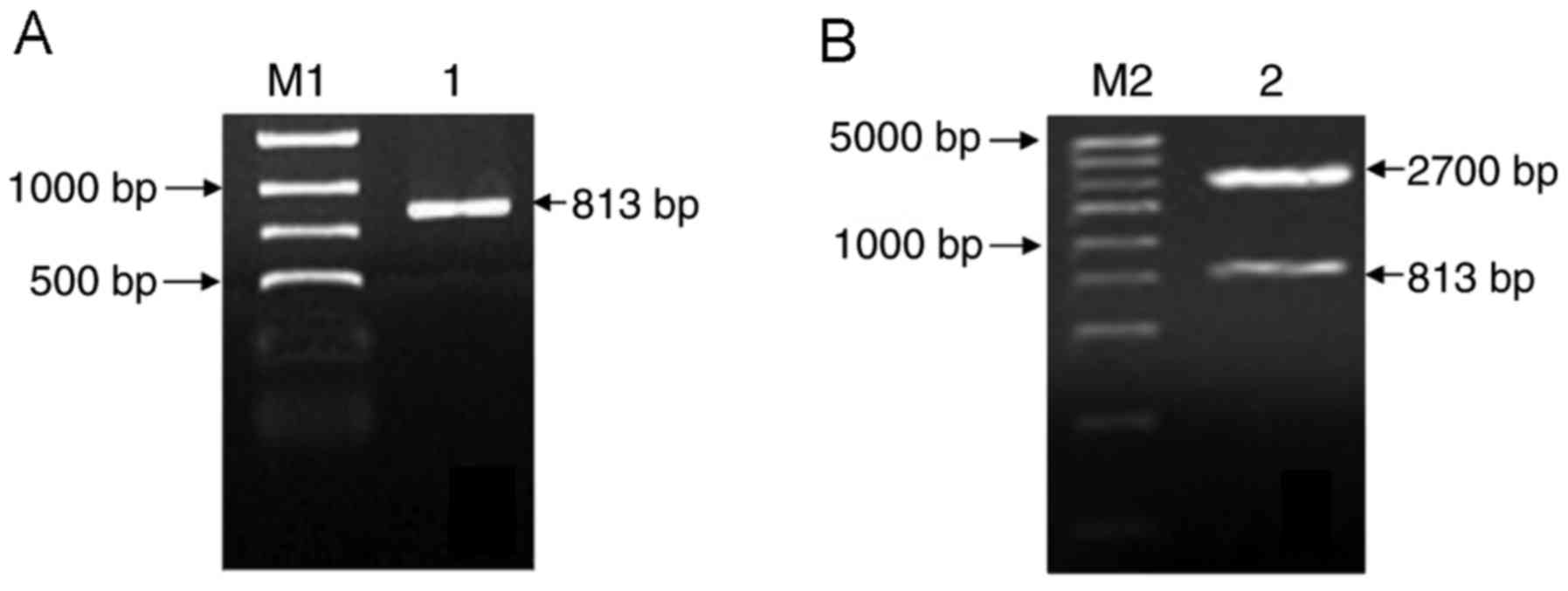
band at 813 base pairs (bp), as presented in Fig. 1A.
Enzyme cutting identification of
recombinant T cloning vector
The PCR production of the Hes1 promoter was purified
and connected to the pMD18-T vector to establish the recombinant
vector pMD18-T-Hes1-promoter. Subsequent to transformation the
plasmids were extracted for SacI and HindIII dual
enzyme digestion. The electrophoresis results demonstrated that
there were two specific bands at 813 and 2,700 bp, as presented in
Fig. 1B.
Sequence detection of recombinant T
cloning vector
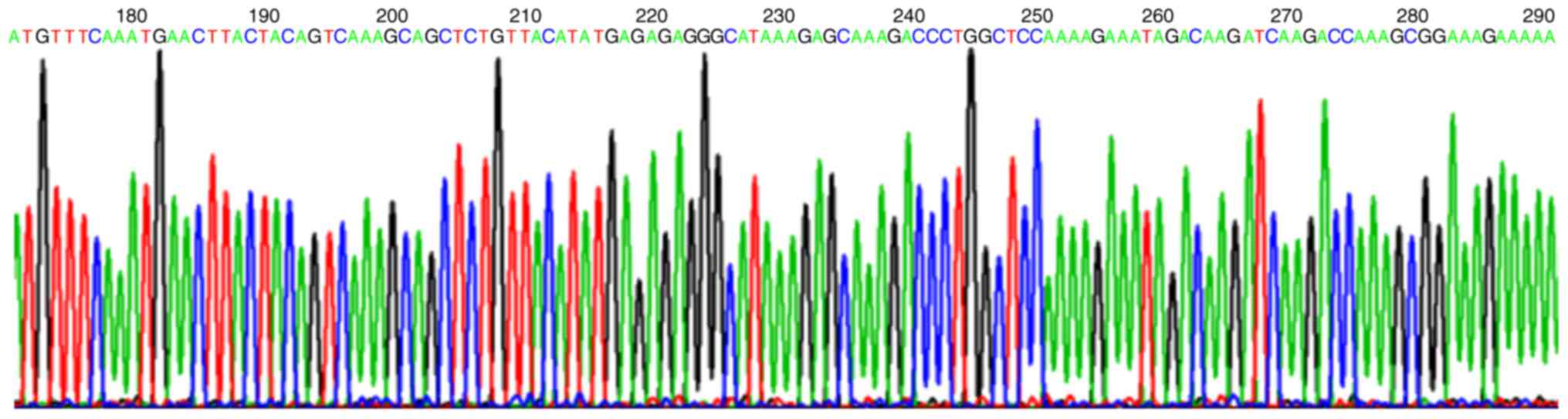
DNA sequence analysis demonstrated that compared
with the Hes1 gene promoter sequence from GenBank, the 5′ and 3′
ends of the cloned target segment of recombinant T cloning vector
were located upstream −747 bp and downstream +66 bp of ATG, the
size was 813 bp as presented in Fig.
2.
Electrophoresis results of recombinant
expression vector pGL3-Hes1-promoter after dual enzyme
digestion
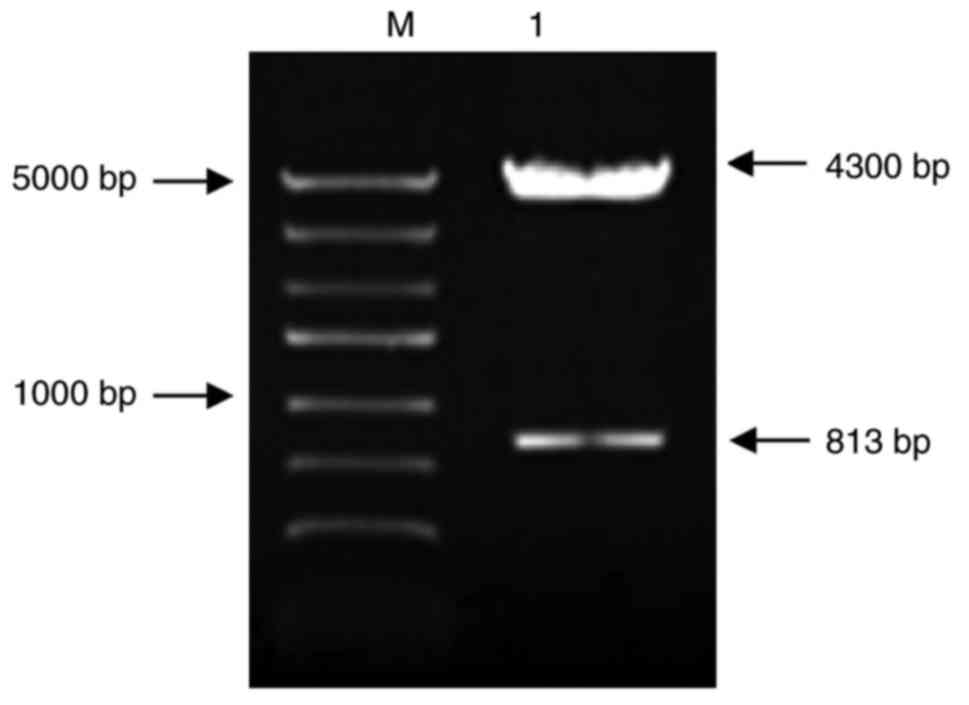
The electrophoresis of recombinant expression vector
pGL3-Hes1-promoter after SacI and HindIII dual-enzyme
digestion demonstrated that there were specific bands at 813 and
4,300 bp, as presented in Fig.
3.
Sequencing results of recombinant
expression vector
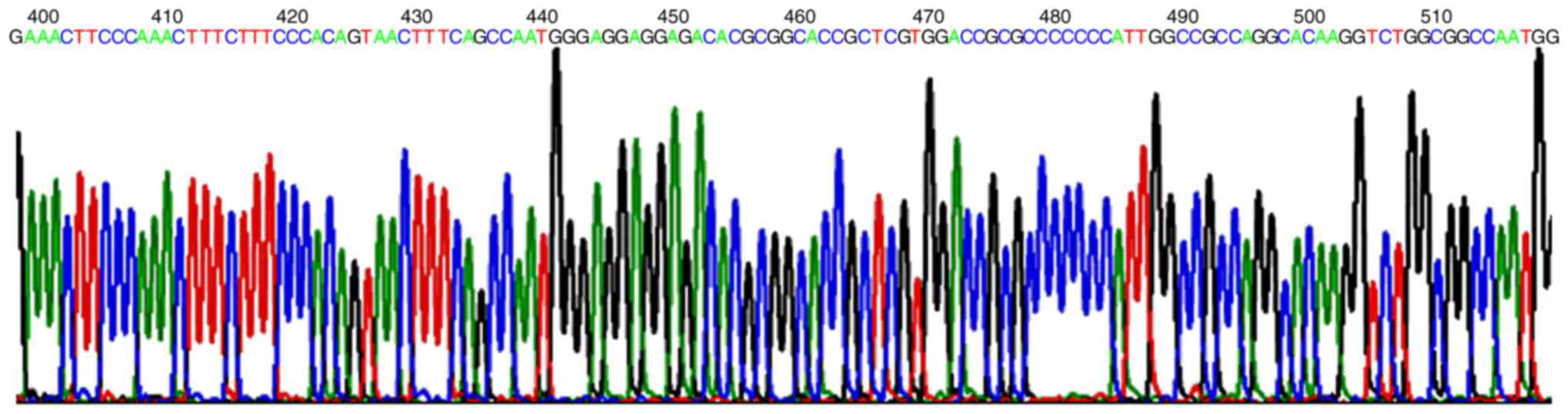
DNA sequencing results demonstrated that compared
with the Hes1 promoter sequence from GenBank, the 5′ and 3′ end of
the target segment sequence of cloned expression vector
pGL3-Hes1-promoter located at upstream −747 bp and downstream +66
bp of ATM, the size was 813 bp, as presented in Fig. 4.
Activity analysis of recombinant
expression vector following HeLa cell transfection
At 48 h after HeLa cell transfection by recombinant
expression plasmid pGL3-Hes1-promoter, pGL3-Basic and pGL3-Control,
the luciferase expression was detected. The ratio of first and
second fluorescence was defined as the relative luciferase
activity. The relative luciferase of pGL3-Basic was 1.076, of
pGL3-Control was 100.47 and of pGL3-Hes1-promoter was 34.44, there
was significant difference (P<0.05; Table II).
 | Table II.Analysis of Hes1 promoter activity
following HeLa cell transfection. |
Table II.
Analysis of Hes1 promoter activity
following HeLa cell transfection.
| Group | n | Relative luciferase
activity | F-value | P-value |
|---|
| pGL3-Basic | 3 |
1.076±0.214 | 84.434 | <0.001 |
|
pGL3-Hes1-promoter | 3 |
34.44±13.76 |
|
|
| pGL3-Control | 3 |
100.47±9.13 |
|
|
Discussion
During the normal development of cells, the
proliferation and differentiation remains in dynamic balance, which
is important for normal physical function (21–23).
The pathways to regulate the balance between cell proliferation and
differentiation include the leukemia inhibitory factor-bone
morphogenetic protein pathway, octamer-binding transcription factor
3/4 pathway, Wnt pathway and Notch-Hes pathway (24,25).
These pathways are composed of various cytokines and transcription
factors and among these pathways, the Notch-Hes signaling
transduction pathway is the most important pathway. If there is
abnormity of Notch signaling pathway, the cells cannot
differentiate normally, thus there is possibility of oncogenesis,
and it has been reported that the Notch receptor is abnormally
expressed in numerous human solid tumor types including cervical,
head and neck, endometrial, kidney, lung and breast cancer, pleural
mesothelioma and salivary gland tumors) and hematological
malignancies (26–28).
Cervical cancer is the second most common type of
female malignant cancer, following breast cancer in developing
countries and the most common type of genital cancer in Asia
(29). The morbidity of cervical
cancer is increasing and the mortality is the second highest cause
of cancer-associated mortality in women in Asia (29–31).
In the early stages of cervical cancer, there is usually no
symptoms and the physical signs are not obvious, however early
diagnosis and early treatment are the key methods to increase the
survival rate. Previous studies have demonstrated that the low
degree of cell differentiation is associated with poor prognosis
(32–34). As an important downstream target
gene of Notch signaling pathway, Hes1 genes can directly combine
with specific DNAs to block the pathway activation and inhibit cell
differentiation (35,36). Thus, exploring the regulatory
mechanism of its transcription is significant in the prevention and
treatment of cervical cancer.
The regulation of gene expression in eukaryotic
cells include transcriptional-level regulation and
post-transcriptional regulation according to the sequencing, and
transcriptional-level regulation is the predominant method
(37). Transcriptional-level
regulation is the most important step in the regulation of gene
expression in eukaryote, which mainly depends on the effects of
certain regulatory sequences (including the promoter and enhancer)
and certain protein factors (such as transcription factors) on the
initiation of transcription. The key regulation of gene expression
is on a transcriptional level, and the promoter is the most
important domain in transcriptional regulation. Thus, the study on
Hes1 is important. The luciferase reporter gene vector doesn't
contain the promoter and enhancer, which can clone the target gene
into the expression plasmid vector containing the reporter gene.
The expression of luciferase is associated with the promoter
sequence in the recombinant plasmid. Subsequent to transfection,
the expression of luciferase activity can directly reflect the
activity of the promoter. At present, this method is widely applied
(12,13). The novelty of the current study is
that the promoter sequence was newly designed.
In the current study, the Hes1 luciferase reporter
recombinant vector was successfully established and transfected
into HeLa cells to verify that it had promoter activity, and the
core area of the promoter had several tumor-promoting and tumor
suppressor genes. This provides a basis for the further study of
the regulatory mechanism of Hes1 transcription and translation. In
addition, it can provide novel strategy, target and experimental
evidence for the prevention and treatment of cervical cancer.
References
|
1
|
Roese-Koerner B, Stappert L and Brustle O:
Notch/Hes signaling and miR-9 engage in complex feedback
interactions controlling neural progenitor cell proliferation and
differentiation. Neurogenesis (Austin). 4:e13136472017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saclier M, Theret M, Mounier R and Chazaud
B: Effects of macrophage conditioned-medium on murine and human
muscle cells: Analysis of proliferation, differentiation, and
fusion. Methods Mol Biol. 1556:317–327. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fujimori K, Kadoyama K and Urade Y:
Protein kinase C activates human lipocalin-type prostaglandin D
synthase gene expression through de-repression of notch-HES
signaling and enhancement of AP-2 beta function in brain-derived
TE671 cells. J Biol Chem. 280:18452–18461. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li X, Liu F, Zhang X, Shi G, Ren J, Ji J,
Ding L, Fan H, Dou H and Hou Y: Notch-Hes-1 axis controls
TLR7-mediated autophagic death of macrophage via induction of P62
in mice with lupus. Cell Death Dis. 7:e23412016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Muranishi Y, Terada K, Inoue T, Katoh K,
Tsujii T, Sanuki R, Kurokawa D, Aizawa S, Tamaki Y and Furukawa T:
An essential role for RAX homeoprotein and NOTCH-HES signaling in
Otx2 expression in embryonic retinal photoreceptor cell fate
determination. J Neurosci. 31:16792–16807. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hidalgo-Sastre A, Brodylo RL,
Lubeseder-Martellato C, Sipos B, Steiger K, Lee M, von Figura G,
Grünwald B, Zhong S, Trajkovic-Arsic M, et al: Hes1 Controls
Exocrine cell plasticity and restricts development of pancreatic
ductal adenocarcinoma in a mouse model. Am J Pathol. 186:2934–2944.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kayahara T, Sawada M, Takaishi S, Fukui H,
Seno H, Fukuzawa H, Suzuki K, Hiai H, Kageyama R, Okano H and Chiba
T: Candidate markers for stem and early progenitor cells, Musashi-1
and Hes1, are expressed in crypt base columnar cells of mouse small
intestine. FEBS Lett. 535:131–135. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nakazaki H, Reddy AC, Mania-Farnell BL,
Shen YW, Ichi S, McCabe C, George D, McLone DG, Tomita T and
Mayanil CS: Key basic helix-loop-helix transcription factor genes
Hes1 and Ngn2 are regulated by Pax3 during mouse embryonic
development. Dev Biol. 316:510–523. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barton A and Fendrik AJ: Sustained vs.
oscillating expressions of Ngn2, Dll1 and Hes1: A model of neural
differentiation of embryonic telencephalon. J Theor Biol. 328:1–8.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee JB, Werbowetski-Ogilvie TE, Lee JH,
McIntyre BA, Schnerch A, Hong SH, Park IH, Daley GQ, Bernstein ID
and Bhatia M: Notch-HES1 signaling axis controls hemato-endothelial
fate decisions of human embryonic and induced pluripotent stem
cells. Blood. 122:1162–1173. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sturrock M, Hellander A, Matzavinos A and
Chaplain MA: Spatial stochastic modelling of the Hes1 gene
regulatory network: Intrinsic noise can explain heterogeneity in
embryonic stem cell differentiation. J R Soc Interface.
10:201209882013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sturrock M, Hellander A, Aldakheel S,
Petzold L and Chaplain MA: The role of dimerisation and nuclear
transport in the Hes1 gene regulatory network. Bull Math Biol.
76:766–798. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang K, Zhang YQ, Ai WB, Hu QT, Zhang QJ,
Wan LY, Wang XL, Liu CB and Wu JF: Hes1, an important gene for
activation of hepatic stellate cells, is regulated by Notch1 and
TGF-β/BMP signaling. World J Gastroenterol. 21:878–887. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Min XH, Yu T, Qing Q, Yuan YH, Zhong W,
Chen GC, Zhao LN, Deng N, Zhang LF and Chen QK: Abnormal
differentiation of intestinal epithelium and intestinal barrier
dysfunction in diabetic mice associated with depressed Notch/NICD
transduction in Notch/Hes1 signal pathway. Cell Biol Int.
38:1194–1204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shi Y, Shu B, Yang R, Xu Y, Xing B, Liu J,
Chen L, Qi S, Liu X, Wang P, et al: Erratum to: Wnt and Notch
signaling pathway involved in wound healing by targeting c-Myc and
Hes1 separately. Stem Cell Res Ther. 6:2542015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z, Li Y, Banerjee S and Sarkar FH:
Emerging role of Notch in stem cells and cancer. Cancer Lett.
279:8–12. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Suprasert P, Srisomboon J, Siriaunkgul S,
Khunamornpong S, Phongnarisorn C, Siriaree S, Charoenkwan K,
Cheewakriangkrai C and Kietpeerakool C: Clinical outcomes and
prognostic factors of node-negative cervical cancer patients with
deep stromal invasion or lymphovascular space involvement following
radical hysterectomy. J Med Assoc Thai. 89:1368–1375.
2006.PubMed/NCBI
|
|
18
|
Liu J, Lu WG, Ye F, Cheng XD, Hong D, Hu
Y, Chen HZ and Xie X: Hes1/Hes5 gene inhibits differentiation via
down-regulating Hash1 and promotes proliferation in cervical
carcinoma cells. Int J Gynecol Cancer. 20:1109–1116. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu J, Ye F, Chen H, Lü W, Zhou C and Xie
X: Expression of differentiation associated protein Hes1 and Hes5
in cervical squamous carcinoma and its precursors. Int J Gynecol
Cancer. 17:1293–1299. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tyagi A, Vishnoi K, Mahata S, Verma G,
Srivastava Y, Masaldan S, Roy BG, Bharti AC and Das BC: Cervical
cancer stem cells selectively overexpress HPV oncoprotein E6 that
controls stemness and self-renewal through upregulation of HES1.
Clin Cancer Res. 22:4170–4184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bidaux G, Borowiec AS, Gordienko D, Beck
B, Shapovalov GG, Lemonnier L, Flourakis M, Vandenberghe M,
Slomianny C, Dewailly E, et al: Epidermal TRPM8 channel isoform
controls the balance between keratinocyte proliferation and
differentiation in a cold-dependent manner. Proc Natl Acad Sci USA.
112:pp. E3345–E3354. 2015; View Article : Google Scholar : PubMed/NCBI
|
|
22
|
McIver SC, Katsumura KR, Davids E, Liu P,
Kang YA, Yang D and Bresnick EH: Exosome complex orchestrates
developmental signaling to balance proliferation and
differentiation during erythropoiesis. Elife. 5:e178772016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Monje PV: To myelinate or not to
myelinate: Fine tuning cAMP signaling in Schwann cells to balance
cell proliferation and differentiation. Neural Regen Res.
10:1936–1937. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Brandenberger R, Wei H, Zhang S, Lei S,
Murage J, Fisk GJ, Li Y, Xu C, Fang R, Guegler K, et al:
Transcriptome characterization elucidates signaling networks that
control human ES cell growth and differentiation. Nat Biotechnol.
22:707–716. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Niwa H: Molecular mechanism to maintain
stem cell renewal of ES cells. Cell Struct Funct. 26:137–148. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wall DS, Mears AJ, McNeill B, Mazerolle C,
Thurig S, Wang Y, Kageyama R and Wallace VA: Progenitor cell
proliferation in the retina is dependent on Notch-independent Sonic
hedgehog/Hes1 activity. J Cell Biol. 184:101–112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sun L, Liu M, Sun GC, Yang X, Qian Q, Feng
S, Mackey LV and Coy DH: Notch signaling activation in cervical
cancer cells induces cell growth arrest with the involvement of the
nuclear receptor NR4A2. J Cancer. 7:1388–1395. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu X, Liu W, Tang D, Xiao H, Wu Z, Chen C,
Yao X, Liu F and Li G: Prognostic values of four Notch receptor
mRNA expression in gastric cancer. Sci Rep. 6:280442016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Moore MA and Tajima K: Cervical cancer in
the asian pacific-epidemiology, screening and treatment. Asian Pac
J Cancer Prev. 5:349–361. 2004.PubMed/NCBI
|
|
30
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jemal A, Thomas A, Murray T and Thun M:
Cancer statistics, 2002. CA Cancer J Clin. 52:23–47. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cuschieri K, Brewster DH, Graham C, Nicoll
S, Williams AR, Murray GI, Millan D, Johannessen I, Hardie A and
Cubie HA: Influence of HPV type on prognosis in patients diagnosed
with invasive cervical cancer. Int J Cancer. 135:2721–2726. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He D, Duan C, Chen J, Lai L, Chen J and
Chen D: The safety and efficacy of the preoperative neoadjuvant
chemotherapy for patients with cervical cancer: A systematic review
and meta analysis. Int J Clin Exp Med. 8:14693–14700.
2015.PubMed/NCBI
|
|
34
|
Yu JQ, Zhou Q, Zhu H, Zheng FY and Chen
ZW: Overexpression of astrocyte elevated gene-1 (AEG-1) in cervical
cancer and its correlation with angiogenesis. Asian Pac J Cancer
Prev. 16:2277–2281. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cenciarelli C, Marei HE, Zonfrillo M,
Casalbore P, Felsani A, Giannetti S, Trevisi G, Althani A and
Mangiola A: The interference of Notch1 target Hes1 affects cell
growth, differentiation and invasiveness of glioblastoma stem cells
through modulation of multiple oncogenic targets. Oncotarget.
8:17873–17886. 2017.PubMed/NCBI
|
|
36
|
Phillips NE, Manning CS, Pettini T, Biga
V, Marinopoulou E, Stanley P, Boyd J, Bagnall J, Paszek P, Spiller
DG, et al: Stochasticity in the miR-9/Hes1 oscillatory network can
account for clonal heterogeneity in the timing of differentiation.
Elife. 5:e161182016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Orphanides G and Reinberg D: A unified
theory of gene expression. Cell. 108:439–451. 2002. View Article : Google Scholar : PubMed/NCBI
|