Introduction
Osteoarthritis (OA) is a degenerative joint disorder
characterized by articular cartilage degradation, osteophyte
formation, synovium hyperplasia, subchondral bone cysts and
sclerosis. It is one of the leading causes of pain and dysfunction
in the joints among the aging population (1). Despite the high prevalence of OA, its
pathogenesis remains unclear. At present, the diagnosis of OA
primarily depends on clinical symptoms and radiographic findings.
However, the majority of individuals are clinically asymptomatic at
the early stages of OA, and pathological degradation of the
articular cartilage already exists before the symptoms arise.
Furthermore, radiographs may appear normal at this stage due to
lack of sensitivity in visualizing minimal cartilage lesions. Thus,
the diagnostic methods currently in use have limitations in
detecting preclinical OA and provide an inaccurate assessment of
disease progression. Although multiple translational techniques
detect early OA processes, it remains unknown as to whether they
truly represent the initial events. Therefore, the clinical
diagnosis of early OA is one of the greatest challenges in this
field (2). Currently, there is no
definitely effective medication for the treatment of early OA.
Traditional medications, such as glucosamine and hyaluronic acid,
once thought of as effective, have no longer been recommended by
the latest American Academy of Orthopaedic Surgeons guideline due
to evidence of a lack of efficacy (3). The medications currently used for the
treatment of OA are predominantly aimed at relieving pain, but do
not prevent disease progression. These limitations give rise to the
interest in proteomic studies to identify candidate biomarkers and
drug targets, which may facilitate early diagnosis and intervention
of OA. Additionally, proteomic studies provide greater insight into
OA pathogenesis and original insights into the molecular mechanisms
involved in disease progression.
Advances in proteomic technologies have facilitated
extensive proteomic characterization of various body fluids,
including serum (4) and urine
(5) in OA. However, evaluating the
synovial fluid (SF) proteome is more likely to yield a higher
concentration of potential biomarkers than serum and urine, as SF
is in direct contact with articular cartilage, synovium, ligament,
meniscus and joint capsule. Alterations in metabolism of any of
these tissues during disease progression are reflected as
alterations in the proteomic profile of SF, which is also less
influenced by systematic conditions. Therefore, it is considered
more appropriate to detect the proteomic profile in SF than in
other body fluids. There are also certain studies designed to
investigate the expression of certain proteins in SF from OA
patients (6). These studies
focused on evaluating the expression of certain proteins, but
failed to screen novel proteins that are differentially expressed
in SF. With the use of quantitative proteomic technologies, a
series of novel proteins differentially expressed in SF from OA
patients have been identified in previous studies (7,8).
However, the majority of these investigations were performed using
low resolution mass spectrometry (MS) and with minimal
fractionation of the samples, which limited the depth of coverage.
Isobaric tags for relative and absolute quantification is a
labeling method with high throughput, easy automation, high
resolution and quantification accuracy, which enables quantitative
proteomics analysis of four samples simultaneously under the same
experimental conditions (9). This
technology has also been applied to investigate alterations of the
proteomic profile in OA in previous studies (10,11).
Although a greater number of novel proteins were identified to be
differentially expressed using this method (10,11),
whether these proteins are specifically regulated in OA remains
unclear due to a lack of effective standard controls or
inter-controls.
SWATH-MS is an emerging data-independent acquisition
method, whichgenerates a single file containing fragment ion
spectra of all ionized species of a sample (12). When combined with a targeted data
analysis strategy, SWATH-MS was demonstrated to achieve favorable
accuracy, dynamic range, and reproducibility of selected reaction
monitoring, which is considered to be the golden standard in
quantitative proteomic technology (13). In order to identify differentially
expressed proteins specifically regulated in OA and further
elucidate the disease pathogenesis, SWATH-MS was performed to
compare the SF proteome in OA, rheumatoid arthritis (RA) and
traumatic arthritis (meniscus injury without cartilage defect).
Subsequently, proteins of interest were quantified by enzyme-linked
immunosorbent assay (ELISA). To the best of our knowledge, no
comparative analysis with respect to the SF proteome has been
conducted in these three types of knee disorders using
SWATH-MS.
Materials and methods
Subjects
The present study was initiated following approval
from the Ethical Committee of the Chinese PLA General Hospital and
informed consent was obtained from all the subjects. SF samples
were acquired from 30 patients who had received surgical treatment
for a knee disorder between September 2014 and January 2015 in the
Orthopedics Department of the Chinese PLA General Hospital
(Beijing, China). A total of 10 knee OA patients (4 men and 6
women; age, 64–72 years; mean age, 67 years) undergoing
arthroplasty were defined as the study group, 10 knee RA patients
(4 men and 6 women; age, 45–69 years; mean age, 60 years)
undergoing arthroplasty served as the inter-control group, and 10
patients (7 men and 3 women; age, 23–47 years; mean age, 29 years)
undergoing arthroscopy for traumatic knee arthritis (meniscus
injury without cartilage defect) within 7 days after injury served
as the standard control group. The diagnosis of knee OA and RA was
based upon the criteria of the American College of Rheumatology
(14). The severity of knee OA and
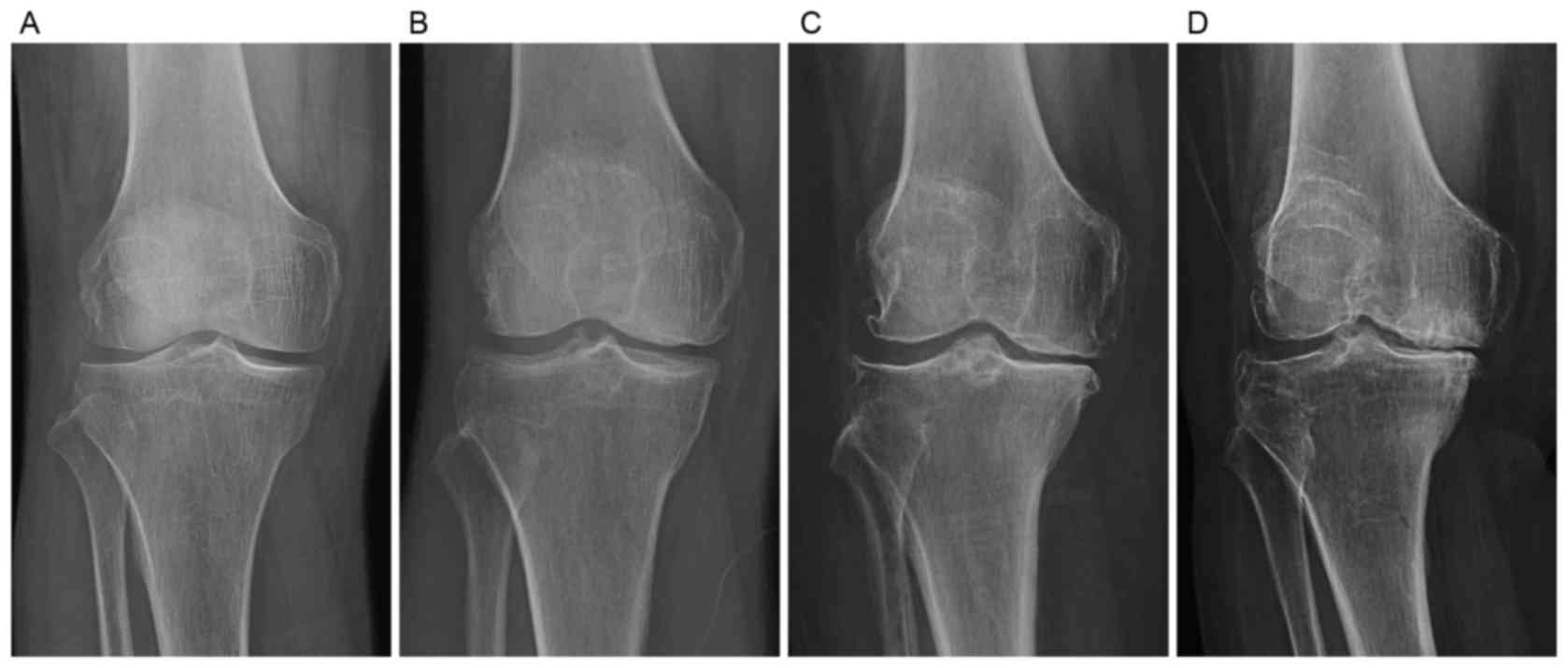
RA was evaluated by the Kellgren-Lawrence (KL) radiographic grading
criteria (Fig. 1), according to
which all the knee OA and RA patients recruited were classified as
KL grade 4. All the patient data were retrieved from our
institutional database through chart review.
Sample preparation and protein
digestion
SF samples (1–2 ml) were aspirated from the affected
knee using a sterile puncture needle prior to surgery and 50 µl
proteinase inhibitor cocktail (Roche Molecular Diagnostics,
Pleasanton, CA, USA) was immediately added to the sample after
collection. The mixture was centrifuged at 19,228 × g for 20 min at
4°C (Hettich MIKRO 22R; Hettich Benelux, Tuttlingen, Germany) to
remove cells and debris. For the comprehensive shotgun analysis,
all the SF samples were depleted of the 14 most abundant proteins
with the multiple affinity removal system (MARS Hu-14 spin
cartridge; Agilent Technologies, Inc., Santa Clara, CA, USA)
according to the manufacturer's instruction. After reduction and
alkylation with 10 mM tris (2-carboxyethyl) phosphine
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and 20 mM
iodoacetamide (Sigma-Aldrich; Merck KGaA), the samples were
subjected to protein quantification using the Bradford Assay
according to the manufacturer's instructions with a commercial
Bradford reagent (Bio-Rad Laboratories, Hercules, CA, USA).
Subsequently, the samples were diluted with 1 M urea and digested
with sequencing-grade trypsin (Promega Corporation, Madison, WI,
USA) at a protease/protein ratio of 1:30 overnight at 37°C.
Following digestion, the samples were stored at −80°C until
use.
Shotgun proteomic measurement
The processed peptides were measured using a Triple
TOF 5600 Plus mass spectrometer (SCIEX, Framingham, MA, USA)
operated in data-dependent acquisition (DDA) mode. The mass
spectrometer was interfaced with a NanoLC Ultra 2D Plus HPLC system
(Eksigent Technologies, Dublin, CA, USA) as previously described
(12,13). Peptides were directly injected onto
a 20-cm PicoFrit emitter and separated using a 120-min gradient
from 2–35% (buffer A: 0.1% Formic acid and 2% acetonitrile; buffer
B: 0.1% Formic acid and 90% acetonitrile) at a flow rate of 300
nl/min. MS1 spectra were collected in the range 350–1,250 m/z for
250 msec. The 20 most intense precursors with charge state 2–5,
which exceeded 250 counts per sec were selected for fragmentation,
and MS2 spectra were collected in the range 100–1,500 m/z for 50
msec. The precursor ions were dynamically excluded from reselection
for 20 sec.
SWATH-MS measurement
SWATH-MS measurements were performed with peptide
mixtures generated by digesting SF samples. The unfractionated,
total peptide samples were analyzed to minimize confounding factors
introduced by sample handling. The same liquid chromatography-MS/MS
system used for DDA measurements was used for SWATH analysis
(12,13). In SWATH-MS mode, the instrument was
specifically tuned to optimize the quadrupole settings for the
selection of 25-m/z wide precursor ion selection windows. Using an
isolation width of 25 m/z, a set of overlapping windows was
constructed, covering the precursor mass range of 400–1,200 m/z.
The effective isolation windows were considered as being 400–425,
424–449, 448–473 m/z, and so on. SWATH-MS2 spectra were collected
from 100–1,500 m/z. The collision energy was optimized for each
window according to the calculation for a charge 2+ion
centered upon the window with a spread of 15 eV. An accumulation
time of 50 msec was used for all fragment ion scans in
high-sensitivity mode, and for each SWATH-MS cycle, a survey scan
in high-resolution mode was acquired for 50 msec. SWATH-MS
measurements were repeated three times on each sample to access the
technical variability. Differentially expressed proteins identified
by SWATH-MS were subjected to gene ontology (GO) and KEGG pathway
annotation.
ELISA
Another 90 SF samples were obtained from 36 knee OA
patients (17 men and 19 women, aged 57–74 years; mean age, 65
years), 36 knee RA patients (15 men and 21 women, aged 41–70 years;
mean age, 58 years) and 18 patients (12 men and 6 women, aged 21–47
years;mean age, 28 years) with traumatic knee arthritis (meniscus
injury without cartilage defect) to detect the level of complement
C1r and that of Dickkopf-related protein 2 (DKK2). According to the
KL radiographic grading criteria, 18 out of the 36 knee OA patients
were classified as KL grade 2, and the other 18 as KL grade 4.
Similarly, 18 out of the 36 knee RA patients were categorized as KL
grade 2, and the other 18 as KL grade 4. The levelsof complement
C1r and that of DKK2 in SF were measured by ELISA (MyBioSource;
R&D Systems, Inc., Minneapolis, MN, USA) according to the
manufacturer's instructions.
Statistical analysis
Statistical analysis was performed with SPSS
software (Version 17.0; SPSS Inc, Chicago, IL, USA). Data were
expressed as the mean ± standard deviation. One way analysis of
variance followed by a post hoc Tukey test for multiple comparisons
was performed to compare the quantitative data from different
groups. Pearson's correlation coefficient was used to analyze the
correlation between protein level and disease severity. P<0.05
was considered to indicate a statistically significant
difference.
Results
SWATH-MS
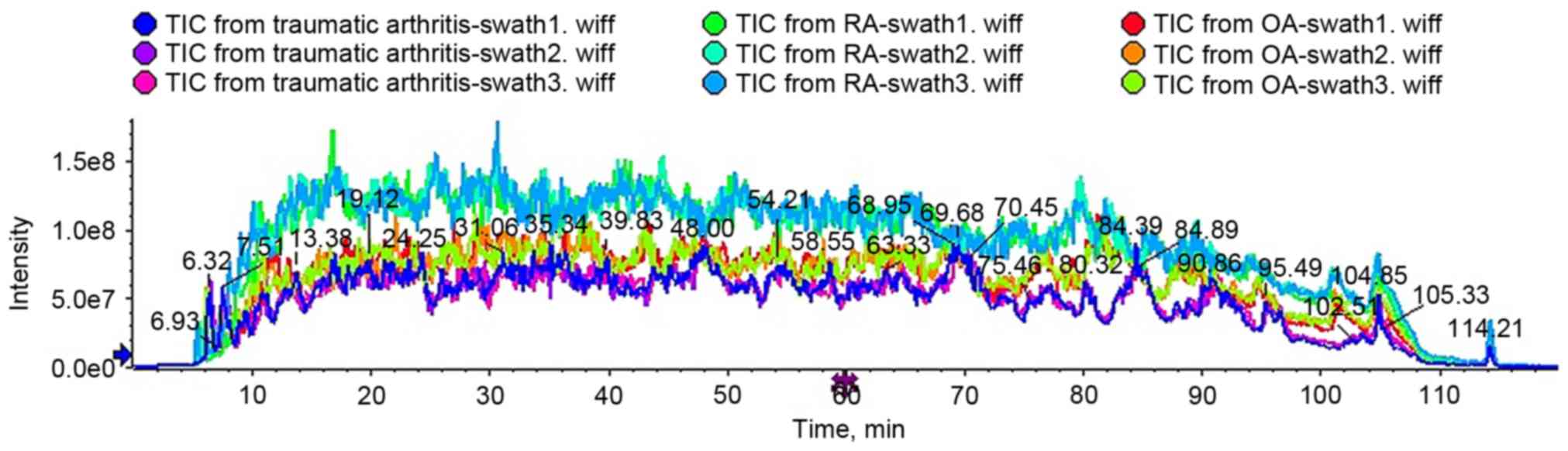
To evaluate technical variability, each sample was
analyzed in triplicate. As presented in Fig. 2, total ion current chromatograms
from triplicates of the same sample exhibited high similarity,
whereas those from different samples displayed obvious variation.
The results revealed small technical variability and excellent
reproducibility of SWATH-MS measurements and great variation in
proteomic profiles of SF from the different groups.
Proteins were regarded to be differentially
expressed when ion intensity between groups was significantly
different (P<0.05) and fold change between groups was >1.5.
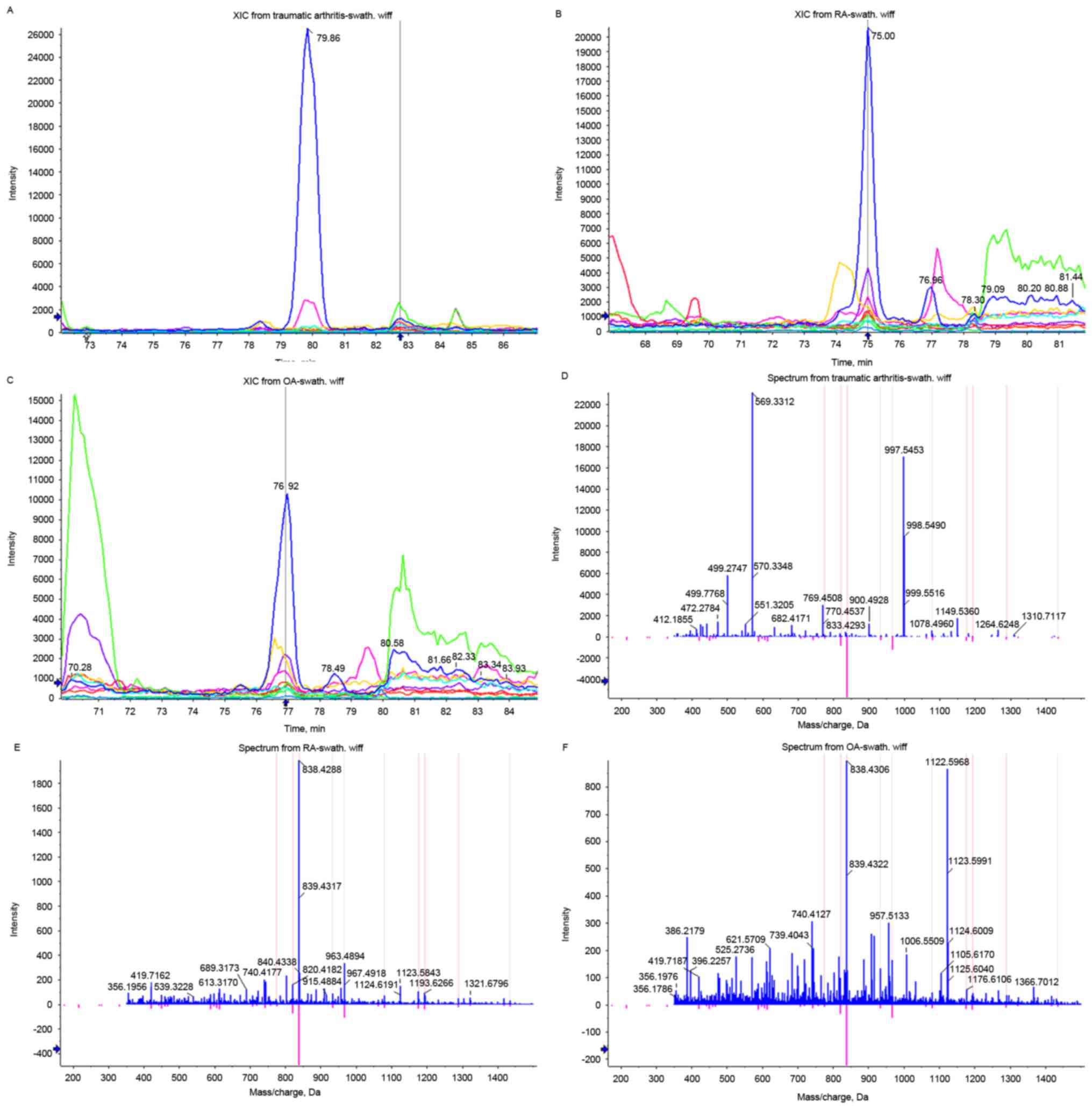
Based on these criteria, the comparison between OA and traumatic
arthritis resulted in the identification of 131 significantly
different proteins, of which 93 corresponded to upregulation and 38
to downregulation in OA. Complement C1r was the most significantly
upregulated protein in OA (Fig.
3). The comparison between RA and traumatic arthritis led to
the identification of 185 significantly different proteins, of
which 152 corresponded to upregulation and 33 to downregulation in
RA. The comparison between OA and RA gave rise to the
identification of 139 significantly different proteins, of which 22
corresponded to upregulation and 117 to downregulation in OA.
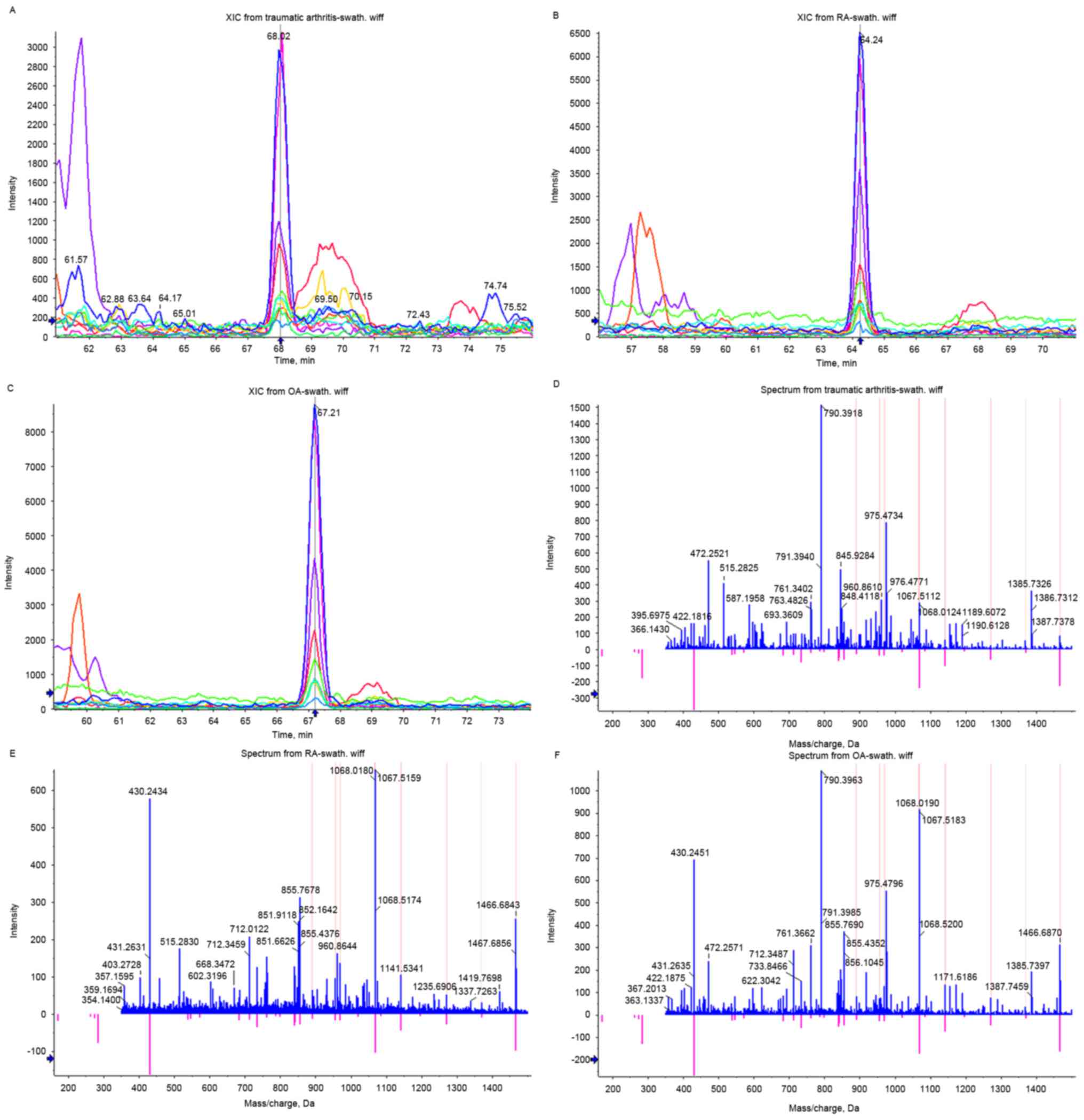
Proteins with a fold change >1.5 in OA and a fold
change <1.5 in RA were defined as differentially expressed
proteins specifically regulated in OA. On the basis of these
criteria, 28 proteins were considered to be specifically regulated
in OA, of which 17 were upregulated and 11 were downregulated
(Table I). DKK2 was one of the
proteins specifically upregulated in OA (Fig. 4). Proteins up-/downregulated with a
fold change >1.5 in OA and down-/upregulated with a fold change
>1.5 in RA were defined as differentially expressed proteins
conversely regulated in OA and RA. According to these criteria, 8
proteins were identified to be conversely regulated in OA and RA,
of which 3 were upregulated in OA and downregulated in RA, 5 were
downregulated in OA and upregulated in RA (Table II).
 | Table I.Differentially expressed proteins
specifically regulated in osteoarthritis identified by SWATH-mass
spectrometry. |
Table I.
Differentially expressed proteins
specifically regulated in osteoarthritis identified by SWATH-mass
spectrometry.
| Protein ID | Accession | Protein name | t-value | P-value | Fold change |
|---|
| 1 | E7ES66 | Glycocalicin | −9.835 | 0.0006 | 5.251 |
| 2 | P02748 | Complement component
C9 | 41.185 |
2.08E-06 | 3.898 |
| 3 | Q15848 | Adiponectin |
7.933 | 0.00137 | 3.575 |
| 4 | P01833 | Polymeric
immunoglobulin receptor | −3.682 | 0.02117 | 3.056 |
| 5 | P10643 | Complement
component C7 | 12.308 | 0.00025 | 2.900 |
| 6 | F8W1Q3 | Biotinidase | −54.461 |
6.81E-07 | 2.676 |
| 7 | O95497 | Pantetheinase | −17.427 |
6.36E-05 | 2.391 |
| 8 | F6SYF8 | Dickkopf-related
protein 2 | −19.531 |
4.05E-05 | 2.178 |
| 9 | P01024 | Complement C3 | 20.642 |
3.25E-05 | 1.981 |
| 10 | P04208 | Ig λ chain V–I
region WAH | −3.730 | 0.0203 | 1.918 |
| 11 | P01019 |
Angiotensinogen | −43.998 |
1.60E-06 | 1.846 |
| 12 | Q5VY30 | Plasma
retinol-binding protein (1–182) | −5.867 | 0.00421 | 1.830 |
| 13 | F5H6I0 |
β2-microglobulin | 30.320 |
7.05E-06 | 1.781 |
| 14 | P02768 | Serum albumin | −51.884 |
8.26E-07 | 1.723 |
| 15 | P02760 | Protein AMBP | −11.934 | 0.00028 | 1.709 |
| 16 | P10599 | Thioredoxin |
3.941 | 0.01694 | 1.647 |
| 17 | P07355-2 | Annexin A2 | 26.293 |
1.24E-05 | 1.622 |
| 18 | P01857 | Ig γ-1 chain C
region |
6.790 | 0.00246 | 0.546 |
| 19 | P04207 | Ig κ chain V–III
region CLL |
8.997 | 0.00084 | 0.534 |
| 20 | P01861 | Ig γ-4 chain C
region |
8.329 | 0.00114 | 0.505 |
| 21 | P02647 | Apolipoprotein
A-I | 66.441 |
3.07E-07 | 0.484 |
| 22 | P58546 | Myotrophin |
4.805 | 0.00862 | 0.456 |
| 23 | C9JIZ6 | Saposin-D |
7.474 | 0.00171 | 0.340 |
| 24 | E9PGN7 | Plasma protease C1
inhibitor | 14.422 | 0.00013 | 0.327 |
| 25 | P60174 | Triosephosphate
isomerase | 15.263 | 0.00011 | 0.322 |
| 26 | Q9H299 | SH3 domain-binding
glutamic acid-rich-like protein 3 | 8.893 | 0.00088 | 0.268 |
| 27 | B7Z6Z4 | Myosin light
polypeptide 6 | 24.641 |
1.61E-05 | 0.151 |
| 28 | Q14624-2 | Isoform 2 of
Inter-α-trypsin inhibitor heavy chain H4 | 75.790 |
1.82E-07 | 0.076 |
 | Table II.Differentially expressed proteins
conversely regulated in OA and RA identified by SWATH-MS. |
Table II.
Differentially expressed proteins
conversely regulated in OA and RA identified by SWATH-MS.
| Protein ID | Accession | Protein name | P-value (OA) | Fold change
(OA) | P-value (RA) | Fold change
(RA) |
|---|
| 1 | P09211 | Glutathione
S-transferase | 0.01097 | 2.051 |
2.87E-06 | 0.527 |
| 2 | P13611-2 | Isoform V1 of
Versican core protein |
5.08E-05 | 1.985 |
2.34E-05 | 0.380 |
| 3 | P10915 | Hyaluronan and
proteoglycan link protein |
1.28E-05 | 1.636 |
4.36E-06 | 0.212 |
| 4 | P37837 | Transaldolase | 0.00826 | 0.577 | 0.00022 | 5.286 |
| 5 | P07737 | Profilin-1 | 0.03004 | 0.572 |
3.57E-05 | 4.777 |
| 6 | P63104 | 14-3-3 protein
zeta/delta |
1.82E-07 | 0.501 |
1.28E-05 | 2.567 |
| 7 | P06727 | Apolipoprotein
A-IV |
1.73E-06 | 0.463 |
1.24E-06 | 1.536 |
| 8 | P01880 | Ig delta chain C
region | 0.02884 | 0.261 |
3.47E-05 | 2.521 |
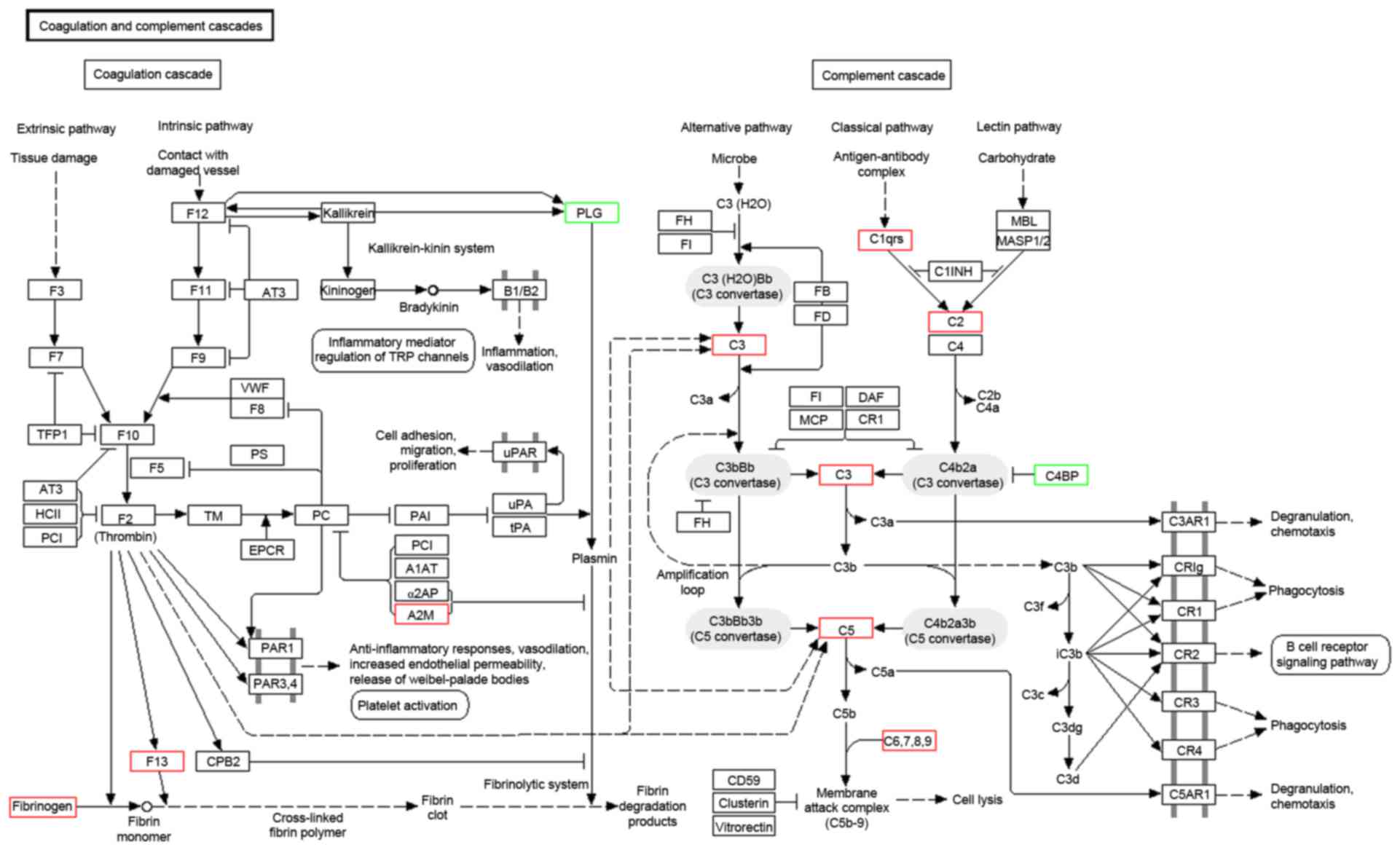
GO annotation with respect to the 131 differentially
expressed proteins in OA demonstrated that the cellular component
of these proteins was predominantly derived from the extracellular
matrix (ECM), cytoplasmic membrane, cytoplasm, secretory granule
and lipoprotein particle. The molecular function of these proteins
primarily included binding, transporter, enzyme activator and
inhibitor activity. The biological process these proteins
participated in predominantly included the acute inflammatory
response, complement activation, immune response, response to
stimulus, regulation of apoptosis and homeostatic process. KEGG
pathway annotation indicated that these proteins were enriched in
complement and coagulation cascades (Fig. 5), ECM-receptor interaction and the
peroxisome proliferator activated receptor signaling pathway.
ELISA
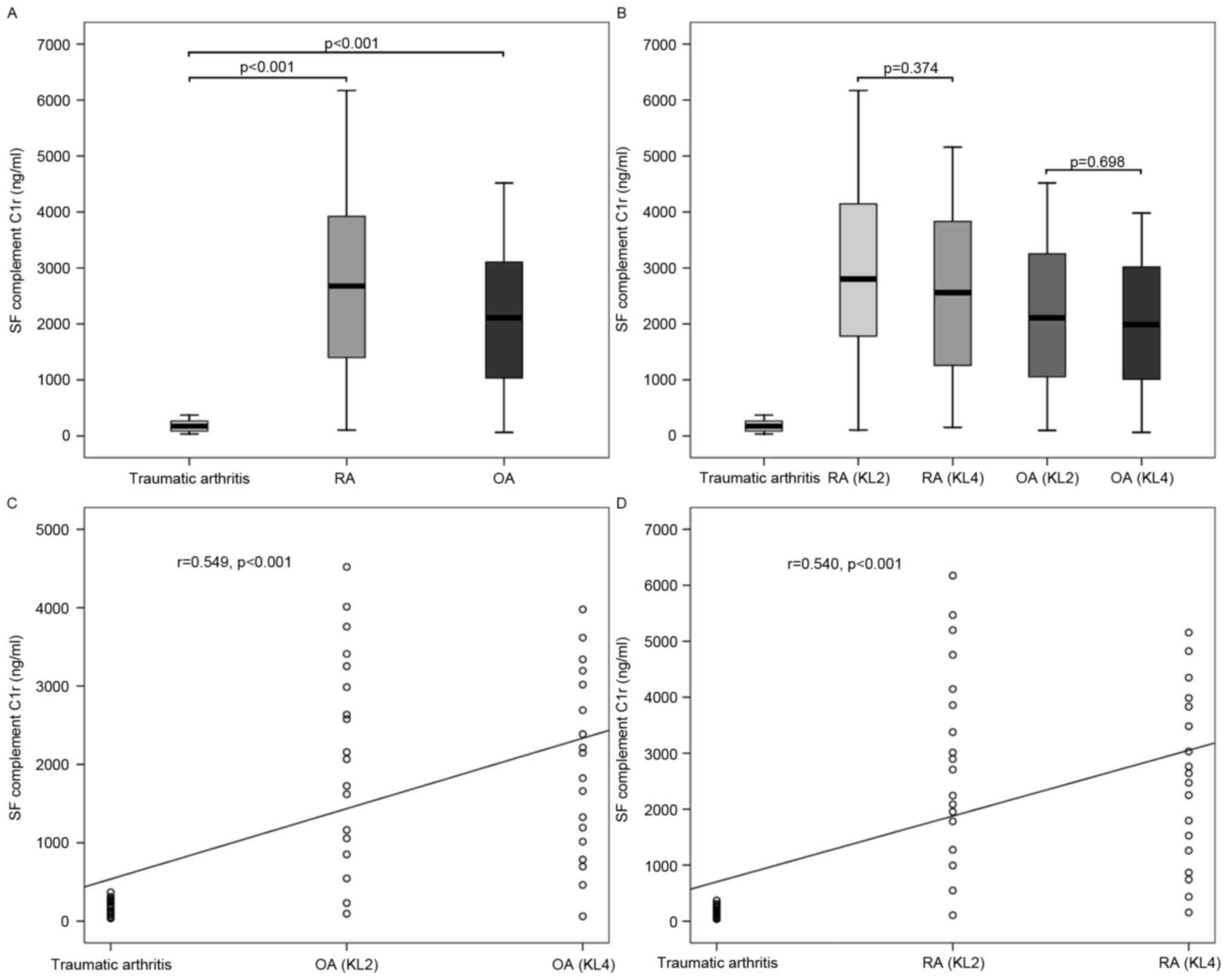
As presented in Fig.
6, the mean level of complement C1r in SF from OA was
2,063.1±1,238.1 ng/ml, that from RA was 2,726.4±1,627.8 ng/ml and
that from traumatic arthritis was 180.6±102.4 ng/ml. The mean level
of complement C1r in SF from RA was significantly higher than those
from OA (P=0.033) and traumatic arthritis (P<0.001), and that
from OA was also significantly higher than that from traumatic
arthritis (P<0.001). According to the KL radiographic grading
criteria, the mean level of complement C1r in SF from OA with KL
grade 2 was 2,147.8±1,337.2 ng/ml, and that from OA with KL grade 4
was 1,978.4±1,163.1 ng/ml. The mean level of complement C1r in SF
from traumatic arthritis was significantly lower than those from OA
with KL grade 2 (P<0.001) and KL grade 4 (P<0.001); however,
no significant difference was identified between the latter two
(P=0.698). The levels of complement C1r in SF from OA were
positively correlated with disease severity (r=0.549, P<0.001).
Similarly, the mean level of complement C1r in SF from RA with KL
grade 2 was 2,920.9±1,743.0 ng/ml, and that from RA with KL grade 4
was 2,531.8±1,528.7 ng/ml. In addition, the mean level of
complement C1r in SF from traumatic arthritis was significantly
lower than those from RA with KL grade 2 (P<0.001) and KL grade
4 (P<0.001); however, no significant difference was identified
between the latter two (P=0.374). The levels of complement C1r in
SF from RA were also positively correlated with disease severity
(r=0.540, P<0.001).
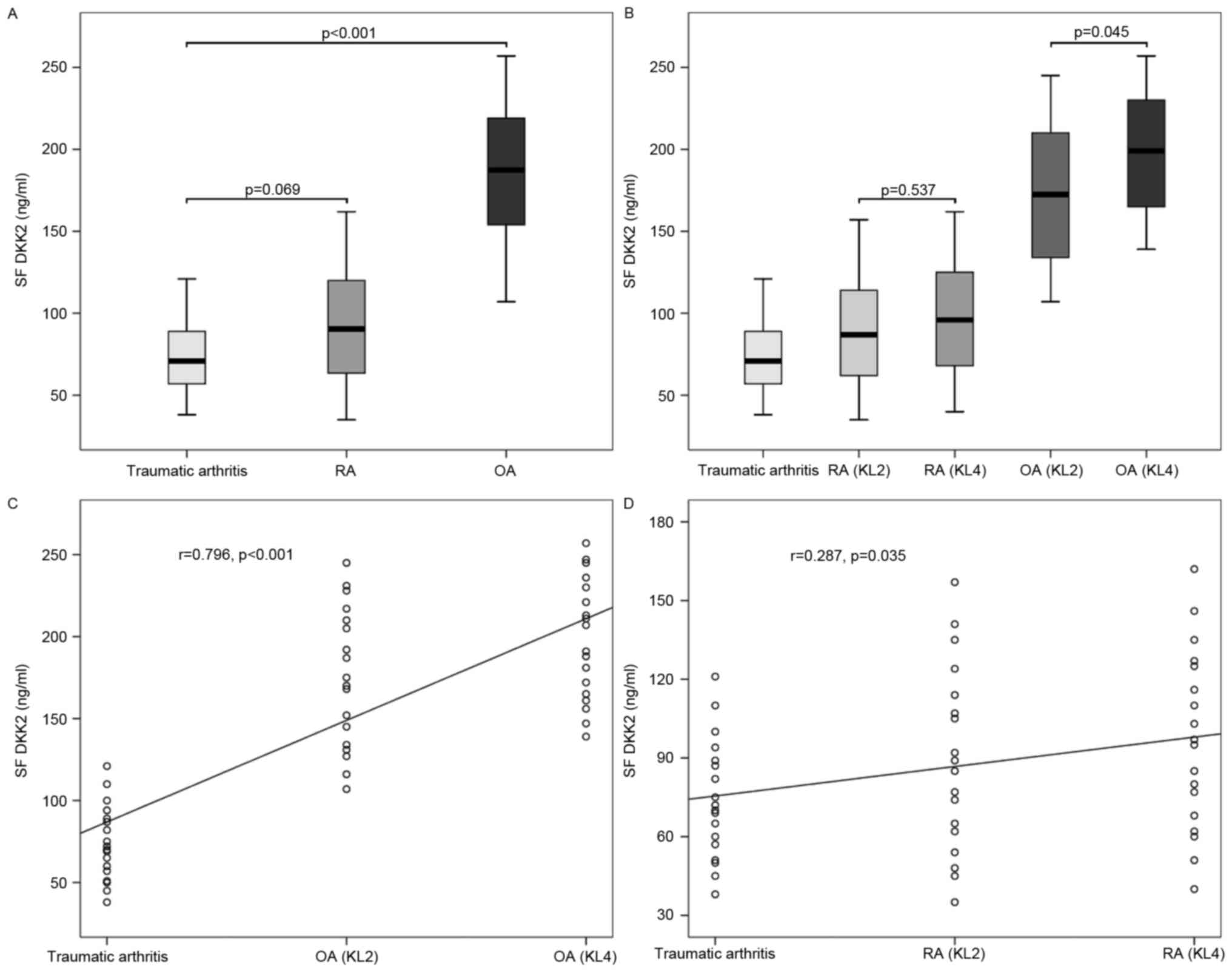
As shown Fig. 7,
the mean level of DKK2 in SF from OA was 186.3±41.0 ng/ml, that
from RA was 93.0±34.5 ng/ml and that from traumatic arthritis was
74.2±22.9 ng/ml. The mean level of DKK2 in SF from OA was
significantly higher when compared with those from RA (P<0.001)
and traumatic arthritis (P<0.001); however, no significant
difference was identified between the latter two (P=0.069).
According to the KL radiographic grading criteria, the mean level
of DKK2 in SF from OA with KL grade 2 was 174.4±42.5 ng/ml, and
that from OA with KL grade 4 was 198.2±36.8 ng/ml. The mean level
of DKK2 in SF from OA with KL grade 4 was significantly higher than
those from OA with KL grade 2 (P=0.045) and traumatic arthritis
(P<0.001), and that from OA with KL grade 2 was also
significantly higher than that from traumatic arthritis
(P<0.001). The levels of DKK2 in SF from OA were positively
correlated with disease severity (r=0.796, P<0.001). Similarly,
the mean level of DKK2 in SF from RA with KL grade 2 was 89.4±35.4
ng/ml, and that from RA with KL grade 4 was 96.6±34.3 ng/ml. No
significant difference was detected between the mean levels of DKK2
in SF from RA with KL grade 2 and traumatic arthritis (P=0.195),
between those from RA with KL grade 4 and traumatic arthritis
(P=0.057) and between those from RA with KL grade 2 and KL grade 4
(P=0.537). In addition, the levels of DKK2 in SF from RA were
positively correlated with disease severity (r=0.287, P=0.035).
Discussion
OA exerts a great influence on physical and
psychological health of the elderly due to its high morbidity and
disability rate (1). The lack of
early diagnostic maneuvers and effective pharmacotherapy for OA is
primarily attributed to the limitations in current understanding of
its pathogenesis, which may be further elucidated by investigating
the SF proteome. However, for a variety of reasons, previous
studies with regard to proteomic analysis of OA were not consistent
in demonstrating the pathogenesis and failed to detect effective
biomarkers or drug targets to facilitate early diagnosis and
intervention of OA. Therefore, this provided the rationale for
conducting the present study. In the current study, with the use of
SWATH-MS, a total of 131 proteins were identified to be
differentially expressed in SF from OA, of which 28 were
specifically regulated in OA and eight were conversely regulated in
OA and RA. The quantity of differentially expressed proteins
identified in the current study is markedly more than in previous
studies analyzing SF proteome using other quantitative proteomic
technologies (7,8). With the use of two-dimensional
polyacrylamide gel electrophoresis, Liu et al (15) identified 10 differential protein
spots between the synovial membranes of the bilateral and
unilateral knee OA in surgery-induced rabbit models. Similarly,
Fernandez-Puente et al (16) developed a multiple reaction
monitoring method with which a panel of 14 protein biomarker
candidates for OA was identified. Differential proteins identified
in the above-mentioned studies, such as lumican (15) and haptoglobin (16) were also identified in the current
study. These indicate that, as a novel proteomic technology,
SWATH-MS provides a more comprehensive quantification result in
delineating the SF proteome and markedly benefits the screening of
biomarkers due to its higher resolution and coverage rate.
The hypothesis that the complement system may
participate in OA pathogenesis is substantiated by the
identification of complement C1r and enrichment of differentially
expressed proteins in complement and coagulation cascades. The
results are consistent with those reported by Wang et al
(17), which indicates abnormally
high expression levels and activation of complement components in
SF and synovial membranes from OA. Complement activation in turn
results in the formation of membrane attack complex (MAC) on
chondrocytes, which either kills the cells or causes them to
produce matrix-degrading enzymes, inflammatory mediators and other
complement effectors, all of which promote joint pathology. A
previous study indicates that MAC mediates collagenase expression
and cartilage destruction primarily via extracellular
signal-regulated kinase-dependent c-Fos induction (18). In addition, complement activation
and MAC assembly also stimulate the cell cycle by inducing
expression of c-Jun, JunD, and c-Fos mRNAs (19), which appears to have a potential
correlation with the pathology of hyperosteogeny and synovial
hyperplasia in OA. These findings provide a rationale for targeting
the complement system as a disease-modifying therapeutic strategy
for OA.
DKK2 is one of the proteins that is specifically
upregulated in OA, the alteration of which has been confirmed by
subsequent ELISA in the present study. As a Wnt signaling
antagonist, the overexpression of DKK2 contributes to the abnormal
phenotype and poor mineralization of OA osteoblasts by altering the
Wnt signaling pathway, which is crucial for osteogenesis and
regulates terminal osteoblast differentiation (20). The upregulation of DKK2 in OA
osteoblasts may be associated with the hypoxic condition and
ischemic episode caused by modification of subchondral bone
vascularization (21). Besides
DKK2, there are many other proteins specifically regulated in OA.
These proteins and their possible underlying mechanism in OA
pathogenesis are as follows: i) Adiponectin is a protein hormone
secreted predominantly by differentiated adipocytes, it increases
intercellular adhesion molecule-1 (ICAM-1) expression levels in OA
synovial fibroblasts via the liver kinase B1/calmodulin-dependent
protein kinase II, AMP-activated protein kinase, c-Jun, and
activator protein 1 signaling pathway. Adiponectin-induced ICAM-1
expression promotes the adhesion of monocytes to OA synovial
fibroblasts, which may be involved in the pathogenesis of OA
(22). Furthermore, adiponectin
enhances production of nitric oxide, interleukin (IL)-6, matrix
metalloproteinase (MMP)-1 and MMP-3 in OA chondrocytes in a
mitogen-activated protein kinase-dependent manner (23). ii) The mechanisms by which
β2-microglobulin induces joint destruction is to induce the release
of MMP-1 without concomitant release of tissue inhibitor of
metalloproteinase-1 (TIMP-1) from OA synovial fibroblasts, leading
to an increase in the MMP-1/TIMP-1 ratio, which is indicative of
uncontrolled collagenolysis. The release of MMP-1 induced by
β2-microglobulin may be inhibited by doxycycline (24). iii) Thioredoxin, known as a
cellular reducing catalyst induced by oxidative stress, is involved
in the redox regulation of transcription factors, such as nuclear
factor (NF)-κB. It appears to accelerate the nuclear translocation
of NF-κB, a major transcriptional regulator for production of IL-6
and IL-8 on stimulation with tumor necrosis factor-a (25). iv) As for annexin A2, a discoidin
domain receptor 2 (DDR-2) binding protein in the DDR-2/MMP
signaling pathway, its phosphorylation by phosphorylated-DDR-2
leads to MMP-13 secretion (26).
Glutathione S-transferase (GST) is one of the
proteins conversely regulated in OA and RA identified in the
present study. The differential expression of GST in OA coincides
with the results documented by Ostalowska et al (27), which indicate aberrant changes in
the activities of antioxidant enzymes, including superoxide
dismutase and GST in SF from OA. GST is critically important for
cellular defense against oxidative stress-induced apoptosis in OA
chondrocytes, possibly by elimination of 4-hydroxynonenal, one of
the most abundant and reactive aldehydes of lipid peroxidation
products (28). Hyaluronan and
proteoglycan link protein is another protein conversely regulated
in OA and RA. It has been reported that LP stabilizes the
proteoglycan aggregates towards dissociation, and helps to protect
them from degradation under conditions where tissue catabolism is
promoted (29).
There were certain limitations of the current study.
Due to ethical restrictions, patients undergoing arthroscopy for
traumatic knee arthritis were selected, rather than healthy
individuals, and served as the standard control group. However, as
SF samples were acquired within 7 days after injury and patients in
this group all exhibited intact articular cartilage under
arthroscopy, it is reasonable for us to consider that these samples
are most similar to those from healthy individuals in the proteomic
profile. Furthermore, to investigate proteins specifically
regulated in OA, the design of the inter-control group requires
more diversification. In addition to RA, other arthropathies due to
systemic lupus erythematosus, ankylosing spondylitis, gout,
psoriasis or haemochromatosis should be included as inter-control
groups.
In conclusion, the findings of the present study
provide an improved understanding of the pathogenesis of OA, and
may facilitate the early diagnosis and treatment of OA following
the identification of various potential biomarkers and drug
targets.
Acknowledgements
The authors would like to thank Dr Yang Liu for
assistance in acquisition of SF samples, and Dr Can Huang for
assistance with collection of patient data.
Glossary
Abbreviations
Abbreviations:
|
DDA
|
data-dependent acquisition
|
|
DDR-2
|
discoidin domain receptor 2
|
|
DKK2
|
Dickkopf-related protein 2
|
|
ECM
|
extracellular matrix
|
|
ELISA
|
enzyme-linked immunosorbent assay
|
|
GO
|
gene ontology
|
|
GST
|
glutathione S-transferase
|
|
ICAM-1
|
intercellular adhesion molecule-1
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
LP
|
link protein
|
|
MAC
|
membrane attack complex
|
|
OA
|
osteoarthritis
|
|
RA
|
rheumatoid arthritis
|
|
SF
|
synovial fluid
|
|
SWATH-MS
|
SWATH-mass spectrometry
|
|
TIMP-1
|
tissue inhibitor of
metalloproteinase-1
|
References
|
1
|
Neogi T: The epidemiology and impact of
pain in osteoarthritis. Osteoarthritis Cartilage. 21:1145–1153.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arnscheidt C, Meder A and Rolauffs B:
Early diagnosis of osteoarthritis: Clinical reality and promising
experimental techniques. Z Orthop Unfall. 154:254–268.
2016.PubMed/NCBI
|
|
3
|
Jevsevar DS: Treatment of osteoarthritis
of the knee: Evidence-based guideline, 2nd edition. J Am Acad
Orthop Surg. 21:571–576. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jiao Q, Wei L, Chen C, Li P, Wang X, Li Y,
Guo L, Zhang C and Wei X: Cartilage oligomeric matrix protein and
hyaluronic acid are sensitive serum biomarkers for early cartilage
lesions in the knee joint. Biomarkers. 21:146–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nemirovskiy O, Li WW and Szekely-Klepser
G: Design and validation of an immunoaffinity LC-MS/MS assay for
the quantification of a collagen type II neoepitope peptide in
human urine: Application as a biomarker of osteoarthritis. Methods
Mol Biol. 641:253–270. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Duan Y, Hao D, Li M, Wu Z, Li D, Yang X
and Qiu G: Increased synovial fluid visfatin is positively linked
to cartilage degradation biomarkers in osteoarthritis. Rheumatol
Int. 32:985–990. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamagiwa H, Sarkar G, Charlesworth MC,
McCormick DJ and Bolander ME: Two-dimensional gel electrophoresis
of synovial fluid: Method for detecting candidate protein markers
for osteoarthritis. J Orthop Sci. 8:482–490. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gobezie R, Kho A, Krastins B, Sarracino
DA, Thornhill TS, Chase M, Millett PJ and Lee DM: High abundance
synovial fluid proteome: Distinct profiles in health and
osteoarthritis. Arthritis Res Ther. 9:R362007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu WW, Wang G, Baek SJ and Shen RF:
Comparative study of three proteomic quantitative methods, DIGE,
cICAT and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res.
5:651–658. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Balakrishnan L, Bhattacharjee M, Ahmad S,
Nirujogi RS, Renuse S, Subbannayya Y, Marimuthu A, Srikanth SM,
Raju R and Dhillon M: Differential proteomic analysis of synovial
fluid from rheumatoid arthritis and osteoarthritis patients. Clin
Proteomics. 11:12014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lourido L, Calamia V, Mateos J,
Fernández-Puente P, Fernández-Tajes J, Blanco FJ and Ruiz-Romero C:
Quantitative proteomic profiling of human articular cartilage
degradation in osteoarthritis. J Proteome Res. 13:6096–6106. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gillet LC, Navarro P, Tate S, Röst H,
Selevsek N, Reiter L, Bonner R and Aebersold R: Targeted data
extraction of the MS/MS spectra generated by data-independent
acquisition: A new concept for consistent and accurate proteome
analysis. Mol Cell Proteomics. 11:O111.0167172012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Collins BC, Gillet LC, Rosenberger G, Röst
HL, Vichalkovski A, Gstaiger M and Aebersold R: Quantifying protein
interaction dynamics by SWATH mass spectrometry: Application to the
14-3-3 system. Nat Methods. 10:1246–1253. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aletaha D, Neogi T, Silman AJ, Funovits J,
Felson DT, Bingham CO III, Birnbaum NS, Burmester GR, Bykerk VP and
Cohen MD: 2010 Rheumatoid arthritis classification criteria: An
american college of rheumatology/european league against rheumatism
collaborative initiative. Arthritis Rheum. 62:2569–2581. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu W, He J, Lin R, Liang J and Luo Q:
Differential proteomics of the synovial membrane between bilateral
and unilateral knee osteoarthritis in surgery–induced rabbit
models. Mol Med Rep. 14:2243–2249. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fernandez-Puente P, Calamia V,
González-Rodríguez L, Lourido L, Camacho-Encina M, Oreiro N,
Ruiz-Romero C and Blanco FJ: Multiplexed mass spectrometry
monitoring of biomarker candidates for osteoarthritis. J
Proteomics. 152:216–225. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Q, Rozelle AL, Lepus CM, Scanzello
CR, Song JJ, Larsen DM, Crish JF, Bebek G, Ritter SY and Lindstrom
TM: Identification of a central role for complement in
osteoarthritis. Nat Med. 17:1674–1679. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Litherland GJ, Elias MS, Hui W, Macdonald
CD, Catterall JB, Barter MJ, Farren MJ, Jefferson M and Rowan AD:
Protein kinase C isoforms zeta and iota mediate collagenase
expression and cartilage destruction via STAT3- and ERK-dependent
c-fos induction. J Biol Chem. 285:22414–22425. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rus HG, Niculescu F and Shin ML: Sublytic
complement attack induces cell cycle in oligodendrocytes. J
Immunol. 156:4892–4900. 1996.PubMed/NCBI
|
|
20
|
Chan TF, Couchourel D, Abed E, Delalandre
A, Duval N and Lajeunesse D: Elevated Dickkopf-2 levels contribute
to the abnormal phenotype of human osteoarthritic osteoblasts. J
Bone Miner Res. 26:1399–1410. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bouvard B, Abed E, Yéléhé-Okouma M,
Bianchi A, Mainard D, Netter P, Jouzeau JY, Lajeunesse D and Reboul
P: Hypoxia and vitamin D differently contribute to leptin and
dickkopf-related protein 2 production in human osteoarthritic
subchondral bone osteoblasts. Arthritis Res Ther. 16:4592014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen HT, Tsou HK, Chen JC, Shih JM, Chen
YJ and Tang CH: Adiponectin enhances intercellular adhesion
molecule-1 expression and promotes monocyte adhesion in human
synovial fibroblasts. PLoS One. 9:e927412014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koskinen A, Juslin S, Nieminen R, Moilanen
T, Vuolteenaho K and Moilanen E: Adiponectin associates with
markers of cartilage degradation in osteoarthritis and induces
production of proinflammatory and catabolic factors through
mitogen-activated protein kinase pathways. Arthritis Res Ther.
13:R1842011. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Moe SM, Singh GK and Bailey AM:
beta2-microglobulin induces MMP-1 but not TIMP-1 expression in
human synovial fibroblasts. Kidney Int. 57:2023–2034. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yoshida S, Katoh T, Tetsuka T, Uno K,
Matsui N and Okamoto T: Involvement of thioredoxin in rheumatoid
arthritis: Its costimulatory roles in the TNF-alpha-induced
production of IL-6 and IL-8 from cultured synovial fibroblasts. J
Immunol. 163:351–358. 1999.PubMed/NCBI
|
|
26
|
Zhao W, Zhang C, Shi M, Zhang J, Li M, Xue
X, Zhang Z, Shu Z, Zhu J and Mu N: The discoidin domain receptor
2/annexin A2/matrix metalloproteinase 13 loop promotes joint
destruction in arthritis through promoting migration and invasion
of fibroblast-like synoviocytes. Arthritis Rheumatol. 66:2355–2367.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ostalowska A, Birkner E, Wiecha M,
Kasperczyk S, Kasperczyk A, Kapolka D and Zon-Giebel A: Lipid
peroxidation and antioxidant enzymes in synovial fluid of patients
with primary and secondary osteoarthritis of the knee joint.
Osteoarthritis Cartilage. 14:139–145. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Vaillancourt F, Fahmi H, Shi Q, Lavigne P,
Ranger P, Fernandes JC and Benderdour M: 4-Hydroxynonenal induces
apoptosis in human osteoarthritic chondrocytes: The protective role
of glutathione-S-transferase. Arthritis Res Ther. 10:R1072008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rodriguez E and Roughley P: Link protein
can retard the degradation of hyaluronan in proteoglycan
aggregates. Osteoarthritis Cartilage. 14:823–829. 2006. View Article : Google Scholar : PubMed/NCBI
|