Introduction
Colorectal cancer (CRC) is one of the most common
cancers worldwide (1,2). In China, CRC is among the five most
prevalent types of cancer, with 376,300 new diagnoses made in 2015
alone (3). Furthermore, CRC is one
of the five leading causes of cancer-associated mortality in China,
with 191,000 CRC-associated mortalities reported in 2015 (3). Despite recent improvements in
diagnostic technologies and clinical treatments, the prognoses for
patients with CRC remain poor, particularly in patients with
advanced stage CRC, due to risks of relapse and metastasis
(4). It has previously been
estimated that ~50% of patients with CRC develop distant
metastases, particularly liver metastases (5). However, the exact molecular
mechanisms underlying CRC progression and metastasis development
remain undetermined.
Rab11 family interacting proteins (Rab11-FIPs),
comprising of Rip11, Rab11-FIP1, Rab11-FIP2, Rab11-FIP3, Rab
coupling protein (RCP) and Rab11-FIP4, have previously been
revealed as regulators of multiple Rab and ADP-ribosylation factor
GTPases (6–9). Rab11-FIP4 was first identified in a
human cDNA project as a gene termed KIAA1821 (10). Rab11-FIP4 shares a common
C-terminal Rab11 binding domain (RBD) with the other FIPs, and
selectively interacts with the GTP-bound form of Rab11 (11). Furthermore, Rab11-FIP4 belongs to
the class II FIPs, and its N-terminus contains an EF-hand
calcium-binding motif (12). In
addition, Rab11-FIP4 localizes to the endosomal recycling
compartment and functions as a downstream modulator of Rab11 to
regulate vesicle trafficking (8,13). A
previous study demonstrated that Rab11-FIP4 interacts with Rab11 in
a GTP-dependent manner, and that overexpression of Rab11-FIP4
condenses the Rab11 positive compartment in HeLa cells (14). In addition, previous studies have
also revealed that Rab11-FIP4 is involved in cell proliferation and
differentiation during retinal development, also in a
Rab11-independent manner (15,16).
Furthermore, a recent study demonstrated that Rab11-FIP4 has a
pro-metastatic function via activation of the mechanistic target of
rapamycin/AKT1 substrate 1 pathway in hepatocellular carcinoma
(HCC) (17). However, the
potential implication of Rab11-FIP4 activity in CRC remains
undetermined. In the present study, the clinical implications,
functions and underlying mechanisms of Rab11-FIP4 in CRC were
investigated. The results of the present study demonstrated that
Rab11-FIP4 could be a promising therapeutic target for CRC
treatment.
Materials and methods
Patients and sample collection
CRC samples and their corresponding non-tumorous
(NT) samples (50 pairs of tissue samples and 100 CRC tissues) were
obtained at The Second Affiliated Hospital and Yuying Children's
Hospital of Wenzhou Medical University (Wenzhou, China) between
January 2009 and October 2015. Pathological diagnosis of CRC was
confirmed by a pathologist without knowledge of patient
characteristics. All patients included in this study provided
signed consent, and the study was approved by the Ethics Committee
of Wenzhou Medical University.
Immunohistochemical staining
Tissue microarrays (TMAs) were constructed, and
immunohistochemistry (IHC) staining and immunostaining scoring were
performed according to previous studies (18). Briefly, TMAs were dewaxed,
rehydrated, and blocked for 90 min with 2% normal goat serum at
37°C (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Vectastain Elite ABC Kit (Vector Laboratories, Inc., Burlingame,
CA, USA) was used in IHC analysis. Tissues sections (5 µm) were
incubated with primary antibodies against Rab11FIP4 (cat. no.
HPA021595; 1:50; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and
hypoxia-inducible factor 1-α (cat. no. MA1-16504; 1:100; Thermo
Fisher Scientific, Inc.) overnight at 4°C in a humidified chamber.
Horseradish peroxidase-conjugated secondary antibodies against
rabbit and mouse immunoglobulins were developed for 20 min at 37°C
with the DAB peroxidase substrate kit (cat. no. K500711-2; 1:1,000;
Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). Cell
nuclei were stained with hematoxylin and eosin (cat. no. 51275;
Sigma-Aldrich; Merck KGaA) for 90 sec at 37°C. Incubations with
pre-immune serum were used as negative controls. Tissue sections
were viewed with a microscope (×400 magnification; Nikon Eclipse
50i; Nikon Corporation, Tokyo, Japan). IOD was calculated using
Image-Pro Plus software (version 6; Media Cybernetics, Inc.,
Rockville, MD, USA) and the mean IOD was calculated from three
images per specimen.
Cell culture and hypoxic
conditions
Human colorectal cancer cell lines (LoVo and HCT116)
were obtained from the Shanghai Cell Bank, Chinese Academy of
Sciences (Shanghai, China). LoVo and HCT116 cells were cultured in
Dulbecco's Modified Eagle's Medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.), which was supplemented with 10% fetal bovine
serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), penicillin (10
units/ml; Thermo Fisher Scientific Inc.) and streptomycin (10
mg/ml; GE Healthcare Life Sciences, Logan, UT, USA). Both cell
lines were incubated in a humidified atmosphere at 37°C. For
hypoxic culture, cells were placed in a hypoxia incubator for 48 h
in an atmosphere consisting of 94% N2, 5% CO2
and 1% O2.
Construction of Rab11-FIP4
overexpression lentivirus
The Rab11-FIP4 opening reading frame sequence
(NM_032932.5) was cloned into the pWPXL vector (Addgene, Inc.,
Cambridge, MA, USA), and the pWPXL-Rab11FIP4 recombinant plasmid
was constructed. Plasmids were purchased from Addgene, Inc.
(Cambridge, MA, USA). Using Lipofectamine 2000™ (Thermo Fisher
Scientific, Inc.), 293T cells were co-transfected with
pWPXL-Rab11FIP4, psPAX2 (the packaging plasmid) and pMD2.G (the
envelope plasmid). Following this, the lentivirus was harvested 48
h post-transfection, and CRC cells were infected with the
lentivirus (MOI=10) in the presence of polybrene (6 µg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells and tissues using
TRIzol reagent (Thermo Fisher Scientific, Inc.), and RT was
performed using PrimeScript™ RT Reagent Kit (Takara Biotechnology
Co., Ltd., Dalian, China) in a total volume of 50 µl. The RT
reaction proceeded for 15 min at 37°C followed by 5 sec at 85°C,
according to manufacturer's instructions. The cDNA was stored at
−20°C. SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.) was
subsequently used to perform qPCR. The PCR conditions were as
follows: 95°C for 15 sec followed by 35 cycles of 95°C for 5 sec
and 60°C for 40 sec. β-actin was used as loading control, and the
relative expression levels were determined by the following
equation: 2−ΔΔCq
(ΔCq=ΔCqtarget-ΔCqβ-actin) (19). The primers used in this study were
as follows: Rab11FIP4 forward, 5′-CTGCTCTCAATGCTGCAAGA-3′ and
reverse, 5′-TCGCAAGAGTCAATGCTGTC'; insulin-like growth factor 1
receptor (IGF1R) forward, 5′-TCGACATCCGCAACGACTATC-3′ and reverse,
5′-CCAGGGCGTAGTTGTAGAAGAG-3′; β-actin forward,
5′-TTGTTACAGGAAGTCCCTTGCC-3′ and reverse,
5′-ATGCTATCACCTCCCCTGTGTG-3′.
Western blot
Western blot was performed according to a previous
study (18). Briefly, tumor cells
and tissues were lysed, and cell debris was removed. Lysates of
cells or tissues were prepared with T-PER tissue protein extraction
reagent (Pierce; Thermo Fisher Scientific, Inc.) supplemented with
proteinase inhibitors (Roche Applied Science, Switzerland) and
phosphatase inhibitors (Roche Diagnostics, Basel, Switzerland). To
investigate the effects of IGF1R inhibition on protein expression,
HCT116-Rab11-FIP4 cells were treated with NVP-AEW541 (5 µM) for 12
h before they were lysed. Protein concentrations were determined
with a bicinchoninic assay kit (Pierce; Thermo Fisher Scientific,
Inc.). A total of 60–80 µg protein/sample was separated on 10%
SDS-PAGE and immobilized to nitrocellulose membranes (Bio-Rad
Laboratories Inc., Hercules, CA, USA). The nitrocellulose membranes
were then blocked for ~2 h with 5% fat-free milk at room
temperature. Specific primary antibodies against Rab11-FIP4 (cat.
no. sc-165331; 1:1,000; Santa Cruz Biotechnology Inc., Dallas, TX,
USA), Rab11 (cat. no. sc-6565; 1:1,000; Santa Cruz Biotechnology
Inc.), hypoxia-inducible factor-1α (HIF-1α; cat. no. sc-10790;
1:1,000; Santa Cruz Biotechnology Inc.), IGF1R (cat. no. sc-7952;
1:1,000; Santa Cruz Biotechnology Inc.), phosphorylated
(p)-extracellular signal-regulated kinase 1/2 (ERK1/2; cat. no.
sc-101760; 1:1,000; Santa Cruz Biotechnology Inc.), total-ERK1/2
(cat. no. sc-514302; 1:1,500; Santa Cruz Biotechnology Inc.), p-AKT
serine/threonine kinase (AKT; cat. no. sc-7985-R; 1:1,000; Santa
Cruz Biotechnology Inc.), total-AKT (cat. no. sc-8312; 1:1,500;
Santa Cruz Biotechnology, Inc.), and β-actin (cat. no. sc-58673;
1:3,000; Santa Cruz Biotechnology, Inc.) were incubated at 4°C
overnight. Following incubation with horseradish
peroxidase-conjugated secondary anti-rabbit or anti-mouse
antibodies (BS12478 or BS13278; 1:5,000; Bioworld Technology, Inc.,
St. Louis Park, MN, USA) for 1 h at room temperature. Bands were
visualized using an enhanced chemiluminescence reagent (ECL Plus;
GE Healthcare, Chicago, IL, USA). ImageLab software (version 4.1;
Bio-Rad Laboratories, Inc.) was used for image acquisition and
densitometric analysis of bands.
Co-immunoprecipitation (Co-IP)
Pierce® Co-IP kit (Thermo Fisher
Scientific, Inc.) was used to perform a Co-IP assay according to
the manufacturer's instructions. In brief, an antibody against
Rab11-FIP4 (cat. no. NBP2-45496; 1:50; Novus Biologicals, LLC,
Littleton, CO, USA) was immobilized to AminoLink Plus Coupling
Resin (Thermo Fisher Scientific, Inc.) at room temperature. Control
resin was used to prevent nonspecific binding and following this,
cell lysates were added to the spin column, which contained the
immobilized antibody resin, and incubated overnight at 4°C.
Following this, gentle elution buffer was used to disassociate the
interacting proteins from the immobilized antibodies.
Dual-luciferase reporter assay
Dual-luciferase reporter assay (Promega Corporation,
Madison, WI, USA) was then used to detect luciferase activity.
HCT116 cells were plated in a 96-well plate. Using Lipofectamine
2000™ (Thermo Fisher Scientific, Inc.), pGL3 (Firefly luciferase)
constructs of the truncated Rab11-FIP4 promoter were co-transfected
with pCMV-HIF1α (200 ng/well) or pCMV-vector (200 ng/well) and
pRL-TK (6 ng/well; Renilla luciferase) in HCT116 cells.
Empty plasmid of pGL3 was defined as pGL3-Basic. The pGL3 and
pRL-TK vectors were purchased from Promega Corporation (Madison,
WI, USA). The Pcmv vector was purchased from Addgene, Inc.
(Cambridge, MA, USA). The plate was washed with PBS 6 h
post-transfection and replaced with fresh culture medium. The
firefly and Renilla luciferase activity was then measured 48
h post-transfection using the Dual-Luciferase® Reporter
Assay System (Promega Corporation). Firefly luciferase activity was
normalized to the Renilla luciferase activity.
Cell proliferation assay
The proliferation of CRC cells was detected using
the Cell Counting Kit-8 (CCK-8) reagent (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan). Briefly, cells
(4×103 per well) were seeded in 96-well culture plates
and then cultured at 37°C for 0, 24, 48 and 72 h after attachment.
CCK-8 assay buffer (100 µl/per well) was then added into the
plates, and 450 nm optical density values were obtained using a
microplate reader (BioTek Instruments, Inc., Winooski, VT, USA)
according to manufacturer's protocol.
Plate colony formation assay
LoVo and HCT116 cells (800 per well) were seeded
into separate 6-well plates for 8 days. Culture medium was changed
every 2 days. Following this, cells were washed with PBS, fixed
with 4% paraformaldehyde for 15 min at 37°C and stained with 1%
crystal violet solution (Merck KGaA) for 5 min. Colonies with
>50 cells were counted using a light microscope (×400
magnification).
Transwell (migration) and Matrigel
(invasion) assays
Transwell chambers (8 µm; BD Biosciences, Franklin
Lakes, NJ, USA) were placed in 24-well plates. In order to perform
the migration assay, CRC cells (2×104) in serum free
DMEM were seeded on the upper chamber of Transwell chambers and
medium supplemented with 15% FBS was then added to the lower
chamber. Following incubation for 20 h, migrated CRC cells were
stained for 5 min with 1% crystal violet and imaged using a CKX41
light microscope (Olympus Corporation, Tokyo, Japan) at ×200
magnification. The mean number of migrated cells was determined by
counting the number of cells in three random fields from three
replicate wells. In order to carry out the invasion assay,
Transwell chambers were coated with Matrigel (BD Biosciences). CRC
cells (4×104) in serum-free DMEM were seeded on the
upper chamber of Transwell chambers, and medium supplemented with
15% FBS was then added to the lower chamber. Following incubation
for 36 h, migrated cells were then stained, imaged and counted as
detailed in the aforementioned migration assay. Images of three
random fields from three replicate wells were obtained, and the
numbers of migrated or invasion cells were then counted. To
investigate the potential effects of ERK and AKT on migration and
invasion, wHCT116-Rab11-FIP4 cells were pretreated with either ERK
inhibitor (U0126; 5 µM) or AKT inhibitor (LY294002; 10 µM) for 6 h,
respectively, before seeding onto the upper chamber of Transwell
chambers.
Human phospho-kinase array
HCT116-vector or HCT116-Rab11FIP4 cells were used in
the phosphokinase array assays, according to the manufacturer's
instructions (Proteome Profiler; cat. no. ARY003B; R&D Systems,
Inc., Minneapolis, MN, USA). Briefly, cell lysates (800 µg) were
mixed with array buffer and incubated with pre-blocked array
membrane at 4 °C overnight. Membranes were then washed and
incubated with the primary antibody cocktail for 2 h, followed by
washing and incubation with the secondary antibody for 30 min at
37°C. Membranes were washed again and subjected to chemiluminescent
detection.
In vivo metastasis assay
The Medical Experimental Animal Care Commission,
Wenzhou Medical University, approved all animal experimentation.
Male BALB/c nude mice (age, 6 weeks; weight, 15–18 g) were
purchased from Shanghai Laboratory Animal Center (Shanghai, China)
and randomly divided into two groups (8 mice per group). A total of
4×106 HCT116 cells/200 µl PBS were injected into the
tail veins of nude mice. All mice were maintained at 25°C and ~60%
humidity with a 12 h light/dark cycle with free access to food and
water. After 7 weeks the mice were sacrificed, and their lung
tissues were dissected and fixed with 10% formalin at room
temperature for 48 h. Tumor sections (5 µm) were subsequently
incubated with Mayer's hematoxylin (Sigma-Aldrich; Merck KGaA) for
1 min at room temperature and washed with running tap water for 3
min. The tissues were immerged within Eosin Y solution
(Sigma-Aldrich; Merck KGaA) for 20 sec at room temperature.
Statistical analysis
GraphPad Prism 5.0 software (La Jolla, CA, USA) was
used to analyze all data in the present study. Data were presented
as the mean ± standard deviation. Multiple group comparisons were
performed with one-way analysis of variance, followed by Tukey's
test. Overall survival was determined by the Kaplan-Meier method
and compared with the log-rank test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Rab11-FIP4 expression is upregulated
in CRC tissues and high expression of Rab11-FIP4 is associated with
poor prognosis of patients with CRC
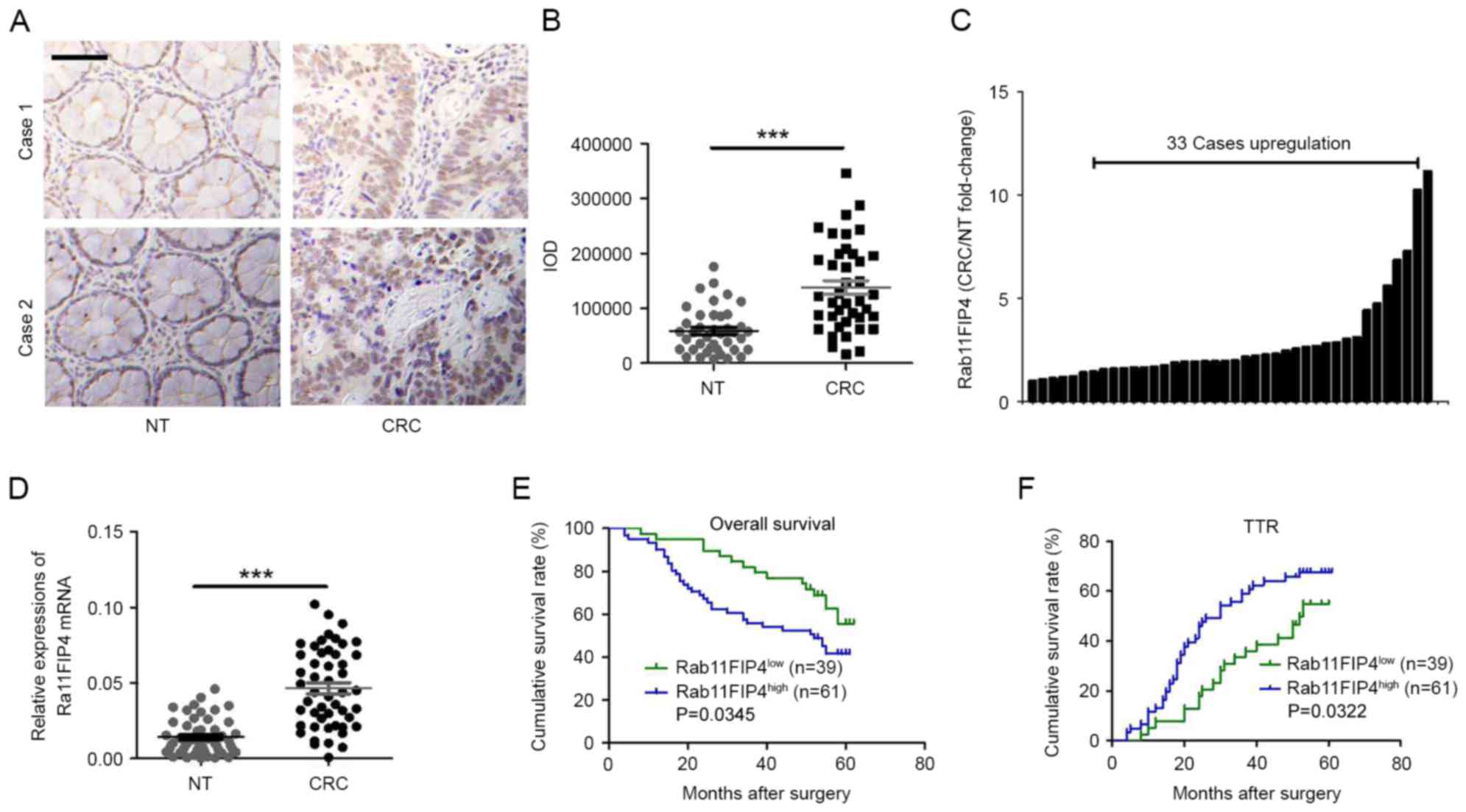
Using IHC analyses, the expression patterns of
Rab11-FIP4 in CRC tissues and corresponding NT tissues in CRC
samples were investigated. As presented in Fig. 1A, the majority of Rab11-FIP4 was
distributed in the cytoplasm and membranes of CRC cells in clinical
samples. The expression level of Rab11-FIP4 was significantly
increased in CRC tissues compared with paired NT tissues in 33 of
the 40 patients with CRC (Fig. 1B and
C). Similar results were revealed by the RT-qPCR analyses
(Fig. 1D). Furthermore, the
clinical significance of Rab11-FIP4 expression was investigated in
patients with CRC. The protein level of Rab11-FIP4 in 100 patients
with CRC was detected via TMAs using IHC analyses. Subsequently,
patients with CRC were divided into two groups based on their
respective IHC scores: High Rab11-FIP4 expression group (n=61) and
low Rab11-FIP4 expression group (n=39). The clinical and
pathological characteristics of patients with CRC included in the
present study are presented in Table
I. Kaplan-Meier survival analyses revealed that the high
Rab11-FIP4 group (n=61) demonstrated a reduced overall survival
time period (P<0.05; Fig. 1E)
and a higher tendency for CRC recurrence (P<0.05; Fig. 1F) compared with the low Rab11-FIP4
group (n=39). These results suggest that Rab11-FIP4 may act as an
oncogene in CRC progression, and that Rab11-FIP4 expression may be
used as a biomarker for the prognostic prediction of patients with
CRC.
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
| Variables | No. of cases (n=100
tumor tissues)a | No. of cases (n=40
tumor pairs)b | No. of cases (n=50
tumor pairs)c |
|---|
| Sex |
|
|
|
|
Male | 53 | 20 | 27 |
|
Female | 47 | 20 | 23 |
| Age |
|
|
|
|
<60 | 10 | 15 | 15 |
|
60–69 | 36 | 10 | 20 |
|
70–79 | 37 | 13 | 13 |
|
≥80 | 17 | 2 | 2 |
| TNM |
|
|
|
|
I–II | 52 | 14 | 21 |
|
III–IV | 48 | 26 | 29 |
| Venous
invasion |
|
|
|
|
Positive | 55 | 32 | 41 |
|
Negative | 45 | 8 | 9 |
| Tumor size |
|
|
|
| ≤5
cm | 43 | 15 | 18 |
| >5
cm | 57 | 25 | 32 |
| Location |
|
|
|
| Right
colon | 40 | 20 | 25 |
| Left
colon | 30 | 12 | 15 |
|
Rectum | 30 | 8 | 10 |
| Histological
type |
|
|
|
|
Adenocarcinoma | 82 | 33 | 38 |
|
Non-adenocarcinoma | 18 | 7 | 12 |
| Tumor
differentiation |
|
|
|
|
Well | 4 | 1 | 2 |
|
Moderate | 75 | 34 | 42 |
|
Poor | 21 | 5 | 6 |
Overexpression of Rab11-FIP4 promotes
the proliferation, migration and invasion of CRC cells in vitro,
and tumor metastasis in vivo
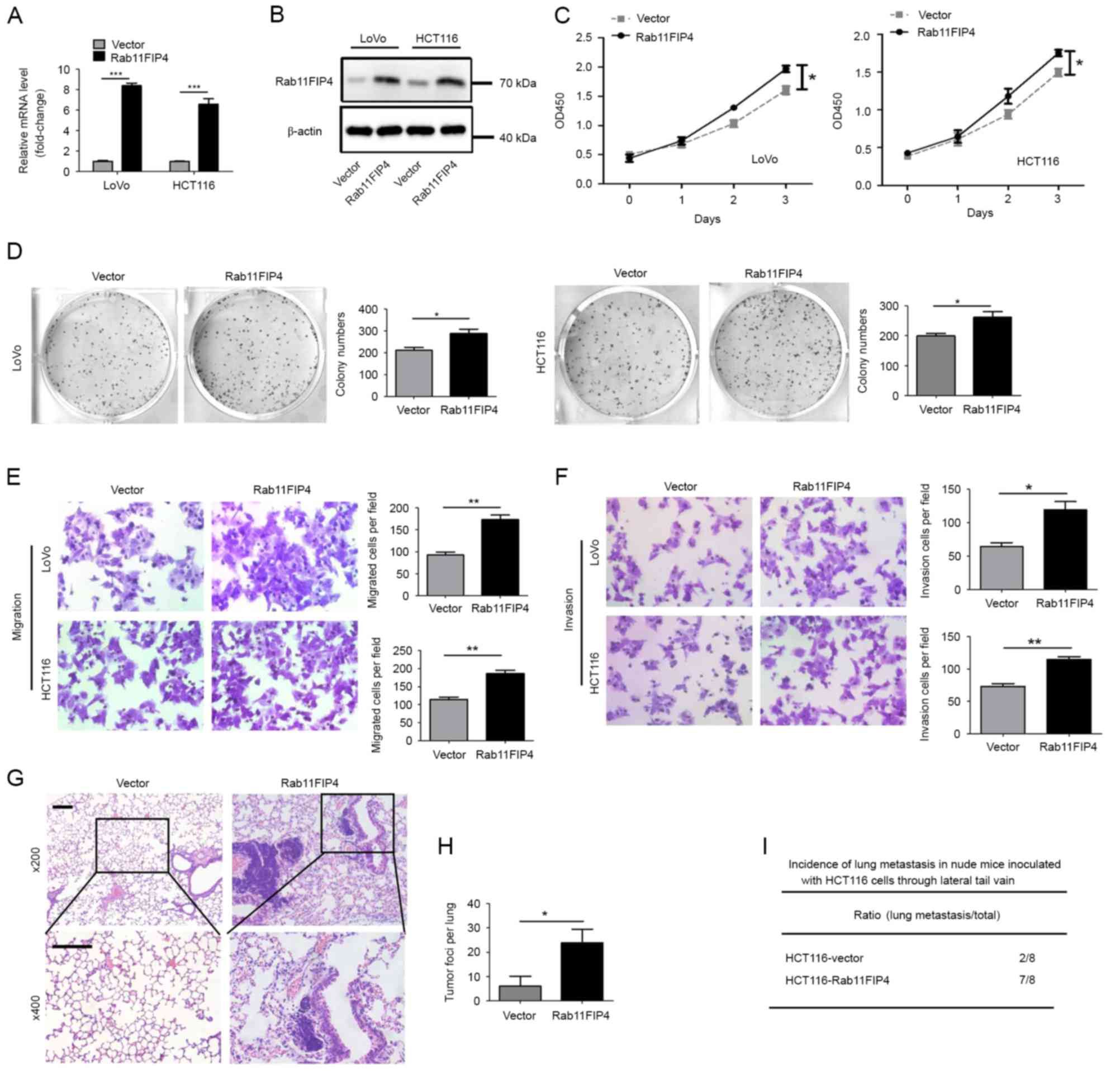
In order to investigate the function of Rab11-FIP4
in CRC cells, two common CRC cell lines, HCT116 and LoVo, were
transfected with a lentivirus that stably expresses Rab11-FIP4. The
results of the RT-qPCR and western blot analyses demonstrated that
mRNA and protein levels of Rab11-FIP4 were significantly
overexpressed in both HCT116 and LoVo cells transfected with the
lentivirus (Fig. 2A and B). The
CCK-8 assay revealed that overexpression of Rab11-FIP4 promoted the
proliferation of CRC cells (Fig.
2C). Furthermore, the clonogenic assay demonstrated that
overexpression of Rab11-FIP4 enhanced colony formation of CRC cells
(Fig. 2D). In addition, Rab11-FIP4
overexpression significantly increased the migration and invasion
of HCT116 and LoVo cells in vitro (Fig. 2E and F). Furthermore, the effect of
Rab11-FIP4 expression on CRC metastasis was investigated in
vivo. HCT116 cells transfected with either the Rab11-FIP4 or
control vector were injected into nude mice via the lateral tail
vein. At 8 weeks post-treatment, increased incidences of lung
metastases and metastatic lesions were observed in the Rab11-FIP4
overexpression group of mice compared with the empty vector group
(Fig. 2G-I).
Overexpression of Rab11-FIP4 increases
the phosphorylation of ERK1/2 and AKT, which is mediated by
IGF1R
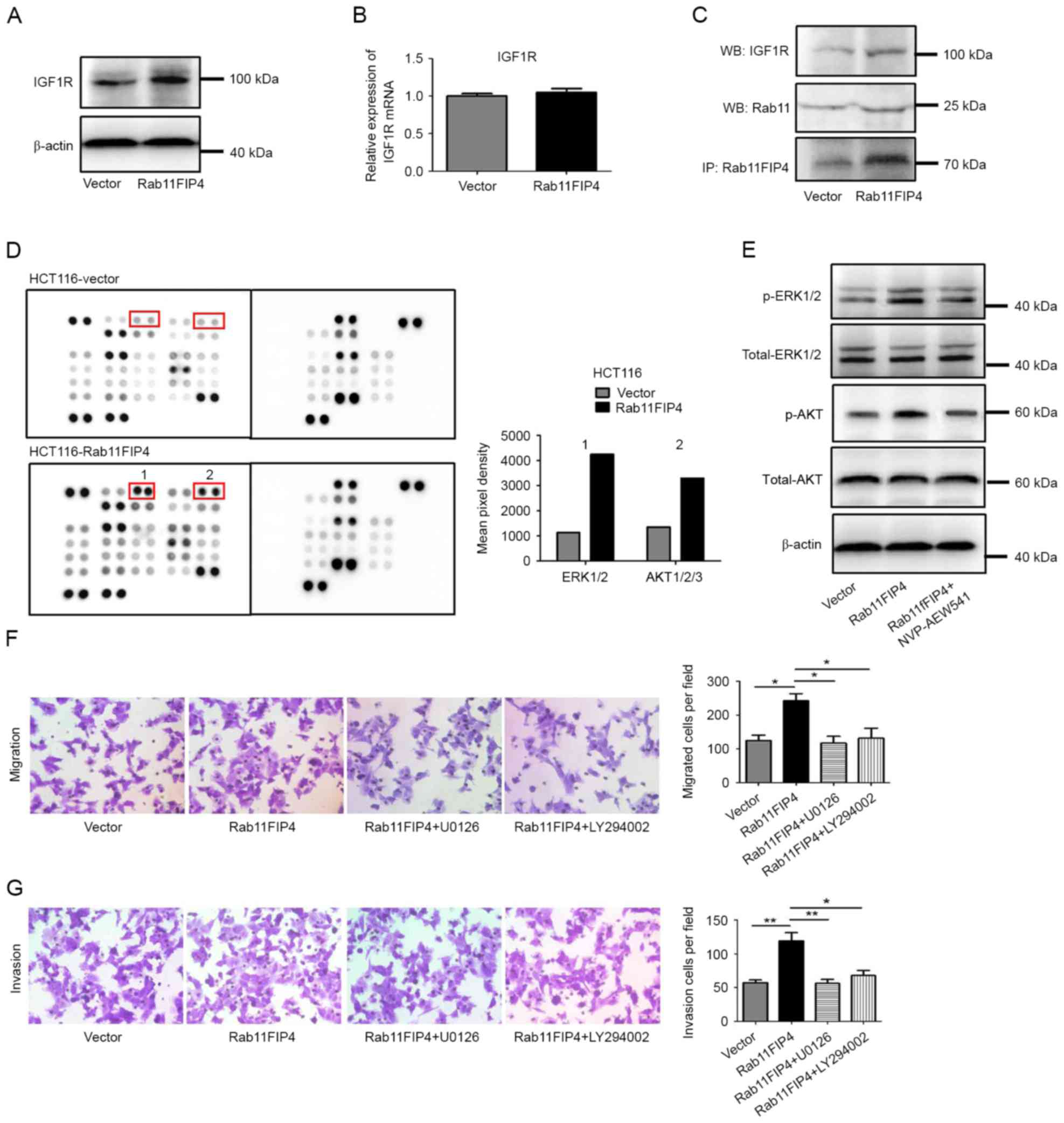
IGF1R is frequently overexpressed in CRC, and
promotes signaling pathways that regulate various functions, such
as cell proliferation, tumor cell motility, invasion and
metastasis. In the present study, it was revealed that Rab11-FIP4
could interact with IGF1R and regulate the expression level of
IGF1R. Firstly, the effect of Rab11-FIP4 overexpression on the
protein level of IGF1R was investigated using western blot
analysis. The results demonstrated that the protein level of IGF1R
was upregulated following Rab11-FIP4 overexpression (Fig. 3A). However, it was revealed that
overexpression of Rab11-FIP4 in HCT116 cells did not result in a
corresponding increase in the level of IGF1R mRNA (Fig. 3B). These results suggest that
Rab11-FIP4 may either protect the IGF1R protein from degradation,
or promote IGF1R recycling. To further investigate this, a Co-IP
assay was performed using HCT116-Rab11-FIP4 cells and corresponding
control cells. The results suggested that Rab11-FIP4 forms a
complex with Rab11 and IGF1R, and increased expression of
Rab11-FIP4 increases the formation of this complex in HCT116 cells
(Fig. 3C). In order to further
investigate the signaling affected by IGF1R in CRC cells with
increased expression of Rab11-FIP4, human phosphokinase array
assays were performed. The results revealed that the
phosphorylation levels of ERK1/2 and AKT were increased following
overexpression Rab11FIP4 in HCT116 cells (Fig. 3D; fold change ≥2.0). To determine
whether these signaling molecules are activated by IGF1R in cells
with increased Rab11-FIP4 expression, an IGF1R inhibitor was used
to treat HCT116 cells. The results demonstrated that p-ERK1/2 and
p-AKT levels were reduced following IGF1R inhibitor treatment
(Fig. 3E). To further confirm the
involvement of ERK1/2 and AKT signaling in the Rab11-FIP4-mediated
process of CRC metastasis, HCT116 cells stably infected with
Rab11-FIP4 were treated with inhibitors of ERK1/2 and AKT,
respectively. The results revealed that CRC cells treated with
ERK1/2 and AKT inhibitors had significantly reduced migration and
invasion rates compared with the untreated cells (Fig. 3F and G). These results suggest that
Rab11-FIP4 promotes CRC migration and invasion via the
phosphorylation of ERK1/2 and AKT, which is regulated by IGF1R.
Expression of Rab11-FIP4 is regulated
by HIF-1α in CRC cells
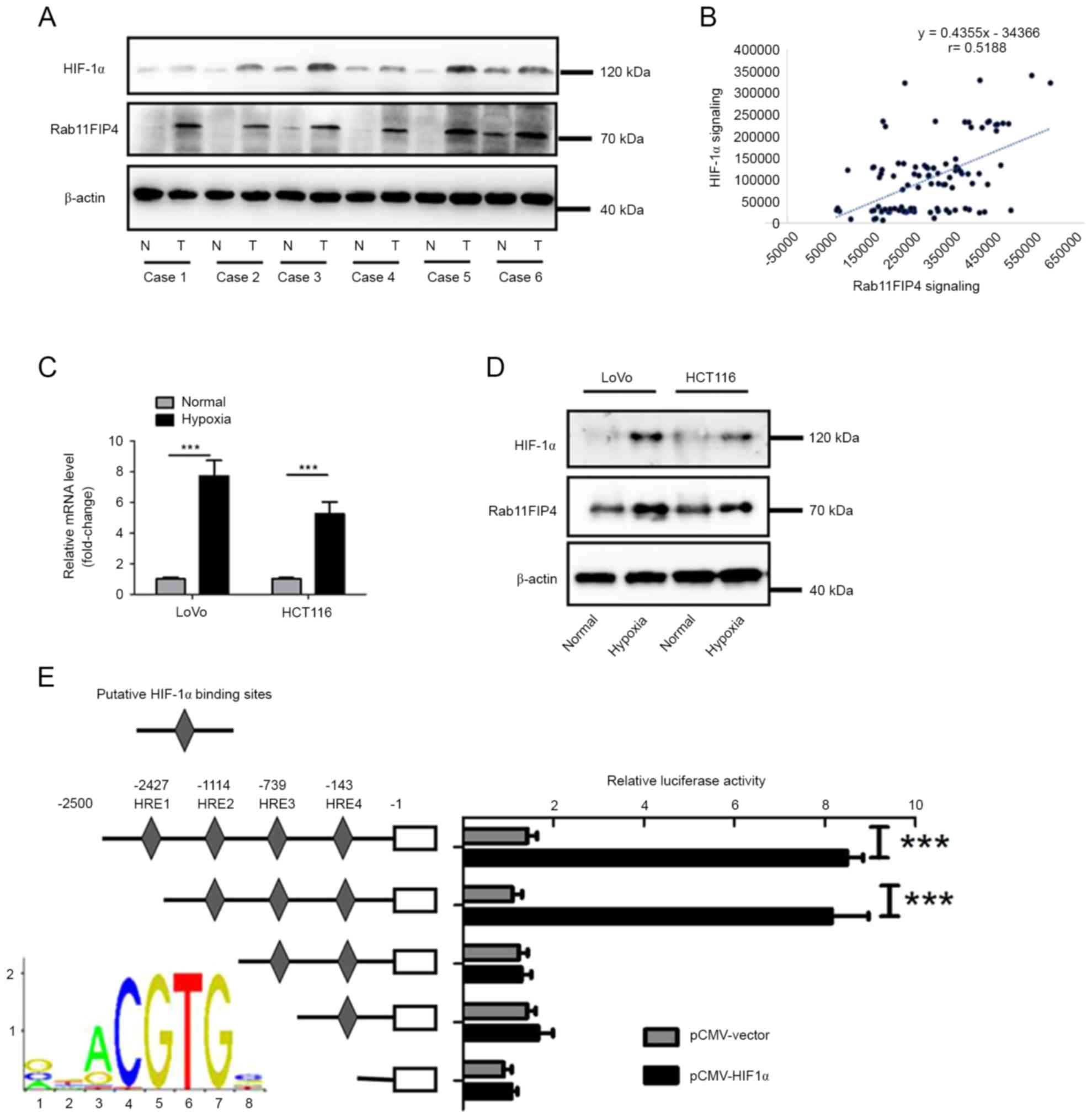
To investigate whether Rab11FIP4 is
transcriptionally regulated by HIF-1α in CRC, the expression levels
of Rab11-FIP4 and HIF-1α in six pairs of CRC tissues and their
corresponding non-tumoral tissues were analyzed. The results
demonstrated that the expression levels of Ratb11-FIP4 and HIF-1α
increased in CRC tissues compared with corresponding NT tissues
(Fig. 4A). Furthermore, tissues
with higher levels of HIF-1α expression exhibited relatively higher
levels of Rab11FIP4 (Fig. 4A). In
addition to using IHC analysis to investigate Rab11-FIP4
expression, the expression of HIF-1α in the same TMA containing 100
cases of CRC samples was also analyzed. Based on analysis of
integrated optical density value, it was revealed that there was a
significant positive correlation between expression of Rab11-FIP4
and HIF-1α (Fig. 4B). In addition,
whether hypoxia could induce Rab11-FIP4 expression in CRC was
investigated. HCT116 and LoVo cells were exposed to hypoxic
conditions for up to 48 h, and the expression level of Rab11-FIP4
was then analyzed. The results demonstrated that mRNA and protein
levels of Rab11-FIP4 were significantly increased under hypoxic
conditions (Fig. 4C and D).
Furthermore, whether Rab11-FIP4 is a transcriptional target of
HIF-1α in CRC cells was investigated. There were four putative
hormone response elements (HREs) located at the transcriptional
start site of Rab11FIP4. Using the dual-luciferase reporter assay,
it was revealed that the deletion of the HRE2 site significantly
decreased the promoter activity of Rab11-FIP4, which was induced by
HIF-1α (Fig. 4E). These results
demonstrate that Rab11-FIP4 is a target gene of HIF-1α in CRC.
Discussion
Cancer invasion and distant metastasis, likely
driven by upregulation of oncogene activity or loss of tumor
suppressors, are the leading causes of cancer-associated mortality
in the majority of cancers, including CRC (4). Due to metastasis, patients with CRC
who undergo surgical resection or chemotherapy still face poor
survival rates (5). Thus far, the
key factors responsible for metastasis in patients with cancer have
not yet been determined. In the present study, it was demonstrated
that the expression levels of Rab11-FIP4 were significantly higher
in CRC tissues compared with corresponding NT tissues, and were
associated with overall survival and time until recurrence of
patients with CRC. High expression levels of Rab11-FIP4 promoted
proliferation, invasion and metastasis of CRC cells. To the best of
our knowledge, this study revealed for the first time that
Rab11-FIP4 may be an oncogene implicated in CRC.
Rab11 small G proteins (Rab11a, Rab11b and Rab25),
members of the Ras superfamily, share high sequence identity and
are regulators of the surface expression of receptors and adhesion
proteins (11,20). Numerous studies have suggested that
Rab11 may regulate the transport of several receptors and adhesion
proteins, including the
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor,
rhodopsin, epidermal growth factor receptor, Toll-like receptor 4,
α5β1 integrin, E-cadherin and N-cadherin (21). Rab11 directly interacts with the
myosin Vb (MyoVb) globular tail domain, while the C-terminal of
MyoVb interacts with the Rab11-FIPs. Furthermore, numerous studies
have revealed that members of Rab11-FIPs interact with Rab11. Using
a high throughput yeast two-hybrid screen, Fukuda et al
(22) demonstrated an interaction
between Rab14 and FIP2. Furthermore, Lall et al (9) suggested that all Rab11-FIPs interact
with Rab14 and the class I FIPs (RCP, FIP2 and Rip11), but not the
class II FIPs (FIP3 and FIP4). In addition, other studies have
demonstrated that FIP2 and FIP3 RBDs in complex with Rab11 form a
heterotetrameric structure with dyad symmetry (23–25).
In the study, it was revealed that increased expression of
Rab11-FIP4 significantly increased the protein level of IGF1R, but
had no effect on the level of IGF1R mRNA. To the best of our
knowledge, this study is the first to demonstrate that Rab11-FIP4
may form a complex with Rab11 and IGF1R, and that increased
expression of Rab11-FIP4 increases the formation of this complex in
HCT116 cells. However, the exact structural, biophysical and
cellular mechanisms underlying this process have not yet been
determined.
Molecular and clinical evidence have suggested that
the insulin-like growth factor (IGF)/IGF1R system, including IGF,
IGF1R and IGF binding proteins, is implicated the in proliferation,
differentiation, migration, invasion and angiogenesis of solid
cancer cells (26). IGF1R is
commonly overexpressed and activated in CRC tissues, and
participates in the progression and metastasis of CRC (27). By binding of its ligands, IGF1 or
IGF2, the intrinsic tyrosine kinase activity of IGF1R is activated,
resulting in its autophosphorylation, and subsequent activation of
AKT and mitogen-activated protein kinase pathways (28,29).
Furthermore, analysis of human phosphokinase array assay results
and the effects of IGF1R inhibitor treatment demonstrated that
levels of p-ERK1/2 and p-AKT were significantly increased following
overexpression of Rab11-FIP4 in HCT116 cells, however, this was
reversed following administration of an IGF1R inhibitor. In
addition, CRC cells treated with either ERK1/2 or AKT inhibitors
exhibited significantly reduced levels of migration and invasion.
These results further suggest that ERK1/2 and AKT signaling are
implicated in the tumor promoting function of Rab11-FIP4.
Rapid growth of solid tumors creates a hypoxic
microenvironment, which can promote the angiogenesis and metastasis
of a tumor. HIF-1α is stabilized by a hypoxic microenvironment, and
induces the expression of a series of target genes involved in CRC
metastasis (30). A recent study
demonstrated that Rab11-FIP4 was a direct target gene of HIF-1α in
HCC (17). In this study, it was
also revealed that there was a significant positive correlation
between expression levels of Rab11-FIP4 and HIF-1α in CRC tissues.
Under hypoxic conditions, levels of Rab11-FIP4 and HIF-1α were
significantly increased. The present study also demonstrated that
HIF-1α could induce Rab11-FIP4 transcription by directly binding to
its HRE site in the promoter. In conclusion, the results suggest
that Rab11-FIP4 is a target gene of HIF-1α. Hypoxia and
hypoxia-mediated signaling has a critical role in solid tumor
progression. The present study provided evidence for a novel
mechanism of hypoxia-mediated oncogenic signaling in CRC
progression and Rab11-FIP4 may be a potential target in therapies
for the prevention and treatment of CRC.
Acknowledgements
The present study was supported by grants from the
Zhejiang Provincial Natural Science Foundation of China (grant no.
LY16H160055) and the Wenzhou Science and Technology Bureau (grant
no. Y20150156).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wakamura K, Kudo Se, Miyachi H, Hayashi S,
Maeda Y, Kouyama Y, Ichimasa K, Toyoshima N, Misawa M, Mori Y, et
al: Mo1695 the prognosis of colorectal cancer patients with
negative fecal immunochemical tests: The long-term follow-up study.
Gastroenterology. 150 Suppl 1:S754–S755. 2016. View Article : Google Scholar
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lai Y, Wang C, Civan JM, Palazzo JP, Ye Z,
Hyslop T, Lin J, Myers RE, Li B, Jiang B, et al: Effects of cancer
stage and treatment differences on racial disparities in survival
from colon cancer: A united states population-based study.
Gastroenterology. 150:1135–1146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vatandoust S, Price TJ and Karapetis CS:
Colorectal cancer: Metastases to a single organ. World J
Gastroenterol. 21:11767–11776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prekeris R, Klumperman J and Scheller RH:
A Rab11/Rip11 protein complex regulates apical membrane trafficking
via recycling endosomes. Mol Cell. 6:1437–1448. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lindsay AJ, Hendrick AG, Cantalupo G,
Senic-Matuglia F, Goud B, Bucci C and McCaffrey MW: Rab coupling
protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol
Chem. 277:12190–12199. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wallace DM, Lindsay AJ, Hendrick AG and
McCaffrey MW: The novel Rab11-FIP/Rip/RCP family of proteins
displays extensive homo- and hetero-interacting abilities. Biochem
Biophys Res Commun. 292:909–915. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lall P, Lindsay AJ, Hanscom S, Kecman T,
Taglauer ES, McVeigh UM, Franklin E, McCaffrey MW and Khan AR:
Structure-function analyses of the interactions between Rab11 and
Rab14 small GTPases with their shared effector rab coupling protein
(RCP). J Biol Chem. 290:18817–18832. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nagase T, Kikuno R and Ohara O: Prediction
of the coding sequences of unidentified human genes. XXII. The
complete sequences of 50 new cDNA clones which code for large
proteins. DNA Res. 8:319–327. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Welz T, Wellbourne-Wood J and Kerkhoff E:
Orchestration of cell surface proteins by Rab11. Trends Cell Biol.
24:407–415. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Horgan CP and McCaffrey MW: The dynamic
Rab11-FIPs. Biochem Soc Trans. 37:1032–1036. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meyers JM and Prekeris R: Formation of
mutually exclusive Rab11 complexes with members of the family of
Rab11-interacting proteins regulates Rab11 endocytic targeting and
function. J Biol Chem. 277:49003–49010. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wallace DM, Lindsay AJ, Hendrick AG and
McCaffrey MW: Rab11-FIP4 interacts with Rab11 in a GTP-dependent
manner and its overexpression condenses the Rab11 positive
compartment in HeLa cells. Biochem Biophys Res Commun. 299:770–779.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Muto A, Arai K and Watanabe S: Rab11-FIP4
is predominantly expressed in neural tissues and involved in
proliferation as well as in differentiation during zebrafish
retinal development. Dev Biol. 292:90–102. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muto A, Aoki Y and Watanabe S: Mouse
Rab11-FIP4 regulates proliferation and differentiation of retinal
progenitors in a Rab11-independent manner. Dev Dyn. 236:214–225.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu F, Deng X, Yang X, Jin H, Gu D, Lv X,
Wang C, Zhang Y, Huo X, Shen Q, et al: Hypoxia upregulates
Rab11-family interacting protein 4 through HIF-1α to promote the
metastasis of hepatocellular carcinoma. Oncogene. 34:6007–6017.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu CL, Wang JZ, Xia XP, Pan CW, Shao XX,
Xia SL, Yang SX and Zheng B: Rab11-FIP2 promotes colorectal cancer
migration and invasion by regulating PI3K/AKT/MMP7 signaling
pathway. Biochem Biophys Res Commun. 470:397–404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prekeris R: Rabs, Rips, FIPs, and
endocytic membrane traffic. Scientific World Journal. 3:870–880.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kelly EE, Horgan CP and McCaffrey MW:
Rab11 proteins in health and disease. Biochem Soc Trans.
40:1360–1367. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fukuda M, Kanno E, Ishibashi K and Itoh T:
Large scale screening for novel rab effectors reveals unexpected
broad Rab binding specificity. Mol Cell Proteomics. 7:1031–1042.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eathiraj S, Mishra A, Prekeris R and
Lambright DG: Structural basis for Rab11-mediated recruitment of
FIP3 to recycling endosomes. J Mol Biol. 364:121–135. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jagoe WN, Lindsay AJ, Read RJ, McCoy AJ,
McCaffrey MW and Khan AR: Crystal structure of rab11 in complex
with rab11 family interacting protein 2. Structure. 14:1273–1283.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shiba T, Koga H, Shin HW, Kawasaki M, Kato
R, Nakayama K and Wakatsuki S: Structural basis for Rab11-dependent
membrane recruitment of a family of Rab11-interacting protein 3
(FIP3)/Arfophilin-1. Proc Natl Acad Sci USA. 103:pp. 15416–15421.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weroha SJ and Haluska P: The insulin-like
growth factor system in cancer. Endocrinol Metab Clin North Am.
41:335–350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shali H, Ahmadi M, Kafil HS, Dorosti A and
Yousefi M: IGF1R and c-met as therapeutic targets for colorectal
cancer. Biomed Pharmacother. 82:528–536. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martinez-Quetglas I, Pinyol R, Dauch D,
Torrecilla S, Tovar V, Moeini A, Alsinet C, Portela A,
Rodriguez-Carunchio L, Solé M, et al: IGF2 is up-regulated by
epigenetic mechanisms in hepatocellular carcinomas and is an
actionable oncogene product in experimental models.
Gastroenterology. 151:1192–1205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jin H, Wang C, Jin G, Ruan H, Gu D, Wei L,
Wang H, Wang N, Arunachalam E, Zhang Y, et al: Regulator of
calcineurin 1 gene isoform 4, down-regulated in hepatocellular
carcinoma, prevents proliferation, migration, and invasive activity
of cancer cells and metastasis of orthotopic tumors by inhibiting
nuclear translocation of NFAT1. Gastroenterology. 153:799–811.e33.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nagaraju GP, Bramhachari PV, Raghu G and
El-Rayes BF: Hypoxia inducible factor-1α: Its role in colorectal
carcinogenesis and metastasis. Cancer Lett. 366:11–18. 2015.
View Article : Google Scholar : PubMed/NCBI
|