Introduction
Osteosarcoma is the most common bone tumor and
accounts for ~6% of all cancers in children and adolescents
(1–3). Prior to the 1970s, the survival rate
of surgical resection for osteosarcoma was 15–20%. Presently,
surgery combined with a neoadjuvant chemotherapy regimen increases
the survival rate of osteosarcoma to ~80% (4). The 5-year disease-free survival rate
in the patients with localized disease can reach 65–70%. However,
this means that there are patients, which relapse within five years
(5). The majority of osteosarcomas
are diagnosed when they are already a high-grade malignant tumor at
diagnosis and prone to early metastasis. The most common sites of
metastases are the lungs, which account for ~90% (6). With improved understanding of
osteosarcoma and its invasion and metastasis mechanisms have become
the focus of current research.
Apoptosis, also termed programmed cell death, is
encoded by genes, which regulate spontaneous cell death process and
cellular growth, differentiation, developmental and pathological
processes. Normal epithelial or endothelial cells have adhesion
dependency. Their survival depends on intercellular signaling and
signaling between cells and the matrix is termed anchor-dependent
(7). Normal epithelial cells or
solid tumor cells, which do not have the ability to shed into the
bloodstream from the extracellular matrix (ECM) will trigger
apoptosis. This type of apoptosis occurs is termed anoikis
(8).
The activating transcription factor (ATF) family
encompasses a large group of transcription factors containing the
leucine zipper basic region. ATF4 is a member of the ATF family.
The influence of various stress factors may increase ATF4
expression levels, such as hypoxicconditions (9,10),
amino acid deprivation, endoplasmic reticulum and oxidative stress
(11), and regulation of growth
factor heregulin. ATF4 is an important protective gene that allows
cells to adapt to the aforementioned stress factors. It has been
previously reported that, compared with normal tissue, primary
tumors in human tissue have a very high ATF4 protein expression
levels. ATF4 may promote tumor growth in xenograft models (12). Myelocytomatosis oncogene (MYC) is
the product of oncogene c-Myc. c-Myc is the first v-myc homolog to
be confirmed to have a transformation effect in the MC29 avian
retroviruses (13). MYC is widely
expressed, particularly in cells and tissues that proliferate
rapidly. Previous studies have determined that the function of MYC
is associated with cell proliferation, differentiation and
tumorigenesis (14–16).
A previous study reported that ATF4 may prevent
anoikis and promote metastasis in human fibrosarcoma cells
(17). The present study
investigated the function of ATF4 and MYC in osteosarcoma cell
lines. It was determined that ATF4 and MYC were highly expressed in
the human osteosarcoma cell lines 143B and ZOS. Suspension culture
of original adherent cells led to an increase in the expression
levels of ATF4 and MYC. Additionally, overexpression of ATF4 and
MYC in normal human cell lines was induced, where ATF4 and MYC
usually have low expression levels. The present study determined
that upregulation of ATF4 and MYC may significantly reduce the
adherent ability of cells and aid them in bypassing anoikis.
Conversely, a knockdown of ATF4 and MYC in human osteosarcoma cell
lines may significantly increase the adherent ability of cells and
increase the rate of anoikis. In these processes, MYC acted as a
transcription factor regulating the expression of ATF4. Chromatin
immunoprecipitation (ChIP) and luciferase reporter assays confirmed
that MYC was able to bind to the promoter of ATF4 to regulate
anoikis resistance.
Materials and methods
Cell culture
HUVEC, CHON-001, MG-63, U-2 OS and 293T cell lines
were obtained from American Type Culture Collection (Manassas, VA,
USA). HUVECs were maintained in EGM-2 medium supplemented with 5
ng/ml rh vascular endothelial growth factor, 5 ng/ml rh EGF, 5
ng/ml rh fibroblast growth factor basic, 15 ng/ml rh IGF-1, 10 mM
L-glutamine, 0.75 U/ml heparin sulfate, 1 ng/ml hydrocortisone
hemisuccinate, 2% fetal bovine serum (FBS) and 50 µg/ml ascorbic
acid (Lonza, Inc., Allendale, NJ, USA). CHON-001 cells were
maintained in Dulbecco's modified Eagle's medium supplemented with
0.1 mg/ml G-418 and 10% FBS (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). MG-63 cells were maintained in Eagle's minimum
essential medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 0.1 mg/ml G-418 and 10% FBS. U-2 OS cells were
maintained in McCoy's 5A medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 0.1 mg/ml G-418 and 10% FBS. 293T cells
were maintained in Dulbecco's modified Eagle's medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. Cells
were cultured in a 37°C incubator with humidified atmosphere of 5%
CO2.
Induction of anoikis
To induce detachment from the ECM, four cell lines
were cultured in a culture dish with a low attachment surface (cat
no. 3473; Corning Incorporated, Corning, NY, USA). Suspension
culture was performed for 24 h. The mRNA and protein expression
level changes of ATF4 and MYC were subsequently quantified.
RNA isolation from cells and reverse
transcription-polymerase chain reaction (RT-PCR) analysis
Total RNA was extracted from the cells using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) and was reverse
transcribed into cDNA using RevertAid First Strand cDNA Synthesis
kit (Thermo Fisher Scientific, Inc.). Taq polymerase (Invitrogen;
Thermo Fisher Scientific, Inc.) was used for the PCR. The following
thermocycling conditions were used: 94°C 5 min; 32 cycles of 94°C
30 sec, 55°C 30 sec, 72°C 1 min; 72°C 10 min. All experiments were
performed in triplicate. Sequences of the primers for ATF4 were
forward (F), 5′-TAGAGAAGTCCCGCCTCATAA-3′ and reverse (R),
5′-TTCACTGCCCAGCTCTAAAC-3′; and MYC F, 5′-TTCTCTCCGTCCTCGGATT-3′
and R, 5′-TGCGTAGTTGTGCTGATGT-3′.
Protein extraction from cells and
western blot analysis
Total RNA was extracted from cells using Laemmli
sample buffer with 5% 2-mercaptoethanol (both from Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The Compat-Able™ BCA
protein assay kit (cat no. 23229; Thermo Fisher Scientific, Inc.)
was used to perform protein quantitation. A total of 40 µg protein
per lane was separated by 10% SDS-PAGE and transferred to
polyvinylidene fluoride membranes. Blocking of the membranes was
performed using 5% non-fat milk or 3% bovine serum albumin (cat no.
A500023-0100; Sangon Biotech Co., Ltd., Shanghai, China). Primary
antibodies used were ATF4 (cat no. ab50546; 1:5,000) and MYC (cat
no. ab32072; 1:5,000) (both from Abcam, Cambridge, UK). GAPDH (cat
no. G9545; 1:5,000; Sigma-Aldrich; EMD Millipore, Billerica, MA,
USA) was used as normalization control. Primary antibodies were
incubated with the membrane for 8 h at 4°C. The membranes were
washed three times with Tris-buffered saline with 0.2% Tween-20 and
incubated with goat anti-mouse (cat no. ab6789; 1:2,000) and goat
anti-rabbit secondary antibodies (cat no. ab6721; 1:2,000) (both
from Abcam) labeled with horseradish peroxidase >1 h at room
temperature, and signals were visualized using film exposure. All
experiments were performed in triplicate.
Construction of overexpression and
knockdown vectors
The coding sequence of ATF4 and MYC was cloned into
pcDNA3.1 vector (plasmid #70219; Addgene, Inc., Cambridge, MA, USA)
to overexpress them in cell lines. The following short hairpin
(sh)RNAs were used to specifically target ATF4 and MYC and were
cloned into pLKO.1 (plasmid #10878; Addgene, Inc.) vector to induce
knockdown of their expression in cell lines. All experiments were
performed in triplicate. shRNA-1 targeted ATF4 sequence,
GCCTAGGTCTCTTAGATGATT; shRNA-2 targeted ATF4 sequence,
CCACTCCAGATCATTCCTTTA; shRNA-1 targeted MYC sequence,
CAGTTGAAACACAAACTTGAA; shRNA-2 targeted MYC sequence,
CCTGAGACAGATCAGCAACAA.
Adhesion assay
Six-well plates were coated with 10 mg/ml
fibronectin (cat no. F2006; Sigma-Aldrich; Merck KGaA) overnight at
4°C. The cell concentration was adjusted to 2×105
cells/ml, and 500 µl was added to each well. The cells were
incubated for 30 min at 37°C, and unattached cells were discarded.
Adhered cells were stained with crystal violet (0.005%) for 10 min,
and the cells were photographed with a microscope under a 40×
objective. The number of adherent cells was averaged from 10
fields.
Cell Death Detection ELISA
The level of anoikis was detected using a Cell Death
Detection ELISAPLUS kit (cat no. 11920685001; Roche
Diagnostics, Basel, Switzerland), according to the manufacturer's
protocol.
ChIP
ChIP assay was performed using an
Imprint® Chromatin Immunopreciptitation kit (cat no.
CHP1; Sigma-Aldrich; Merck KGaA). Cells were crosslinked with 1%
formaldehyde and terminated with 2.5 mM glycine. Briefly, the
scraped cells were lysed in PBS + sodium thiosulfate using a
sonicator, and the lysates were divided into three. One part was
used as a positive control and received no treatment. The second
was used as negative control and was incubated with IgG (cat no.
ab150077; 5 µg/ml; Abcam) and 100 µl Protein G PLUS-Agarose. The
third lysate was incubated with MYC-specific antibody (5 µg/ml) and
Protein A/G PLUS-Agarose at 4°C overnight. Lysates were incubated
with 10 µg RNaseA (cat no. R6148; Sigma-Aldrich; Merck KGaA) for 30
min at 37°C, and 80 µg proteinase K (cat no. P2308; Sigma-Aldrich;
Merck KGaA) for 3 h at 37°C, to remove the RNA and protein. DNA was
subsequently extracted with using phenol-chloroform. The degree of
enrichment was detected using quantitative PCR with the following
thermocycling parameters: 94°C, 5 min; 40 cycles of 94°C for 30
sec, 55°C for 15 sec, 72°C for 1 min; 72°C for 10 min. R1 of the
control was used for normalization. Power SYBR® Green
PCR Master Mix (cat no. 4368708; Applied Biosystems; Thermo Fisher
Scientific, Inc.) was used to perform qPCR. The quantification
performed using the ∆∆Cq method (18) and all experiments were performed in
triplicate. The ChIP primers used to measure enrichment are
detailed in Table I.
 | Table I.ChIP primers. |
Table I.
ChIP primers.
| Primer | Sequence (5′-3′) |
|---|
| R1 forward |
tgtccgtgtgtcatcttggtt |
| R1 reverse |
atactgaatgggcagatga |
| R2 forward |
acataggcaattgctttgagat |
| R2 reverse |
aaagttactcatctttcccca |
| R3 forward |
agtaggtgtttacctttaca |
| R3 reverse |
gcaggcaagtgactagaaggc |
| R4 forward |
gcaaggatactcatcagtagt |
| R4 reverse |
cgcatccaaaagaatcctggct |
| R5 forward |
gctggtccctgaggccactaa |
| R5 reverse |
tctcgggtcgctgctagtcctca |
| R6 forward |
gatcgggaaagcgtagtcgggt |
| R6 reverse |
ttcgtggggcaacagccaagac |
| R7 forward |
gagaaaatggatttgaagga |
| R7 reverse |
aagagatcacaagtgtcatcc |
Luciferase reporter gene
technology
MG-63 (2×105) and U-2 OS
(2×105) cells were plated into a 24-well plate and
transfected with 2 ng of cytomegalovirus (CMV)-Renilla
(Promega Corporation, Madison, WI, USA) and 10 ng of ATF4-dependent
luciferase reporter constructs using Lipofectamine 2000
transfection reagent (Thermo Fisher Scientific, Inc.). Cells were
harvested 36 h after transfection. Luciferase activity was
quantified using the Dual-Luciferase reporter system (Promega
Corporation). All experiments were performed in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS
version 17.0 (SPSS, Inc., Chicago, IL, USA). Statistical
differences were determined using a Student's t-test or one-way
analysis of variance followed by Tukey's honest significant
difference test. P<0.05 was considered to indicate a
statistically significant difference.
Results
ATF4 and MYC are highly expressed in
human osteosarcoma cell lines
ATF4 is a transcription factor associated with
several types of cancer progression, such as breast cancer, lung
cancer and melanoma (19–21). It has been determined that ATF4
expression is induced by stress stimulation, such as endoplasmic
reticulum stress, anoxia/hypoxia and amino acid deprivation. MYC is
also a transcription factor that has been identified as a
proto-oncogene. MYC is expressed with relatively high probability
in tumor cells to regulate proliferation, apoptosis,
differentiation and cell cycle progression. However, the
interaction between these two proteins remains to be elucidated.
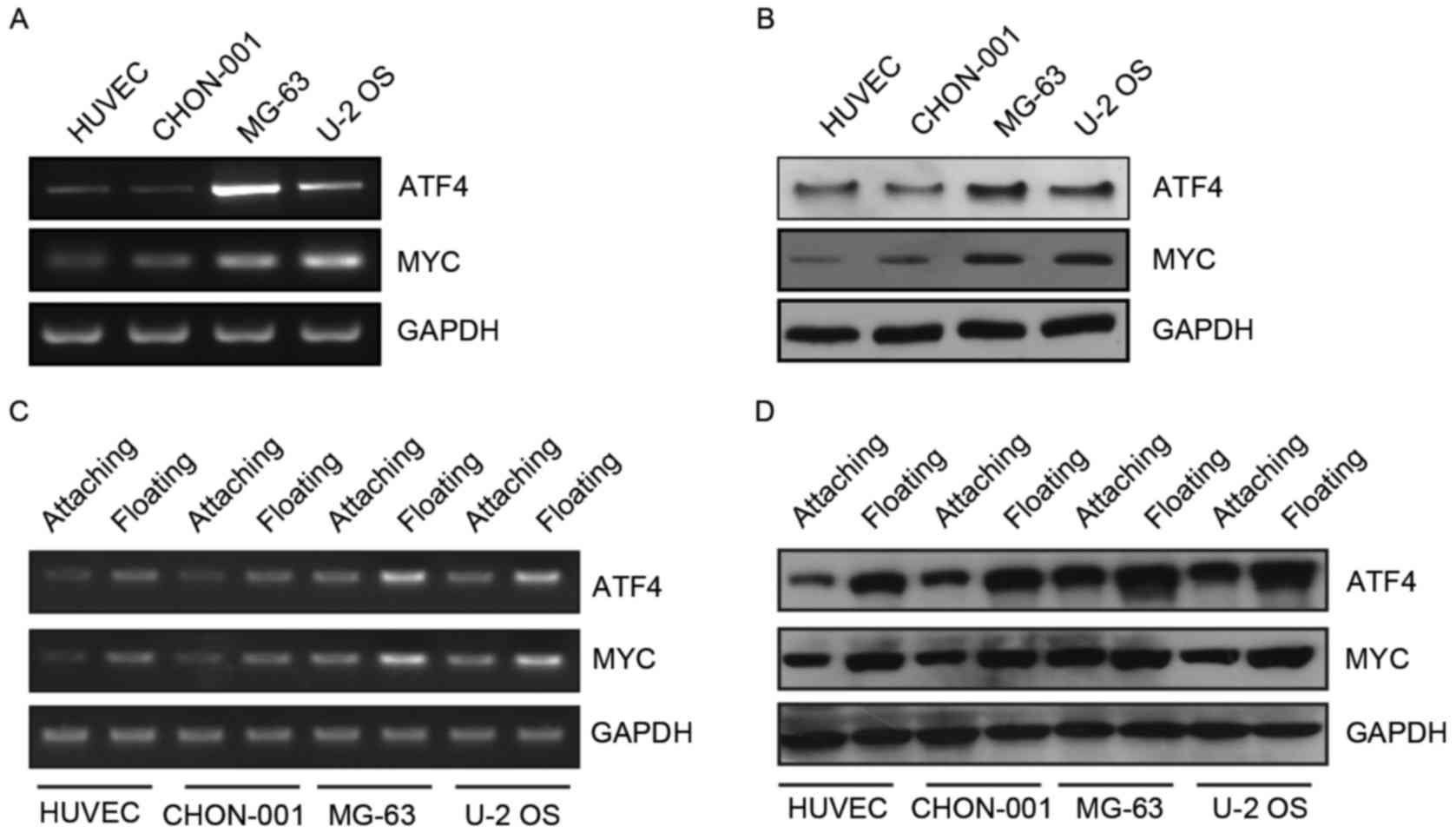
The present study determined that ATF4 and MYC mRNA and protein
expression levels were high in MG-63 and U-2 OS human osteosarcoma
cell lines (Fig. 1A and B).
Therefore, ATF4 and MYC may have an important role in the
progression of osteosarcoma. The present study focused on the
association between these two genes and anoikis. It was determined
that the expression levels of ATF4 and MYC markedly increased
following the suspension culture (Fig.
1C and D). Therefore, ATF4 and MYC may have an important
function in anoikis resistance observed in human osteosarcoma
cells.
Increased expression of ATF4 and MYC
allows normal human cell lines to escape anoikis
Dey et al (17) have determined that ATF4 prevented
anoikis in human fibrosarcoma cell lines. Disease-associated
fibronectin matrix fragments trigger anoikis of human primary
ligament cells through suppression of c-myc (22). The present study investigated
whether increased expression levels of ATF4 and MYC allowed cells
to bypass anoikis in human normal cell lines. Initially, a
lentiviral packaging plasmid and target plasmid overexpressing ATF4
or MYC were transfected into 293T cells. Subsequently, lentiviral
vectors were collected and used to infect HUVEC and CHON-001 human
normal cell lines. Subsequently ATF4 and MYC protein expression
levels were detected (Fig. 2A and
B).
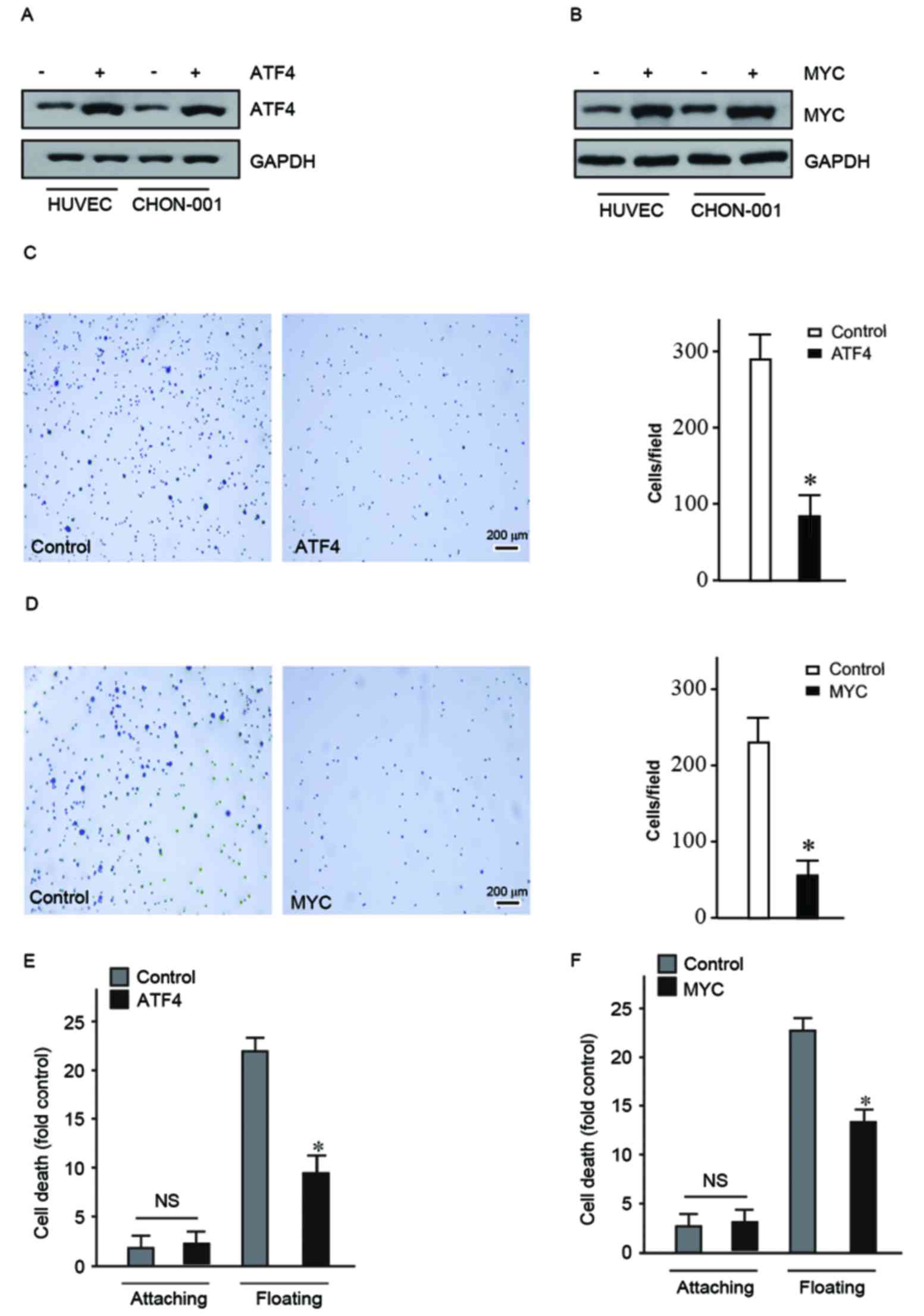
HUVEC and CHON-001 cells overexpressing ATF4 and MYC
were used to conduct adhesion assay. This assay was used to detect
the ability of cells to escape the ECM by observing the number of
cells adhered on the dish coated with fibronectin. The present
study determined that the adhesion ability of HUVEC and CHON-001
cells overexpressing ATF4 or MYC decreased significantly (Fig. 2C and D). This suggested that
overexpression of ATF4 and MYC may promote cells to become detached
and initiate metastasis.
As ATF4 and MYC may facilitate detachment of cells
from the ECM they may also aid cells in bypassing anoikis. The Cell
Death Detection ELISAPLUS kit to quantify cell death
following suspension culture for 24 h. The mortality of HUVEC and
CHON-001 floating cells overexpressing ATF4 and MYC was
significantly reduced compared with the control (Fig. 2E and F). These findings indicated
that ATF4 and MYC contributed to the cells' ability to become
detached and bypass anoikis. Therefore, ATF4 and MYC may have a
crucial role in the carcinogenesis process.
Knockdown of ATF4 and MYC in human
osteosarcoma cells promotes anoikis
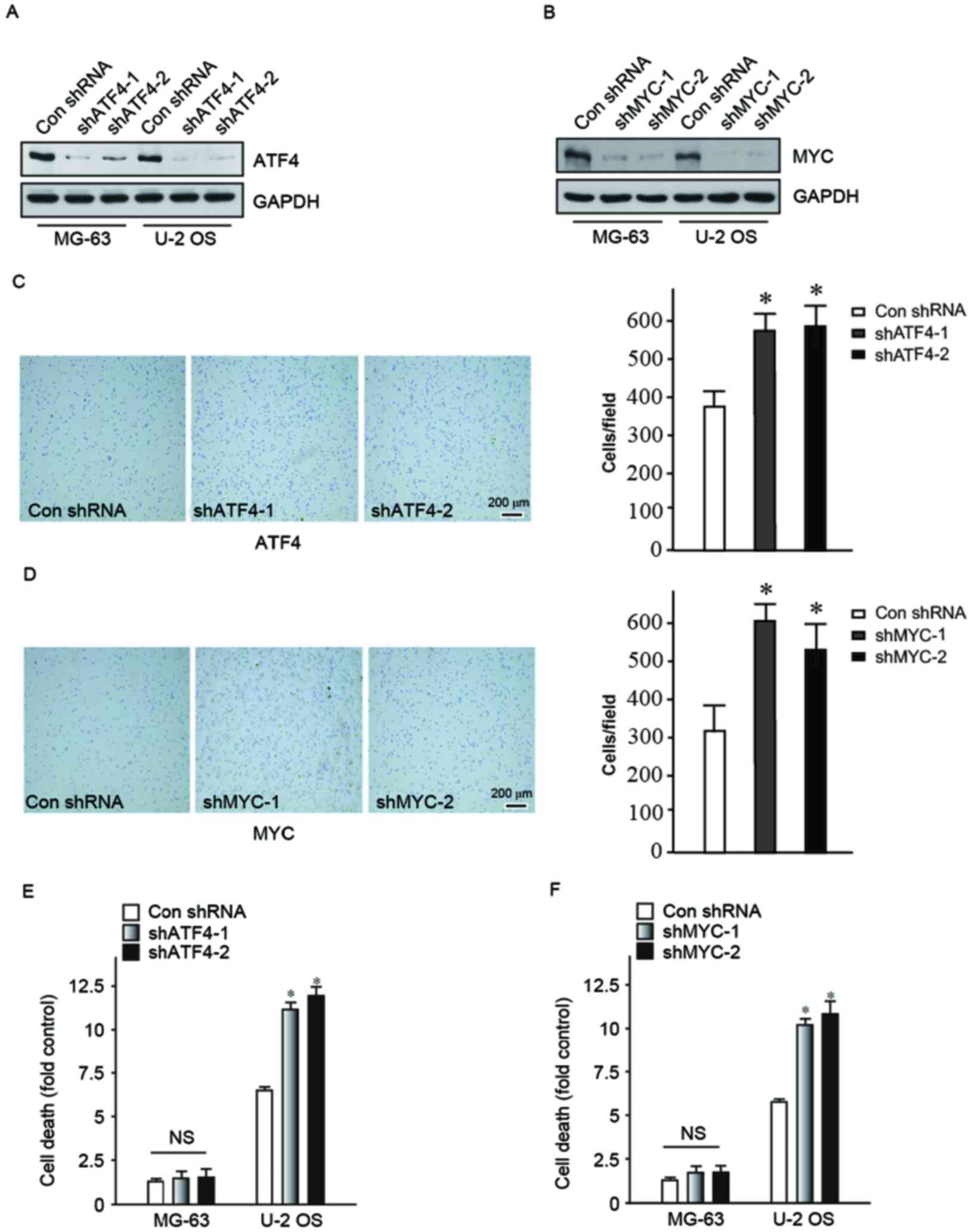
In order to confirm that ATF4 and MYC have a role in
improving cell viability following detachment from ECM during
carcinogenesis, shRNA was used to reduce ATF4 and MYC expression
levels in MG-63 and U-2 OS human osteosarcoma cells. Two clones
were designed for two different target sequences of ATF4 and MYC
respectively, vector-shATF4-1, vector-shATF4-2, vector-shMYC-1 and
vector-shMYC-2. The ATF4 and MYC protein expression levels in the
transfected cell lines were determined (Fig. 3A and B). Subsequently, adhesion
assay and cell death detection ELISA were performed to confirm that
MG-63 and U-2 OS cells, following knock-down of ATF4 and MYC had
reduced detachment abilities and unable to bypassing anoikis. Cells
preferred to adhere to the bottom of the dish (Fig. 3C and D) in the knock-down of ATF4
and MYC groups. Following a suspension culture for 24 h, cells in
the knock-down groups had significantly increased mortality
compared with the control group (Fig.
3E and F). Therefore, it was evident that the downregulation of
ATF4 and MYC expression may increase cell adhesion and induce
apoptosis due to loss of cell-matrix interactions. This confirmed
that ATF4 and MYC contribute to the process of cancer metastasis,
particularly in the aspect of bypassing anoikis.
Expression of ATF4 changes following
alteration of MYC expression
The present study aimed to determine how ATF4 and
MYC function together in the carcinogenesis process. Therefore, the
association between ATF4 and MYC has been further investigated.
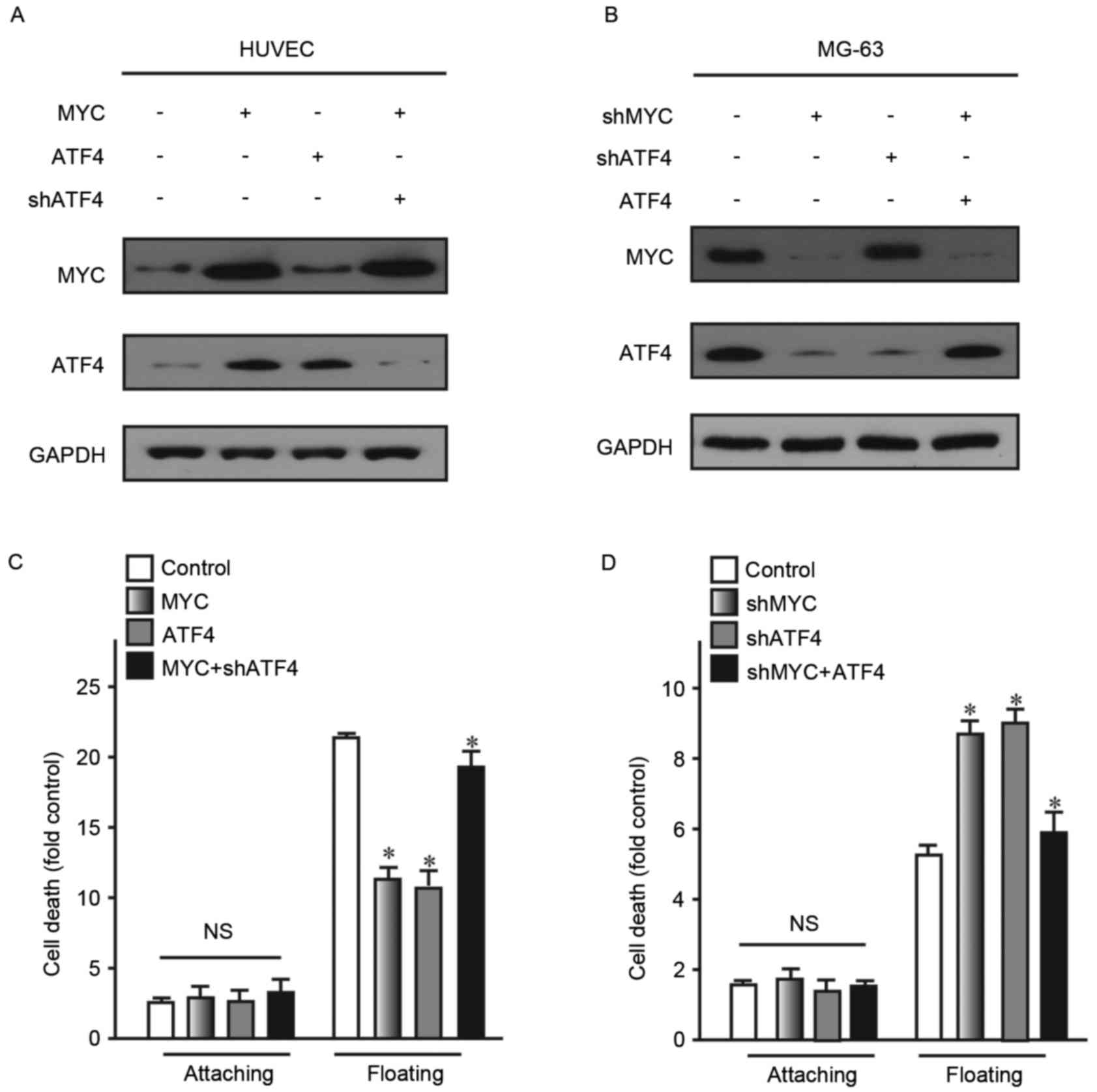
To detect this association, ATF4 and MYC were
overexpressed in human normal cell lines HUVEC and CHON-001.
Subsequently, their expression levels were detected. The findings
revealed that overexpressing MYC may lead to significantly
increased expression levels of ATF4. However, overexpression of
ATF4 did not lead to alteration of MYC expression levels in HUVEC
cells (Fig. 4A). Similarly,
knockdown of MYC did not alter the expression of ATF4 (Fig. 4B). As two the two different vectors
used for the aforementioned ATF4 and MYC knock-down experiments
exhibited a similar effect, shATF4-1 and shMYC-1 were selected for
this experiment. These findings demonstrated that MYC may act as an
upstream regulator of ATF4 expression.
The present study also examined whether anoikis
resistance ability of cells was restored following partial
upregulation of ATF4 in cells with upregulation of MYC expression.
Following 24 h suspension culture, cells with restored ATF4
exhibited reduced cell death, compared with the control (Fig. 4C and D). These findings confirmed
that ATF4 and MYC were associated with anoikis. Additionally, MYC
may act as a transcription factor regulating the expression of ATF4
and ATF4 may activate downstream signaling pathways associated with
anoikis.
MYC binds to the promoter of ATF4 to
regulate its expression
The present study has demonstrated that MYC may act
as an upstream regulatory factor of ATF4 expression, further
affecting cell sensitivity to anoikis-mediated cell death. ChIP was
used to further investigate the specific mechanism of the
underlying molecular process where MYC regulates ATF4, in order to
determine where MYC may bind onto the promoter region of ATF4.
Encyclopedia of DNA Elements at UCSC database (genome.ucsc.edu/ENCODE/) predicted that there was
the possible binding site for MYC in the promoter region of ATF4.
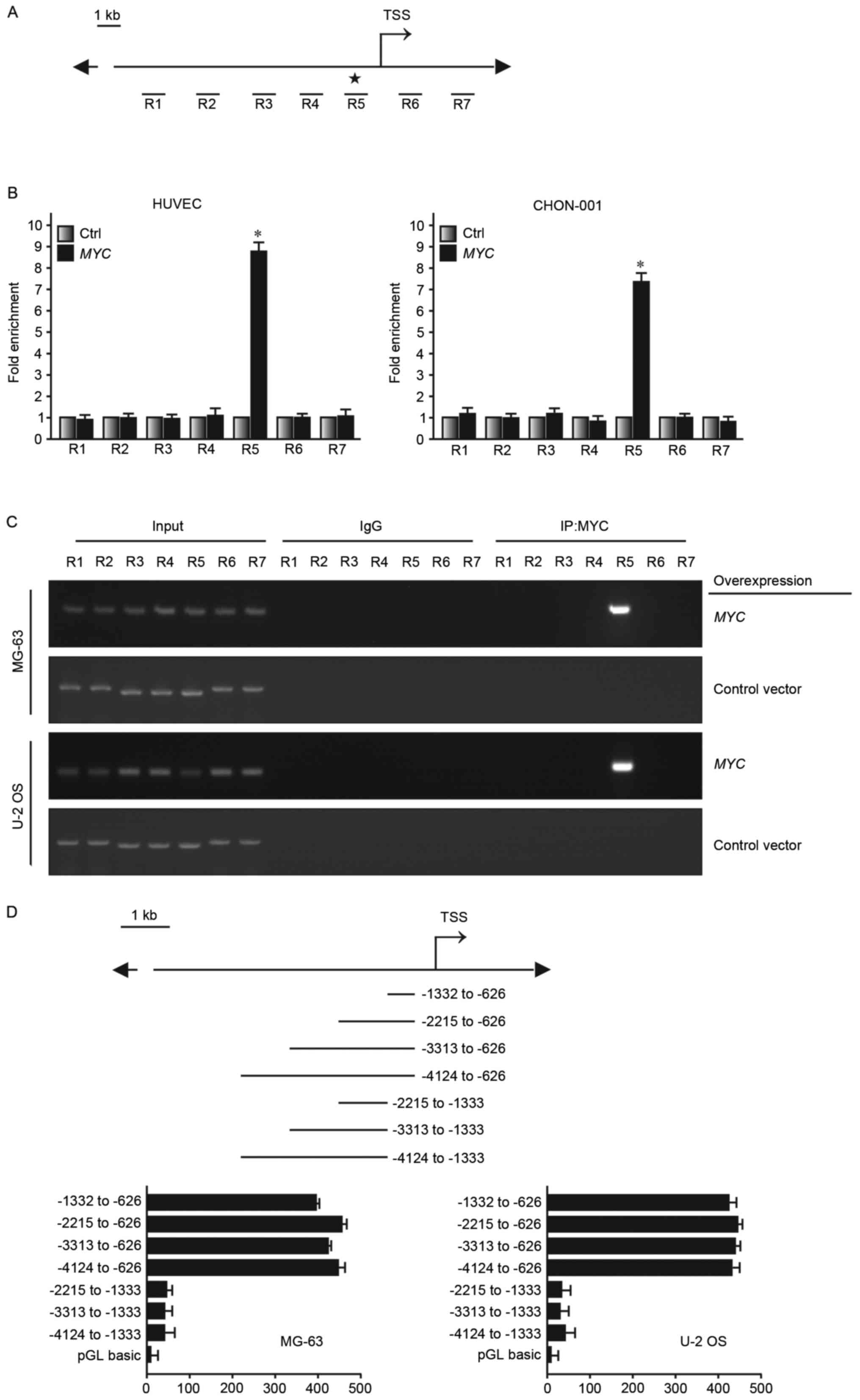
Subsequently, 7 regions were selected (R1~R7) near the
transcription starting site. They were ~1 kb apart from one another
(Fig. 5A). HUVEC and CHON-001
cells overexpressing MYC or transfected with a control plasmid were
used to conduct ChIP. DNA regions associated with MYC would be
pulled down together with via MYC affinity with the MYC monoclonal
antibody. R5 was determined to be the enrichment region of MYC in
the ATF4 promoter region (Fig. 5B and
C). Luciferase reporter was transfected into MG-63 and U-2 OS,
which express MYC at high levels. It was determined that high
luciferase reporter signal was detected at the −1,332 to −626 bp
fragment (Fig. 5D). Therefore, the
findings of the present study demonstrated that MYC may bind to the
ATF4 promoter region and regulate its expression level, thus
altering anoikis resistance in human osteosarcoma cells.
Discussion
Osteosarcoma is a primary malignant bone tumor,
which originates in mesenchymal cells. Typical osteosarcoma
clinical is rare and its incidence is ~300/1,000, accounting for
0.2% of malignant tumors and 15% of primary bone tumors (23). Prior to 1970, the standard
treatment for osteosarcoma was amputation. However, 80% of patients
have been determined to have micrometastasis at the time of
diagnosis (24). In recent years,
limb salvage treatment of osteosarcoma has gradually replaced
amputation, with the development of chemical treatments, surgical
techniques and improved treatment methods of bone reconstruction. A
previous study reported that >80% of patients preferred limb
salvage surgery (25).
Nevertheless, the metastasis mechanism of osteosarcoma remains
fully elucidated.
Anoikis as a specific type of programmed cell death,
which is important role for development, organism homeostasis,
disease and tumor metastasis. It has been previously reported that
suppression of anoikis was closely associated with metastasis and
survival of tumor cells (26).
Previous studies determined that expression of cancer-associated
genes, such as Ras, may prevent anoikis in normal epithelial cells
(27–29). Subsequent studies have reported
that inhibition of anoikis was associated with the malignant degree
of breast, colon and lung cancer (30–32).
The Fas cell surface death receptor and its adapter molecule
Fas-associated via death domain have been determined to be involved
in anoikis (33). Integrins have
also been identified to be closely associated with anoikis
(34). Cell adhesion and anoikis
may be mediated by integrins depending on the cell type and anoikis
model selected (35).
Additionally, adhesion and detachment between cell and matrix alter
Bcl-2 expression levels. Therefore, adjusting the balance of
anti-apoptotic and apoptosis signals determines cell survival or
death by anoikis (36). A previous
study determined that after cells detach from the ECM, FAK does not
bind to PI3K and inhibits its activity, eventually, promotes cells
anoikis (37).
The present study determined that MYC may bind to
the promoter region of ATF4 and regulated its expression to adjust
anoikis resistance in human osteosarcoma cells. A previous study
determined that ATF4-dependent induction of heme oxygenase 1
prevents anoikis and promotes metastasis in a human fibrosarcoma
cell line (17). The effect of MYC
on anoikis has also been previously reported as mitochondrial DNA
depletion prevented anoikis through upregulation of PI3K, which led
to the phosphorylation of downstream substrates Myc (38). The present study determined that
ATF4 has the same function in anoikis resistance different cell
lines. However, the current study also identified a completely
different mechanism where MYC acts as a transcription factor, not a
downstream gene, which regulated ATF4 to trigger anoikis
resistance.
To the best of our knowledge, this study is the
first to confirm that ATF4 and MYC promote anoikis resistance in
human osteosarcoma. Additionally, it was revealed that MYC affects
anoikis resistance through regulation of ATF4. ATF4 and MYC are
likely to become potential targets for future cancer therapy.
However, the underlying mechanism of ATF4-induced anoikis
resistance remains to be elucidated. Furthermore, the role of ATF4
and MYC in anoikis resistance requires verification in other types
of cancer.
Acknowledgements
The present study was supported by Science Computing
and Intelligent Information Processing of GuangXi Higher Education
Key Laboratory (grant no. GXSCIIP201502).
References
|
1
|
Damron TA, Ward WG and Stewart A:
Osteosarcoma, chondrosarcoma, and Ewing's sarcoma: National cancer
data base report. Clin Orthop Relat Res. 459:40–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rainusso N, Wang LL and Yustein JT: The
adolescent and young adult with cancer: State of the art-bone
tumors. Curr Oncol Rep. 15:296–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sweetnam R: Osteosarcoma. Br J Hosp Med.
28:112, 116–121. 1982.
|
|
4
|
Sun L, Li Y, Li H, Zhang J, Li B and Ye Z:
Analysis of chemotherapy dosage and dosage intensity and survival
outcomes of high-grade osteosarcoma patients younger than 40 years.
Clin Ther. 36:567–578. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hayden JB and Hoang BH: Osteosarcoma:
Basic science and clinical implications. Orthop Clin North Am.
37:1–7. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krishnan K, Khanna C and Helman LJ: The
biology of metastases in pediatric sarcomas. Cancer J. 11:306–313.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cheng TL, Symons M and Jou TS: Regulation
of anoikis by Cdc42 and Rac1. Exp Cell Res. 295:497–511. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Frisch SM and Francis H: Disruption of
epithelial cell-matrix interactions induces apoptosis. J Cell Biol.
124:619–626. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ameri K, Lewis CE, Raida M, Sowter H, Hai
T and Harris AL: Anoxic induction of ATF-4 through
HIF-1-independent pathways of protein stabilization in human cancer
cells. Blood. 103:1876–1882. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Blais JD, Filipenko V, Bi M, Harding HP,
Ron D, Koumenis C, Wouters BG and Bell JC: Activating transcription
factor 4 is translationally regulated by hypoxic stress. Mol Cell
Biol. 24:7469–7482. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Harding HP, Zhang Y, Zeng H, Novoa I, Lu
PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al: An
integrated stress response regulates amino acid metabolism and
resistance to oxidative stress. Mol Cell. 11:619–633. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bi M, Naczki C, Koritzinsky M, Fels D,
Blais J, Hu N, Harding H, Novoa I, Varia M, Raleigh J, et al: ER
stress-regulated translation increases tolerance to extreme hypoxia
and promotes tumor growth. Embo J. 24:3470–3481. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vennstrom B, Sheiness D, Zabielski J and
Bishop JM: Isolation and characterization of c-myc, a cellular
homolog of the oncogene (v-myc) of avian myelocytomatosis virus
strain 29. J Virol. 42:773–779. 1982.PubMed/NCBI
|
|
14
|
Schuhmacher M and Eick D: Dose-dependent
regulation of target gene expression and cell proliferation by
c-Myc levels. Transcription. 4:192–197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wall M, Poortinga G, Hannan KM, Pearson
RB, Hannan RD and McArthur GA: Translational control of c-MYC by
rapamycin promotes terminal myeloid differentiation. Blood.
112:2305–2317. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
O'Donnell KA, Yu D, Zeller KI, Kim JW,
Racke F, Thomas-Tikhonenko A and Dang CV: Activation of transferrin
receptor 1 by c-Myc enhances cellular proliferation and
tumorigenesis. Mol Cell Biol. 26:2373–2386. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dey S, Sayers CM, Verginadis II, Lehman
SL, Cheng Y, Cerniglia GJ, Tuttle SW, Feldman MD, Zhang PJ, Fuchs
SY, et al: ATF4-dependent induction of heme oxygenase 1
preventsanoikis and promotes metastasis. J Clin Invest.
125:2592–2608. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-(Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ferguson J, Smith M, Zudaire I, Wellbrock
C and Arozarena I: Glucose availability controls ATF4-mediated MITF
suppression to drive melanoma cell growth. Oncotarget.
8:32946–32959. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hsu HY, Lin TY, Lu MK, Leng PJ, Tsao SM
and Wu YC: Fucoidan induces Toll-like receptor 4-regulated reactive
oxygen species and promotes endoplasmic reticulum stress-mediated
apoptosis in lung cancer. Sci Rep. 7:449902017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan X, Kho D, Xu J, Gajan A, Wu K and Wu
GS: ONC201 activates ER stress to inhibit the growth of
triple-negative breast cancer cells. Oncotarget. 8:21626–21638.
2017.PubMed/NCBI
|
|
22
|
Dai R, Iwama A, Wang S and Kapila YL:
Disease-associated fibronectin matrix fragments trigger anoikis of
human primary ligament cells: p53 and c-myc are suppressed.
Apoptosis. 10:503–512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bishop MW, Janeway KA and Gorlick R:
Future directions in the treatment of osteosarcoma. Curr Opin
Pediatr. 28:26–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rosen G, Tan C, Sanmaneechai A, Beattie EJ
Jr, Marcove R and Murphy ML: The rationale for multiple drug
chemotherapy in the treatment of osteogenic sarcoma. Cancer. 35 3
Suppl:S936–S945. 1975. View Article : Google Scholar
|
|
25
|
Jaffe N: Osteosarcoma: Review of the past,
impact on the future. The American experience. Cancer Treat Res.
152:239–262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Frisch SM and Screaton RA: Anoikis
mechanisms. Curr Opin Cell Biol. 13:555–562. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Debnath J: p66 (Shc) and Ras: Controlling
anoikis from the inside-out. Oncogene. 29:5556–5558. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Halim H, Luanpitpong S and Chanvorachote
P: Acquisition of anoikis resistance up-regulates caveolin-1
expression in human non-small cell lung cancer cells. Anticancer
Res. 32:1649–1658. 2012.PubMed/NCBI
|
|
29
|
Hu Y, Chen H, Duan C, Liu D, Qian L, Yang
Z, Guo L, Song L, Yu M, Hu M, et al: Deficiency of Erbin induces
resistance of cervical cancer cells to anoikis in a STAT3-dependent
manner. Oncogenesis. 2:e522013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mahauad-Fernandez WD and Okeoma CM:
Cysteine-linked dimerization of BST-2 confers anoikis resistance to
breast cancer cells by negating proapoptotic activities to promote
tumor cell survival and growth. Cell Death Dis. 8:e26872017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chieng CK and Say YH: Cellular prion
protein contributes to LS 174T colon cancer cell carcinogenesis by
increasing invasiveness and resistance against doxorubicin-induced
apoptosis. Tumour Biol. 36:8107–8120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yao X, Pham T, Temple B, Gray S, Cannon C,
Chen R, Abdel-Mageed AB and Biliran H: The anoikis effector Bit1
inhibits EMT through attenuation of TLE1-mediated repression of
E-cadherin in lung cancer cells. PloS One. 11:e01632282016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aoudjit F and Vuori K: Matrix attachment
regulates Fas-induced apoptosis in endothelial cells: A role for
c-flip and implications for anoikis. J Cell Biol. 152:633–643.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Frisch SM and Ruoslahti E: Integrins and
anoikis. Curr Opin Cell Biol. 9:701–706. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Grossmann J: Molecular mechanisms of
‘detachment-induced apoptosis-anoikis’. Apoptosis. 7:247–260. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Matter ML and Ruoslahti E: A signaling
pathway from the alpha5beta1 and alpha (v)beta3 integrins that
elevates bcl-2 transcription. J Biol Chem. 276:27757–27763. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Monteiro HP, Silva EF and Stern A: Nitric
oxide: A potential inducer of adhesion-related apoptosis-anoikis.
Nitric Oxide. 10:1–10. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moro L, Arbini AA, Yao JL, di Sant'Agnese
PA, Marra E and Greco M: Mitochondrial DNA depletion in prostate
epithelial cells promotes anoikis resistance and invasion through
activation of PI3K/Akt2. Cell Death Differ. 16:571–583. 2009.
View Article : Google Scholar : PubMed/NCBI
|