Introduction
Lung cancer is one of the main causes of
cancer-associated mortality worldwide, based on histological
investigations into small cell and non-small cell lung cancer. The
latter can be further sorted into several types, among which
adenocarcinoma accounts for ~50% (1). Despite the administration of targeted
treatments for lung adenocarcinoma (LUAD), the survival rates
associated with this disease have not improved (2). In order to develop novel therapeutic
treatments for LUAD, further investigation into the molecular
mechanism underlying lung carcinogenesis is required.
MicroRNAs (miRNAs/miRs) are molecules ~22
nucleotides long that serve important roles in the regulation of
target gene expression to subsequently modulate cellular function
(3). Irregular miRNA expression
has been detected within LUAD samples, which may suggest the
interplay of these molecules in the initiation and development of
cancer (4). However, further
investigation is required to identify which of the aberrantly
expressed miRNAs have the potential to serve as therapeutic targets
in the development of effective treatment. A recent study reported
that miR-197-3p is markedly upregulated within LUAD samples
compared with in adjacent noncancerous tissues; as an oncomiR,
downregulation of miR-197-3p induces lung cancer apoptosis and
inhibits proliferation (5).
However, the putative functions and mRNA targets of miR-197-3p have
yet to be investigated.
Lysine 63 deubiquitinase (CYLD) is a cytoplasmic
protein with three cytoskeletal-associated
protein-glycine-conserved domains, which functions as a
deubiquitinating enzyme (6,7). A
previous study demonstrated that CYLD is downregulated within LUAD
tissues compared with in adjacent noncancerous tissues (8); further investigation is required to
understand the detailed mechanisms of CYLD dysregulation in
LUAD.
In the present study, the expression levels of
miR-197-3p within LUAD and adjacent noncancerous tissues were
analyzed to investigate the effects of miR-197-3p on cell
proliferation and apoptosis. A bioinformatics analysis suggested
that CYLD may be a target of miR-197-3p; the interaction between
miR-197-3p and CYLD was confirmed via dual-luciferase assays.
Additionally, the potential mechanism underlying cell proliferation
and apoptosis mediated by miR-197-3p was investigated.
Materials and methods
Clinical specimen collection, RNA
extraction and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR)
A total of 32 fresh LUAD tissues (cancer group) and
adjacent noncancerous lung tissues (2 cm from the margin of
resection which had been confirmed as R0 resection; control group)
were excised from patients hospitalized at The First Affiliated
Hospital of China Medical University (Shenyang, China) between
August 2015 and August 2016. Patients did not receive chemotherapy
or radiotherapy, and tissues were frozen in liquid nitrogen prior
to use. The present study was approved by the Human Research Ethics
Committee of The First Affiliated Hospital of China Medical
University (Shenyang, China; IRB Approval 2012-40-2). Informed
consent was obtained from all patients prior to enrolment in the
present study.
RNA extraction was performed using the SV Total RNA
Isolation system (Promega Corporation, Madison, WI, USA) according
to the manufacturer's protocol. RT-qPCR analysis of miR-197-3p was
carried out as previously described (9). The mRNA expression levels of CYLD
were analyzed by RT-qPCR using SYBR Green qPCR Master Mix (Takara
Biotechnology Co., Ltd. Dalian, China) and an ABI 7500 Fast System
thermocycler (Applied Biosystems; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). Initial denaturation was at 95°C for 30 sec,
followed by annealing at 95°C for 5–10 sec and extension at 60°C
for 30–34 sec for 40 cycles. Protein expression was calculated
using the 2−ΔΔCq method (10) and normalized to U6.
Cell culture
The normal lung tissue cell line HBE and human LUAD
cell lines, HCC827 and Calu-3 were obtained from the Cell Bank of
the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). Cells were cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.), 100 IU/ml penicillin and
100 µg/ml streptomycin at 37°C with 5% CO2.
Synthetic RNA oligonucleotides and
transient transfection
miRNA inhibitors, mimics and a miRNA negative
control (NC) were synthesized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). The 5′-3′ sequences of the three miRNAs were as
follows: Mimics, CGGGUAGAGAGGGCAGUGGGAGG and
UUCACCACCUUCUCCACCCAGC; inhibitor, AAGUGGUGGAAGAGGUGGGUCG; and NC,
CAGUACUUUUGUGUAGUACAA. CYLD overexpression vectors and the NC
vector were obtained from Shanghai GeneChem Co., Ltd. (Shanghai,
China). Cells (1–1.5×105/well) in a 6-well plate were
transiently transfected with miRNAs (5 µg/well) using
JetPrime® (Polyplus-transfection SA, Illkirch, France),
according to the manufacturer's protocol. Cells were collected for
analysis 48 h post-transfection.
RNA extraction from cell lines and
RT-qPCR
Total RNA was extracted from transfected cells using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.); RT-qPCR
analyses of miR-197-3p and CYLD were performed in triplicate,
according to the aforementioned protocols. The primer sequences
used were as follows: CLYD forward, 5′-TGCCTTCCAACTCTCGTCTTG-3′ and
reverse, 5′-AATCCGCTCTTCCCAGTAGG-3′; GAPDH forward,
5′-CATGTTCGTCATGGGTGTGAACC-3′ and reverse,
5′-GGTCATGAGTCCTTCCACGATACC-3′. The mRNA expression levels of
miR-197-3p and CYLD within the control group were set to 1.
Protein extraction and western blot
analysis
Following lysis with radioimmunoprecipitation assay
buffer (Beyotime Institute of Biotechnology, Haimen, China),
transfected HCC827 cells were harvested and total proteins were
extracted with a protein extraction kit (Beyotime Institute of
Biotechnology) via centrifugation at 12,000 × g, 4°C for 20 min. A
bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology) was used to determine protein concentration and 40
µg of protein per lane was separated by 10% SDS-PAGE and
subsequently transferred onto a nitrocellulose membrane at a
constant electric current of 400 mA for 1 h. Membranes were blocked
for 1 h at room temperature with 5% bovine serum albumin (Thermo
Fisher Scientific, Inc.) and were incubated with the following
primary antibodies: Anti-CYLD (1:2,000; ab137524; Abcam, Cambridge,
UK) and anti-GAPDH (1:2,500; ab9485; Abcam) overnight at 4°C. The
membranes were then washed four times in tris buffered saline with
Tween 20 and were incubated with anti-rabbit horseradish
peroxidase-conjugated secondary antibody (1:3,000; ab6721; Abcam)
for 30 min at 37°C. Proteins were visualized by enhanced
chemiluminescence plus western blotting substrate (Thermo Fisher
Scientific, Inc.).
Cell proliferation assays
MTT assays were conducted to assess the
proliferative ability of transfected HBE, HCC827 and Calu-3 cells.
A total of 5×103 cells/well per cell group were seeded
in 96-well plates. At 0, 24, 48 and 72 h, 10 µl MTT solution was
applied to the wells; plates were subsequently incubated at 37°C
for 3 h. Following MTT incubation, 150 µl dimethyl sulfoxide was
added to the wells and the plates were agitated at low speed for 10
min. Cell viability was measured at 490 nm using a microplate
reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Caspase-3/7 activity analysis
A total of 3–5×103 transfected HCC827 or
Calu-3 cells were seeded in 96-well plates. After 48 h, cells were
treated with Caspase-Glo® 3/7 reagent (Promega
Corporation) according to the manufacturer's protocol; following
agitation for 30 sec, cells were incubated for 2 h at room
temperature. Fluorescence activity was analyzed using a
GloMax® 96 Microplate Luminometer system (Promega
Corporation). Relative fluorescent activity was quantified by
setting the blank control to 1.
miR-197-3p target prediction
To investigate the association between miR-197-3p
and cell function, two independent databases, TargetScan
(http://www.targetscan.org/vert_71/)
and miRanda (http://www.microrna.org/microrna/getGeneForm.do) were
employed to predict the targets of miR-197-3p; predicted genes were
collated. From the results of cell apoptosis and proliferation
assays, CYLD was selected to investigate the effects of miR-197-3p
on cell function.
Dual-luciferase assays
To confirm whether miR-197-3p can interact with CYLD
mRNA, a Dual-Luciferase miRNA Target Expression system was used.
The following reporter plasmids: pGLO-wild-type (wt)-CYLD and
pGLO-mutant (mut)-CYLD of miR-197-3p were constructed by Shanghai
GeneChem Co., Ltd. There are two regions in pGLO-wt-CYLD that
miR-197-2 may bind to. Thus, reporter plasmids containing site 1
and 2 were separately constructed to investigate the effect of each
site on CYLD expression. The mutant sequences of the CYLD
miR-197-3p binding region were as follows: Site 1, position 92–114
of CYLD 3′-UTR, 5′-GCAAGTTCTGTCTTTTGTTGTCT-3′; and site 2, position
1469–1491 of CYLD 3′-UTR, 5′-AAGTGCTGTTTTGGGTGTTGTCG-3′. The
firefly luciferase reporter plasmids including wild-type or mutant
CYLD plasmids and the internal control, Renilla luciferase
plasmid pRL-TK (at a ratio of 10:1) were co-transfected with
miR-197-3p mimics or NC into HCC827 cells using
JetPRIME®. A total of 48 h following co-transfection,
relative luciferase activity compared with Renilla
luciferase activity was assessed using the
Dual-Luciferase® Reporter Assay system (Promega
Corporation) according to the manufacturer's protocol. For
comparisons, values for cells with NC + mut-CYLD 3′-UTR group were
set equal to 1.
Statistical analysis
SPSS 23.0 software (IBM Corp., Armonk, NY, USA) and
GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA)
were used for all statistical analyses. Data are presented as the
mean ± standard deviation of at least 3 independent experiments.
Student's t-test were employed to compare the differences between
cancerous and noncancerous tissues. The analysis of variance and
Dunnett-t test were used to compare which specific groups were
significantly different in cell assays. Spearman's correlation
analysis was used to assess the association between miR-197-3p and
CYLD mRNA expression. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-197-3p inhibition suppresses the
proliferative ability of HCC827 and Calu-3 cells
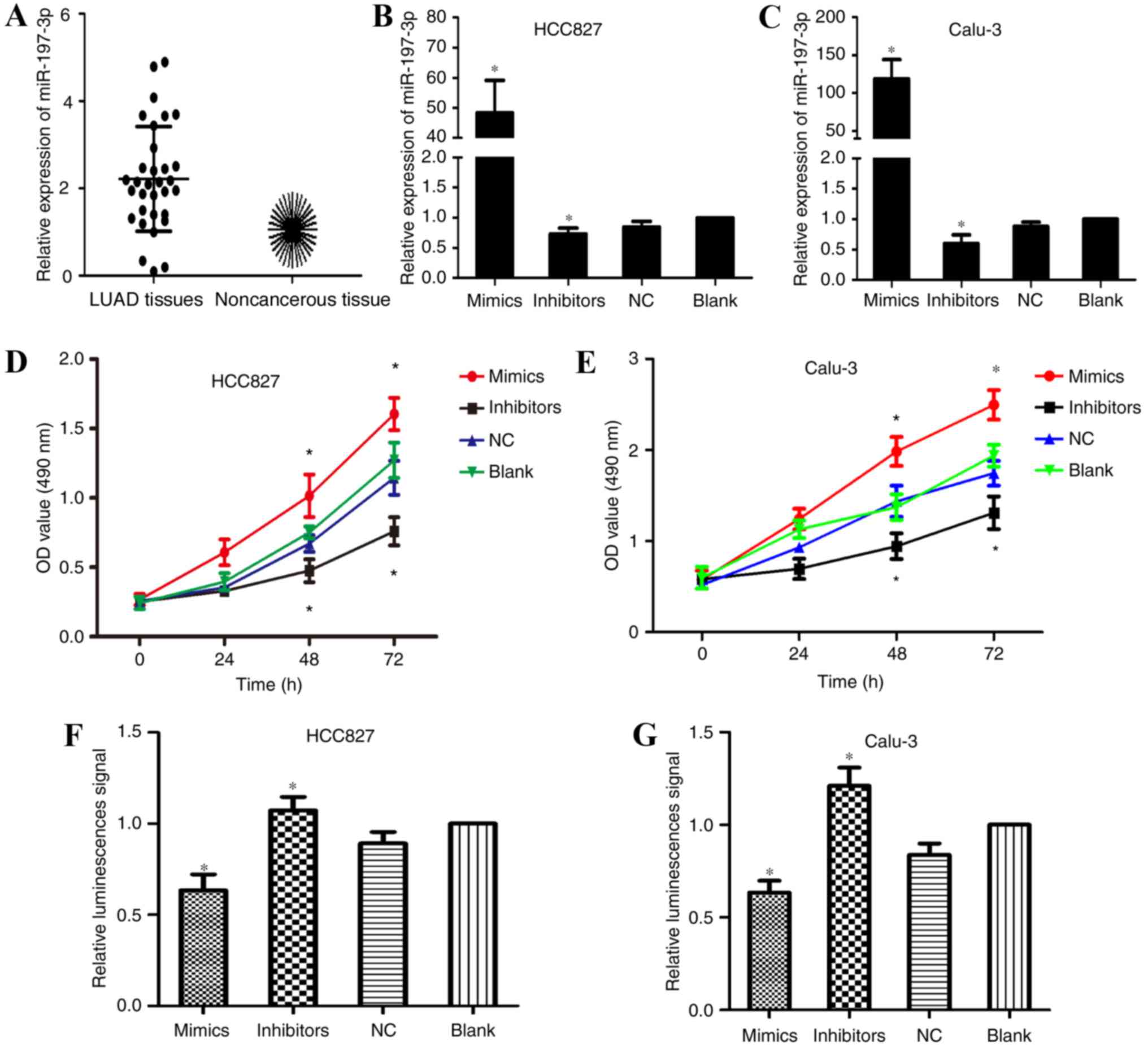
qPCR was employed to detect the expression levels of
miR-197-3p in 32 paired samples of LUAD and adjacent noncancerous
tissues. The results of the present study demonstrated that miR-197
expression was markedly upregulated within LUAD tissues (Fig. 1A). To investigate the effects of
miR-197-3p within LUAD, scrambled miR-197-3p (NC), and miR-197-3p
mimics and inhibitors were transfected into the human LUAD cell
lines, HCC827 and Calu-3. Transfection efficiency was confirmed
using qPCR (Fig. 1B and C). MTT
assays were employed to investigate the effects of miR-197-3p on
HCC827 and Calu-3 cell viability. Cells transfected with miR-197-3p
inhibitors exhibited a decrease in proliferative ability; however,
an increase in proliferative ability was observed within the
miR-197-3p mimic-transfected cells compared with in the control
group (Fig. 1D and E). These
results indicated that the inhibition of miR-197-3p reduced the
proliferative ability of LUAD cells in vitro; therefore,
miR-197-3p may serve a role in the tumorigenesis of LUAD.
Inhibition of miR-197-3p enhances LUAD
cell apoptosis
MTT assays were conducted to investigate the
association between miR-197-3p and decreased cell proliferation.
Caspase-3/7 apoptosis reagents were employed to analyze apoptosis
of transfected HCC827 and Calu-3 cells. As presented in Fig. 1F and G, fluorescent activity of the
miR-197-3p inhibitor-transfected group was significantly increased;
however, cells transfected with miR-197-3p mimics exhibited reduced
fluorescence compared with the blank and NC groups. These results
indicated that downregulation of miR-197-3p enhanced LUAD cell
apoptosis via caspase-3/7 activity.
miR-197-3p promotes tumorigenesis in
HBE cells
miR-197-3p mimics were transfected into HBE cells to
further analyze the oncogenic effects of miR-197-3p, and
transfection efficiency was confirmed by RT-qPCR (Fig. 2A). In addition, the proliferative
ability of transfected cells was analyzed via an MTT assay. The
results of the present study demonstrated that miR-197-3p
overexpression promoted HBE cell proliferation, but inhibited
apoptosis; therefore, miR-197-3p may serve a role in oncogenesis
(Fig. 2B and C).
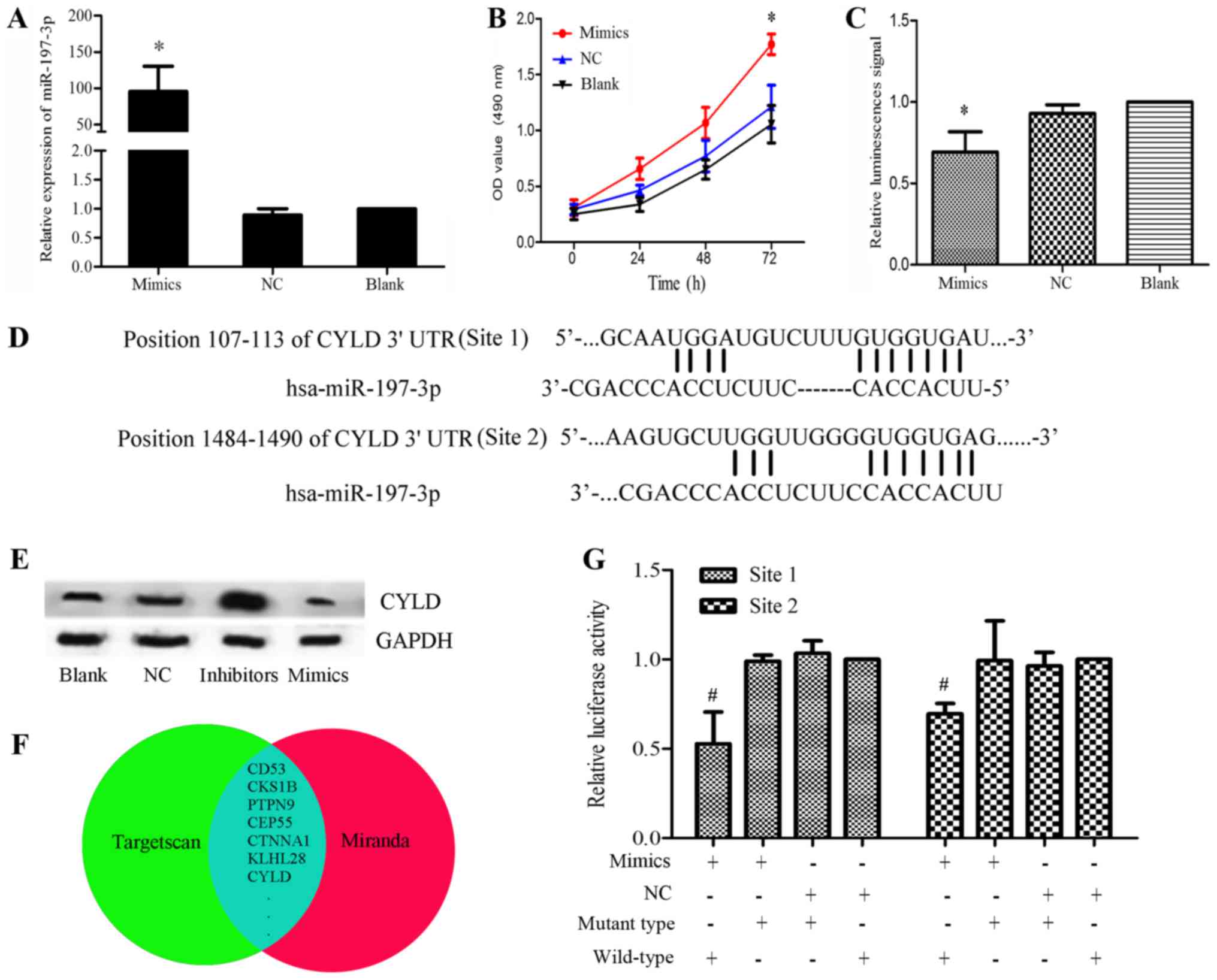 | Figure 2.(A) Relative expression levels of
miR-197-3p were significantly elevated in the mimics group compared
with in the control groups. (B) MTT assay of HBE cells transfected
with miR-197-3p mimics. Proliferation was significantly higher in
the mimics group. *P<0.05 vs. blank group. (C) Apoptotic
analysis of HBE cells transfected with miR-197-3p mimics. Caspase
activity was decreased in the mimics group. *P<0.05 vs. blank
group. (D) Putative miR-197-3p binding sites in the 3′-UTR of CYLD
mRNA. (E) Effects of miR-197-3p mimics and inhibitors on CYLD
expression in HCC827 cells, as detected using western blotting, 48
h post-transfection. GAPDH was used as a loading control. The
expression levels of CYLD were markedly lower in the mimics group
and higher in the inhibitors group compared with in the control
groups (blank or NC groups). (F) Potential target genes in the
overlap of the two gene sets. (G) Dual-luciferase reporter assays
using vectors encoding putative miR-197 target sites in the CYLD
3′-UTR for both wt and mut type. In the pGLO-mut-CYLD groups, no
significant difference was observed between the relative luciferase
activity of the cells co-transfected with the miR-197-3p mimics and
cells co-transfected with NC. Conversely, the relative luciferase
activity of the groups transfected with pGLO-wt-CYLD + miR-197-3p
was markedly lower than in the pGLO-wt-CYLD + NC group. *P<0.05
vs. NC + wild-type group Normalized data were calculated as
Renilla/firefly luciferase activity. CYLD, lysine 63
deubiquitinase; miR, microRNA; mut, mutant; NC, negative control;
OD, optical density; UTR, untranslated region; wt, wild-type. |
miR-197-3p targets CYLD by binding to
its 3′-UTR
To understand the observed miR-197-3p-induced
tumorigenesis of LUAD cells, a bioinformatics analysis was
performed using TargetScan and miRanda, after which the results
were confirmed using western blotting and dual-luciferase reporter
assays (Fig. 2D-G). Overlap
analysis revealed 278 genes within the two gene sets. In order to
narrow the scope of the predictions, genes were selected based on
the total context++ score (<-0.2) and total sites (≥2) predicted
by TargetScan (Fig. 2F). Among
these genes, CYLD was selected, as previous studies have reported
the association of CYLD downregulation with carcinogenesis
(7). The potential target sites
for miR-197-3p in the CYLD mRNA 3′-UTR are presented in Fig. 2D. To investigate the interaction
between miR-197-3p and CYLD mRNA, CYLD protein expression levels
were analyzed within HCC827 cells transfected with miR-197-3p
mimics or miR-197-3p inhibitors. The results of western blotting
suggested that the expression of CYLD was markedly reduced within
the mimics group and elevated within the inhibitors group compared
with in the control groups (blank or NC; Fig. 2E). The dual-luciferase reporter
assays were conducted to confirm the results. Dual-luciferase
reporter vectors containing either the mutant or wt 3′-UTR of CYLD
mRNA were constructed, and co-transfected with the miR-197-3p
mimics or NC in HCC827 cells. In the pGLO-mut-CYLD groups, no
significant difference was observed between the relative luciferase
activity of the cells co-transfected with the miR-197-3p mimics and
the cells co-transfected with NC. Conversely, the relative
luciferase activity of the cells co-transfected with pGLO-wt-CYLD +
miR-197-3p mimics was markedly lower than that of the pGLO-wt-CYLD
+ NC group (P<0.05; Fig. 2G).
These results revealed that miR-197-3p interacted directly with the
3′-UTR of CYLD mRNA, inversely regulating CYLD expression.
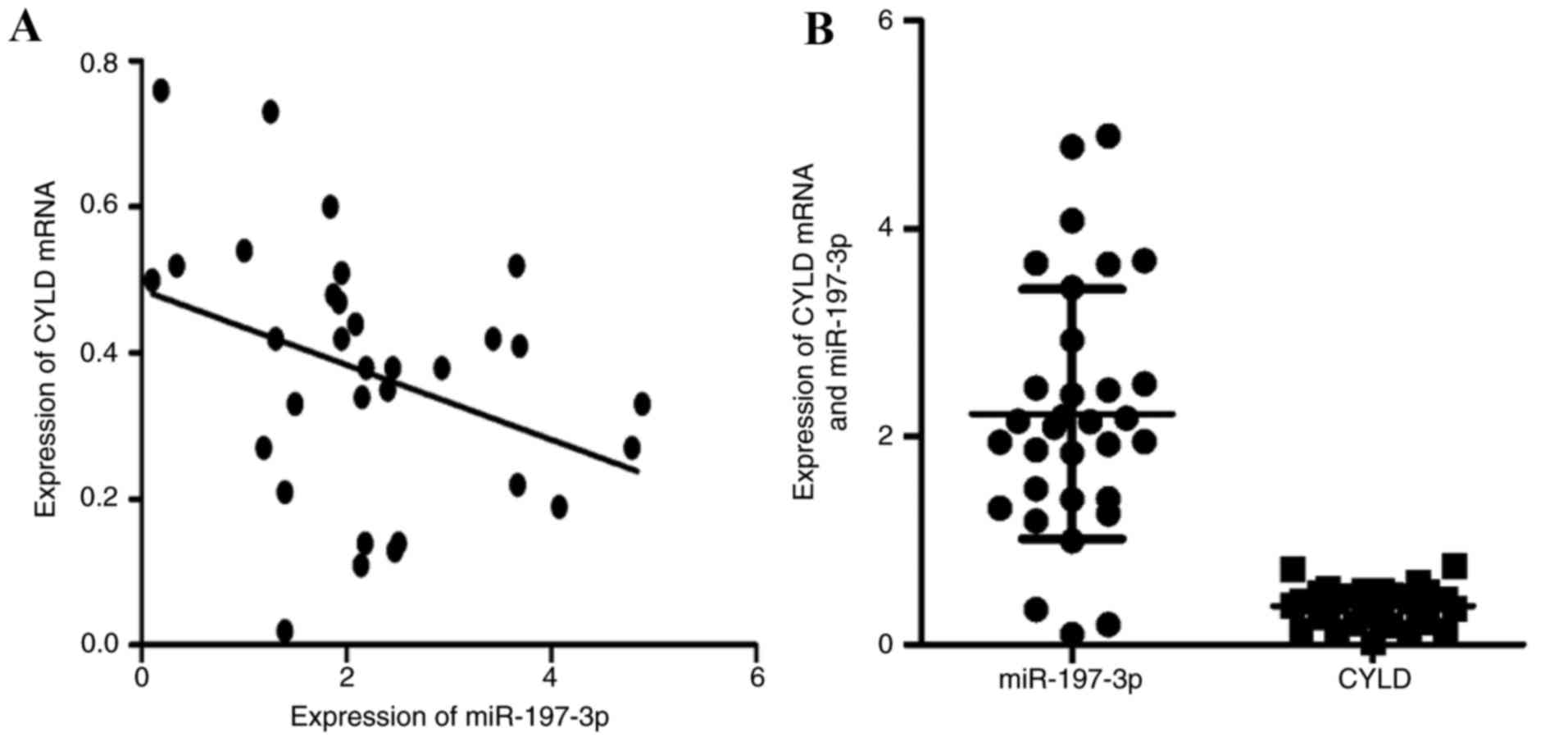
miR-197-3p is negatively associated
with CYLD mRNA expression within LUAD specimens
The association between CYLD mRNA and miR-197-3p
expression within LUAD specimens is presented in Fig. 3A and B. Spearman's correlation
analysis indicated that miR-197-3p was inversely associated with
CYLD mRNA expression (r=−0.436, P<0.05).
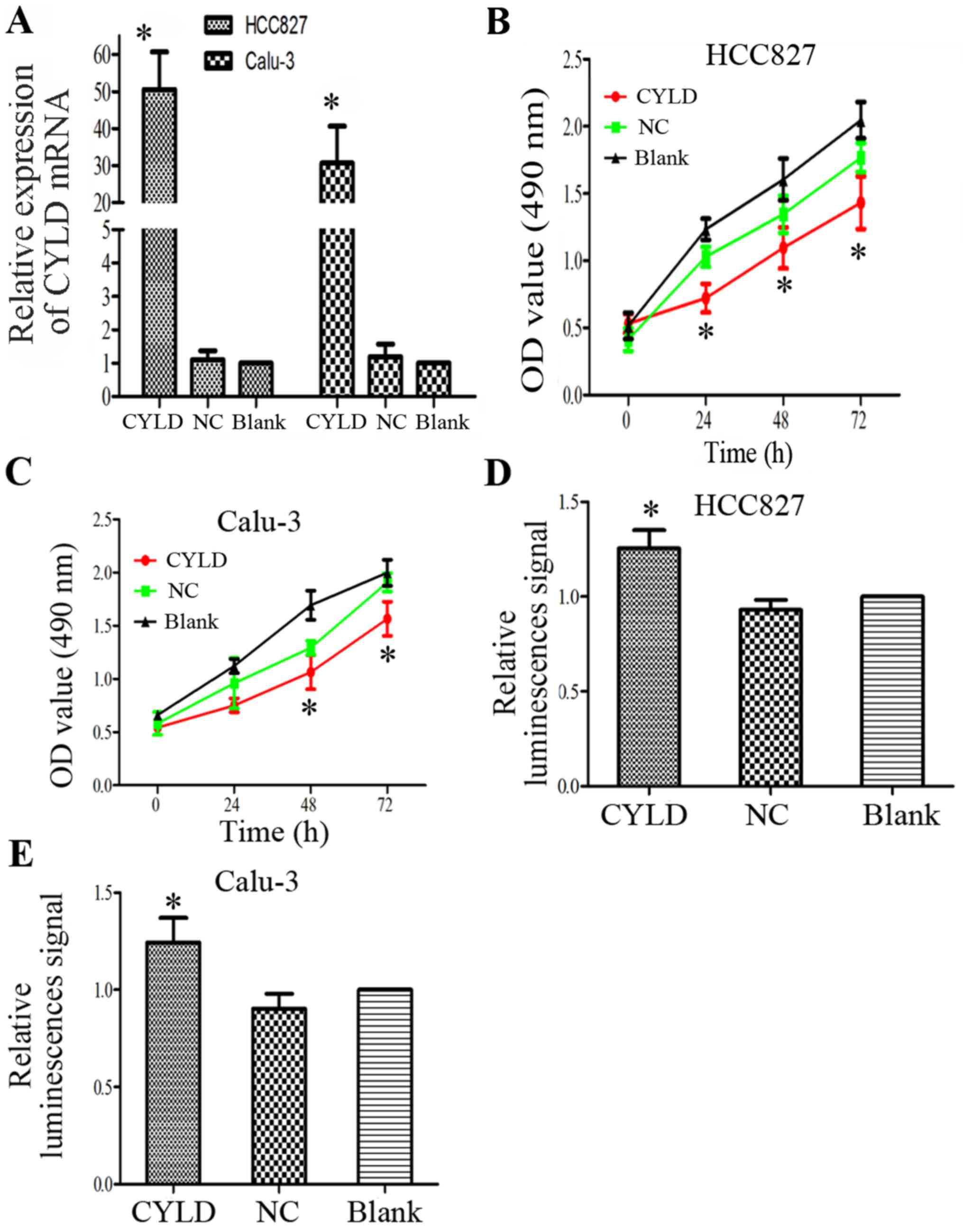
CYLD is a LUAD suppressor gene
The present study investigated whether abnormal
ectopic CYLD expression is sufficient to promote the proliferation
of HCC827 and Calu-3 cells. The transfection efficiency of the CYLD
overexpression vector was confirmed by RT-qPCR (Fig. 4A). Proliferative ability of the
LUAD cells was inhibited in response to CYLD overexpression
(Fig. 4B and C). In addition, CYLD
overexpression markedly increased apoptosis of HCC827 and Calu-3
cells (Fig. 4D and E). These
results indicated that CYLD overexpression and miR-197-3p
suppression may inhibit cell proliferation and enhance apoptosis;
therefore, CYLD overexpression may rescue the effects of miR-197-3p
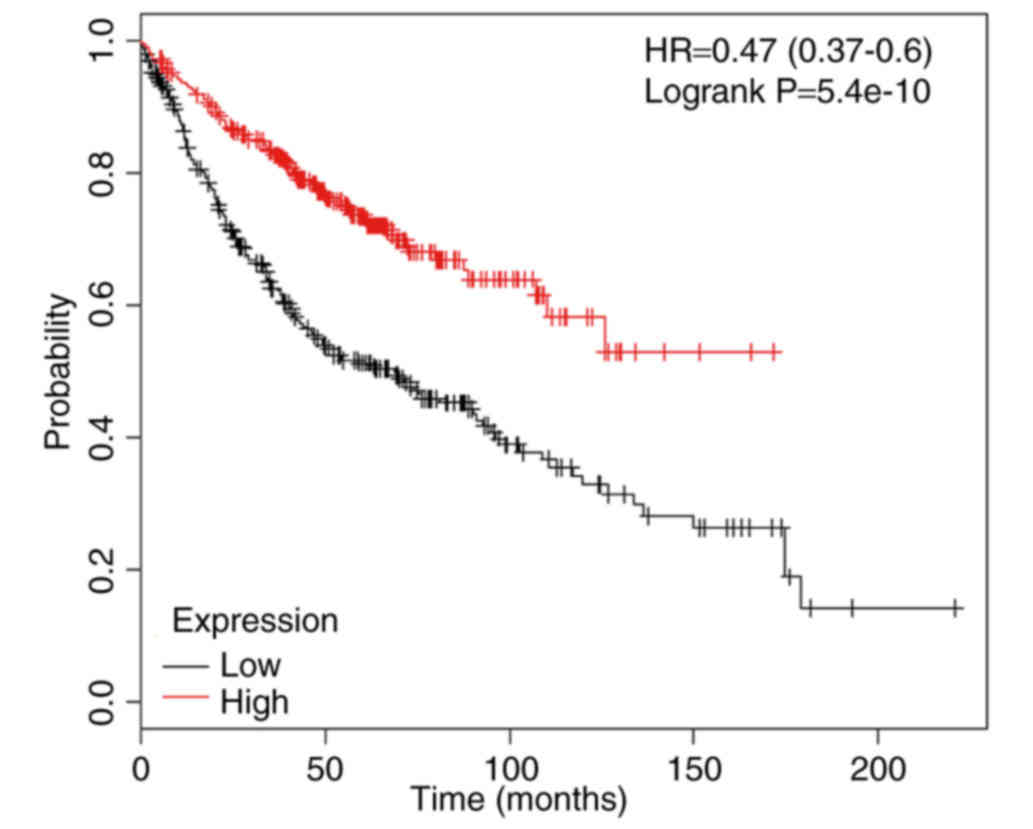
on cell function. The association between the mRNA expression
levels of CYLD and LUAD prognoses of 866 patients from the
Kaplan-Meier plotter database was analyzed using the Kaplan-Meier
plotter database (http://kmplot.com/). The results
demonstrated that the mRNA expression levels of CYLD were
associated with overall survival (Fig.
5).
Discussion
Several LUAD clinical therapies are currently
available; however, the 5-year survival rate of patients with LUAD
has yet to be improved (11). In
an effort to improve prognosis, it is necessary to develop novel
efficient therapeutic strategies. Downregulation of miR-197-3p has
been reported to participate in the progression and development of
numerous types of cancer (5,12,13).
However, further investigation is required to understand the
expression and biological function of miR-197-3p within LUAD0. In
the present study, miR-197-3p was revealed to be overexpressed
within LUAD tissues compared with adjacent noncancerous tissues.
The two cells lines employed in the present study are the most
common models of LUAD available. The results of the present study
revealed that miR-197-3p downregulation reduced proliferative
ability and enhanced apoptosis of the LUAD cell lines via
caspase-3/7 activation. Conversely, upregulation of miR-197-3p
promoted HBE cell tumorigenesis, thus suggesting that miR-197-3p
exerts oncogenic effects. A bioinformatics analysis indicated that
two miR-197-3p binding sites are present within the 3′UTR of CYLD
mRNA. This prediction was applied to investigate the regulatory
role of miR-197-3p on proliferation and apoptosis. Subsequently,
luciferase reporter assays were conducted; the luciferase activity
exhibited by the pGLO-wt-CYLD group was reduced in response to
transfection with miR-197-3p mimics, thus providing confirmation
that CYLD mRNA is a target of miR-197-3p. In the present study,
cells were transfected with a CYLD overexpression vector, and its
role as a tumor suppressor gene was confirmed. The results of the
present study indicated that miR-197-3p downregulation may partly
inhibit LUAD cell proliferation via CYLD upregulation, thus
resulting in inhibition of LUAD progression.
CYLD is a crucial enzyme involved in
deubiquitination. CYLD has been reported to regulate numerous
signaling pathways, including transforming growth factor (TGF)-β,
Wnt/β-catenin and nuclear factor (NF)-κB signaling, and therefore
affects tumorigenesis (14–17).
Furthermore, the downregulation of CYLD has been confirmed within
various malignant tumors (18–21).
In the present study, the data suggested that aberrant expression
of CYLD may participate in the progression and development of LUAD,
and CYLD may serve as a prognostic predictor of LUAD.
The main limitation of the present study is that a
relatively small number of patients were involved in the analysis;
in addition, detailed information for these individuals was not
collected. In addition, in vivo assays and in-depth
investigation into the mechanisms underlying the effects of CYLD
and miR-197-3p are also required. Therefore, further studies are
essential to investigate the regulation of TGF-β, Wnt and NF-κB
signaling pathways by miR-197-3p in the future. In conclusion, the
findings of the present study indicated that miR-197-3p regulated
the biological behaviors of LUAD via CYLD downregulation. In
addition, the expression of CYLD may be significantly associated
with the prognosis of LUAD. These conclusions suggested that the
miR-197-3p/CYLD interaction may be applied in the development of
novel LUAD-targeted treatments.
Acknowledgements
The present study was supported by the Liaoning
Province Natural Science Foundation (grant no. 2013021041).
References
|
1
|
Miller KD, Siegel RL, Lin CC, Mariotto AB,
Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A: Cancer
treatment and survivorship statistics, 2016. CA Cancer J Clin.
66:271–289. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Diaz-Garcia CV, Agudo-López A, Perez C,
López-Martín JA, Rodríguez-Peralto JL, de Castro J, Cortijo A,
Martínez-Villanueva M, Iglesias L, García-Carbonero R, et al:
DICER1, DROSHA and miRNAs in patients with non-small cell lung
cancer: Implications for outcomes and histologic classification.
Carcinogenesis. 34:1031–1038. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lau NC, Lim LP, Weinstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yanaihara N, Caplen N, Bowman E, Seike M,
Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, et
al: Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fiori ME, Barbini C, Haas TL, Marroncelli
N, Patrizii M, Biffoni M and De Maria R: Antitumor effect of
miR-197 targeting in p53 wild-type lung cancer. Cell Death Differ.
21:774–782. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bignell GR, Warren W, Seal S, Takahashi M,
Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al:
Identification of the familial cylindromatosis tumour-suppressor
gene. Nat Genet. 25:160–165. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kovalenko A, Chable-Bessia C, Cantarella
G, Israel A, Wallach D and Courtois G: The tumour suppressor CYLD
negatively regulates NF-kappaB signalling by deubiquitination.
Nature. 424:801–805. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin X, Chen Q, Huang C and Xu X: CYLD
promotes TNF-α-Induced cell necrosis mediated by RIP-1 in human
lung cancer cells. Mediators Inflamm. 2016:15427862016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang YY, Wu ZY, Wang GC, Liu K, Niu XB, Gu
S and Meng JS: LINC00312 inhibits the migration and invasion of
bladder cancer cells by targeting miR-197-3p. Tumour Biol.
37:14553–14563. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang YY, Wu ZY, Wang GC, Liu K, Niu XB, Gu
S and Meng JS: LINC00312 inhibits the migration and invasion of
bladder cancer cells by targeting miR-197-3p. Tumour Biol.
37:14553–14563. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen X, Xu Y, Cao X, Chen Y, Jiang J and
Wang K: Associations of Il-1 Family-Related polymorphisms with
gastric cancer risk and the role of Mir-197 In Il-1f5 Expression.
Medicine (Baltimore). 94:e19822015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lim JH, Jono H, Komatsu K, Woo CH, Lee J,
Miyata M, Matsuno T, Xu X, Huang Y, Zhang W, et al: CYLD negatively
regulates transforming growth factor-β-signalling via
deubiquitinating Akt. Nat Commun. 3:7712012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tauriello DV, Haegebarth A, Kuper I,
Edelmann MJ, Henraat M, Canninga-van Dijk MR, Kessler BM, Clevers H
and Maurice MM: Loss of the tumor suppressor CYLD enhances
Wnt/beta-catenin signaling through K63-linked ubiquitination of
Dvl. Mol Cell. 37:607–619. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pannem RR, Dorn C, Ahlqvist K, Bosserhoff
AK, Hellerbrand C and Massoumi R: CYLD controls c-MYC expression
through the JNK-dependent signaling pathway in hepatocellular
carcinoma. Carcinogenesis. 35:461–468. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang WY, Lim JH and Li JD: Synergistic and
feedback signaling mechanisms in the regulation of inflammation in
respiratory infections. Cell Mol Immunol. 9:131–135. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gautheron J and Luedde T: A novel player
in inflammation and cancer: The deubiquitinase CYLD controls HCC
development. J Hepatol. 57:937–939. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Font-Burgada J, Seki E and Karin M: CYLD
and HCC: When being too sensitive to your dirty neighbors results
in self-destruction. Cancer Cell. 21:711–712. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Urbanik T, Köhler BC, Boger RJ, Wörns MA,
Heeger S, Otto G, Hövelmeyer N, Galle PR, Schuchmann M, Waisman A
and Schulze-Bergkamen H: Down-regulation of CYLD as a trigger for
NF-κB activation and a mechanism of apoptotic resistance in
hepatocellular carcinoma cells. Int J Oncol. 38:121–131.
2011.PubMed/NCBI
|
|
21
|
Hayashi M, Jono H, Shinriki S, Nakamura T,
Guo J, Sueta A, Tomiguchi M, Fujiwara S, Yamamoto-Ibusuki M,
Murakami K, et al: Clinical significance of CYLD downregulation in
breast cancer. Breast Cancer Res Treat. 143:447–457. 2014.
View Article : Google Scholar : PubMed/NCBI
|