Introduction
Colorectal cancer (CRC) is a common gastrointestinal
malignancy and is the third-leading cause of cancer death in the
world (1). Molecular pathogenesis
of CRC is complicated and poorly understood. Several risk factors
are associated with CRC progression, including aging, genetic
aberrations, and chronic intestinal inflammation (2). Although CRC-related oncogenic factors
have been extensively studied, the underlying mechanisms remains to
be elucidated.
MicroRNAs (miRNA), 19–22 nucleotide non-coding RNA
molecules, have been identified as key negative regulators of gene
expression by binding to the 3′-untranslated regions (UTR) of the
target mRNAs of protein-coding genes, resulting in mRNA cleavage or
the inhibition of mRNA translation (3). Accumulating evidence suggested that
miRNAs regulate the expression of genes involved in several
important cancer-related processes including cell adhesion,
proliferation, apoptosis and tumorigenesis, including colorectal
cancer (2). Zhang et al has
reported that miR-520a-3p could suppressed colorectal cancer cell
migration via the regulation of EGFR expression (4). Song et al has reported miR-582
could enhanced the proliferation and migration ability of the CRC
cells by decreasing PTEN expression (5). Exploration of deregulated miRNAs in
carcinogenesis and metastasis of CRC, may reveal novel therapeutic
targets for the effective treatment of CRC (6).
miR-1273g-3p, a length of 21 nt non-coding RNA
molecule, is coded in an intron of the SCP2 gene, and was first
discovered in 2011 years (7).
Previous study has reported that miR-1273g-3p participates in acute
glucose fluctuation-induced proliferation attenuation in human
umbilical vein endothelial cells (8). And miR-1273g-3p could modulate
activation and apoptosis of hepatic stellate cells by directly
targeting PTEN in HCV-related liver fibrosis (9). In addition, recent study has
identified that miR-1273g-3p is upregulated in lung cancer A549
cells and promotes A549 cells migration by targeting CNR1 (10). However, the expression levels and
biological roles of miR-1273g-3p in CRC and underlying mechanism
for its functional remains not fully elucidated.
Cannabinoid receptor 1 (CNR1), the gene encoding
cannabinoid receptors (CB1), a member of the endocannabinoid
system, is highly expressed in the central and peripheral nervous
systems (PNS) in mammalians (11,12).
CNR1 expression outside the brain has been associated with cell
migration and tumor procession (1). For example, PAX3-FOXO1 induces
Cannabinoid receptor 1 to enhance cell invasion and metastasis
(13). In addition, CNR1 is
coupled to several signaling pathways directly involved in cell
survival, migration, proliferation and apoptosis, including MAPK,
cyclic AMP, and PI3K-Akt pathways (14,15).
In the present study, we identified miR-1273g-3p was
frequently up-regulated in CRC cell lines LoVo. We confirmed that
miR-1273g-3p works as a tumor initiator during proliferation,
migration and invasion of LoVo cells. Moreover, CNR1 was identified
as direct and functional target of miR-1273g-3p. Furthermore, our
results showed that miR-1273g-3p promotes proliferation, migration
and invasion of LoVo cells via CNR1 may be through the activation
of ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Materials and methods
Cell culture
Human colorectal cancer cell line LoVo, obtained
from American Type Culture Collection (Manassas, VA, USA), was
cultured in dulbecco modified Eagle medium (DMEM; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) containing high glucose,
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), 2 mM L-glutamine, 100 U/ml
penicillin, and 100 µg/ml streptomycin (P/S; Invitrogen; Thermo
Fisher Scientific, Inc.) and incubated in a 5% CO2
high-humidity atmosphere at 37°C with saturated humidity. The
normal colon epithelial NCM460 cell was cultured at the same
condition.
Oligonucleotide transfection
miR-1273g-3p mimic, miR-1273g-3p inhibitor
(anti-miR-1273g-3p, chemically modified antisense oligonucleotides
designed to target specifically against mature miR-1273g-3p) and
their corresponding controls (mimic control and inhibitor control)
were purchased from Guangzhou RiboBio Co., Ltd., (miR20022742-1-5;
Guangzhou, China). CNR1siRNA was synthesized Shanghai GenePharma
Co., Ltd., (Shanghai, China). When the LoVo cells was grown to
70–80% confluence. miR-1273g-3p mimic or inhibitor, CNR1siRNA was
transfected into cell using Lipofectamine 2000 reagents
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The culture media was replaced with fresh
medium 4 h later.
MTT assay
The influence of miR-1273g-3p on LoVo cell
proliferation was tested using the MTT (3-(4,
5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide)
(Sigma-Aldrich; Merck KGaA) assay. LoVo cells were seeded in
96-well plates at a density of 5×103 per well and
cultured at 37°C in an atmosphere of 5% CO2 for 24 h.
According to Lipofectamin™ 2000 manual, LoVo cells were
transfected with miR-1273g-3p inhibitor, inhibitor control, or
co-transfected with miR-1273g-3p inhibitor and CNR1 siRNA for 24,
48 or 72 h. Then adding 20 µl 5 mg/ml MTT solution to each well and
incubation for 4 h at 37°C. The medium was removed and 150 µl
dimethyl sulfoxide was added to each well, the absorbance was
measured at 570 nm on a microplate reader (Multiskan Spectrum;
Thermo Fisher Scientific, Inc.). Each assay was performed with 5
replicate wells for each condition.
Wound-healing assays
For cell migration assay, LoVo cells were seeded at
1.5×105 per well into 6 well plates in complete medium,
and cultured at 37°C in an atmosphere of 5% CO2 for 24
h. According to Lipofectamin™ 2000 manual, LoVo cells
were transfected with miR-1273g-3p inhibitor, inhibitor control, or
co-transfected with miR-1273g-3p inhibitor and CNR1 siRNA. At 24 h
after transfection, a linear wound was carefully made by a 20 µl
sterile pipette tip across the confluent cell monolayer to leave a
scratch of approximately 0.4–0.5 mm in width, and the cell debris
was removed by washing with phosphate-buffered saline and incubated
with a fresh serum free culture medium. The wounded monolayers were
then photographed at 0, 24 h after wounding.
Cell invasion assay
For cell invasion assay, matrigel coated transwells
(BD Biosciences, Franklin, NJ, USA) containing 8 µm pores was used
(BD Biosciences, Mountain View, CA, USA). LoVo cells were
transfected with miR-1273g-3p inhibitor, inhibitor control, or
co-transfected with miR-1273g-3p inhibitor and CNR1 siRNA. The
transfected cells were seeded into the upper chamber at
5×104 cells per well. DMEM supplemented with containing
10% FBS was added to the bottom chambers. LoVo cells was allowed to
incubate for 24 h at 37°C. After the incubation period, the filter
was removed, and non-invasion cells on the upper side of the filter
were detached using a cotton swab. Then the filter was fixed in 4%
formaldehyde. Cell were stained with 0.1% crystal violet for 20 min
and counted from three random fields.
Western blot analysis
After transfection for 24 h, total proteins were
extracted from the LoVo cells with radio immunoprecipitation assay
(RIPA) lysis buffer (Pierce; Thermo Fisher Scientific, Inc.).
Protein samples were quantified with the Pierce BCA Protein assay
kit (Pierce; Thermo Fisher Scientific, Inc.) The total protein (80
µg/well) from each sample was separated by 6 and 8% sodium dodecyl
sulfate-polyacrylamide gelelectrophoresis (SDS/PAGE) and
transferred to a polyvinylidene difluoride (PVDF) membrane (EMD
Millipore, Billerica, MA, USA). After blocking, the PVDF membranes
were incubated with the following primary antibodies: ERBB4 (cat.
no. AP7631a; dilution, 1:1,000; Abgent, Inc., San Diego, CA, USA),
S6K2 (cat. no. AP8009c; dilution, 1:1,000; Abgent, Inc.), PIK3R3
(cat. no. AP8025a; dilution, 1:1,000; Abgent, Inc.), mTOR (cat. no.
S2481; dilution, 1:1,000; Abgent, Inc.), and GAPDH (cat. no. 97166;
dilution, 1:1,000; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
overnight at 4°C, then incubated with horseradish
peroxidase-conjugated anti-mouse IgG (cat. no. A9044; dilution,
1:2,500; Sigma-Aldrich; Merck KGaA) and horse radish
peroxidase-conjugated anti-rabbit IgG (cat. no. A0545; dilution,
1:2,500; Sigma-Aldrich; Merck KGaA) as the secondary antibodies for
1 h at room temperature. The immunoreactive bands were visualized
with an enhanced chemiluminescence kit (20158; Thermo Fisher
Scientific, Inc.). Images were captured with a Fuji LAS3000-mini
imaging system (Fujifilm, Tokyo, Japan), and immunoreactive bands
were quantified.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacture's protocol. First-strand cDNA was synthesized with the
PrimeScript RT reagent kit (Roche Applied Science, Pleasanton, CA,
USA). qRT-PCR analyse was carried out to detect mRNA expression
using SYBR Premix Ex Taq (ROX; Roche Applied Science). GAPDH mRNA
levels was used as a reference control. The primer sequences are as
follows: CNR1 5′-TCCACTTCTTTTCCGCCTCC-3′ (forward), CNR1
5′-AATCTCTTTGCCCCTTCGCA-3′ (reverse); ERBB4:
5′-CCAAGAGGACAGTAGCACCC-3′ (forward),ERBB4:
5′-CTGGATTCAGGTATTCTTGTTTGGG-3′ (reverse); PIK3R3:
5′-CTTGCTCTGTGGTGGCCGAT-3′ (forward), PIK3R3:
5′-GACGTTGAGGGAGTCGTTGT-3′ (reverse); mTOR:
5′-ATGCAGCTGTCCTGGTTCTC-3′ (forward), mTOR:
5′-AATCAGACAGGCACGAAGGG-3′ (reverse); S6K2:
5′-TGACTCACAGCAGCAAGATGT-3′ (forward), S6K2:
5′-AGTACTCCCACAGCCAGGAA-3′ (reverse); GAPDH
5′-ACCACAGTCCATGCCATCAC-3′ (forward), GAPDH
5′TCACCACCCTGTTGCTGTA-3′ (reverse).
To quantify the expression of miR-1273g-3p, qRT-PCR
was performed using TaqMan microRNA assays (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and U6 snRNA was used as an
endogenous control for miR-1273g-3p. The thermocycling parameters
were 95°C for 10 min, followed by 40 cycles of 95°C for 10 sec and
by 60°C for 30 sec. The relative expression level of CNR1 and
miR-1273g-3p were cultured using the comparative delta Cq
(2−ΔΔCq) method (16).
Each sample was analyzed in triplicate.
Luciferase reporter assay
To construct CNR1 3′UTR plasmid, the full-length
3′UTR of human CNR1 mRNA containing the putative miR-1273g-3p
binding sequence (CNR1 3′-UTR-WT) was cloned into the pGL3 promotor
vector (Promega Corporation, Madison, WI, USA). Mutant-type
CNR13′-UTR (CNR1 3′-UTR-MUT) was amplifed using site-directed
mutagenesis kit (Enzynomic, Daejeon, Korea), with CNR1 3′-UTR-WT as
a template. For the dual luciferase assay, LoVo cells were cultured
in 24-well plates (5×104 cells per well) for 24 h and
co-transfected with 100 ng of CNR1-3′UTR-WT or CNR1-3′UTR MUT and
50 nM of miR-1273g-3p mimics or mimics control, using Lipofectamine
2000 transfection reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. After 6 h, the
Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) transfection
medium was replaced with DMEM supplemented with 10% FBS. Cells were
harvested and assayed with the Luciferase Assay System (Promega
Corporation) at 24 h after transfection. The Renilla luciferase
activity was normalized to the firefly luciferase activity. The
experiment was conducted in triplicate.
Statistical analysis
Statistical analysis were performed using SPSS v17.0
software (SPSS, Inc., Chicago, IL, USA). Quantitative variables
were presented as means ± SEM. Data were analyzed by one-way
analysis of variance followed with the Schefffe post hoc test. *,
**, ##, ***manifested P<0.05, P<0.01, P<0.001,
respectively. P<0.05 was considered to indicate a statistically
significant difference.
Results
LoVo cells miR-1273g-3p is
dramatically upregulated in LoVo cells
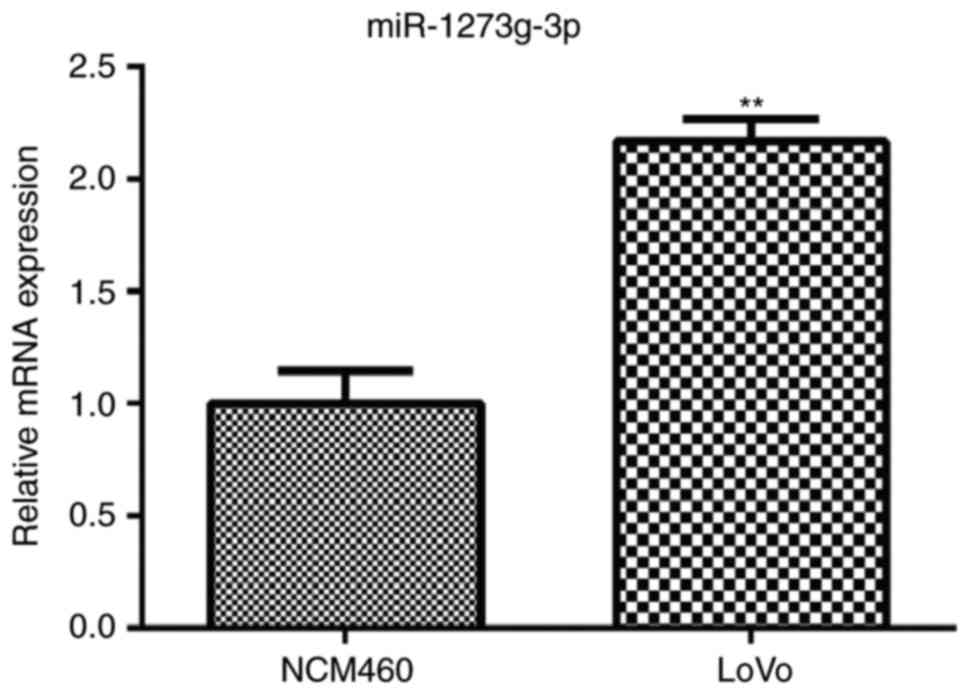
To assess the role of miR-1273g-3p in colorectal
cancer, the expression of miR-1273g-3p in human colorectal cancer
LoVo cells was examined. qRT-PCR analysis showed that the levels of
miR-1273g-3p was markedly upregulated in the LoVo cells compared
with the normal colon epithelial NCM460 cell (Fig. 1). The results indicated that
miR-1273g-3p may be involved in the malignant progression of
colorectal cancer.
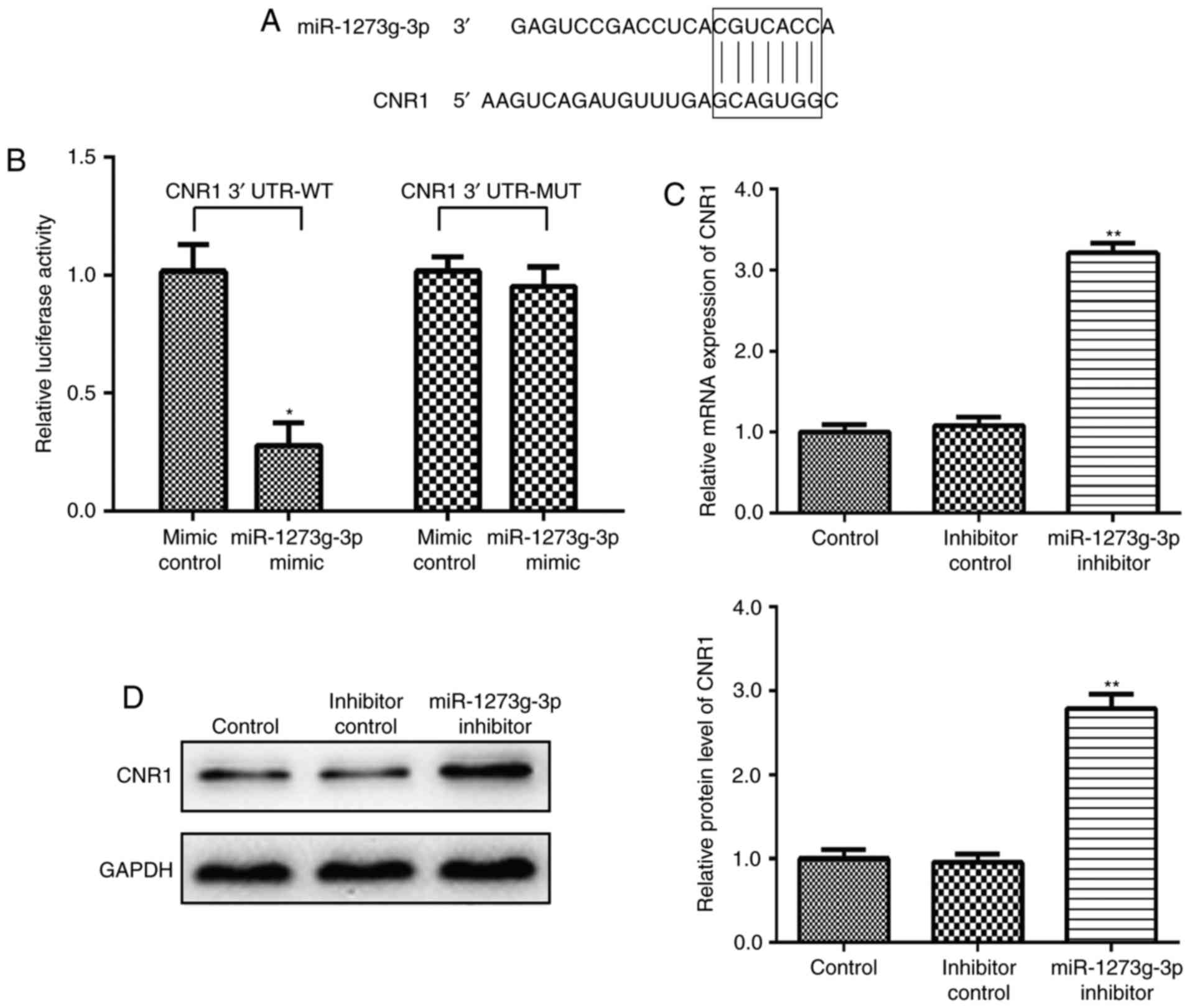
CNR1 is a direct target gene of
miR-1273g-3p in LoVo cells
Firstly, the targets of miR-1273g-3p were analyzed
using miRecords resource from three independent databases: PicTar
(http://pictar.mdc-berlin.de/),
TargetScan (http://www.targetscan.org/vert_71/), and miRBase
(http://www.mirbase.org/search.shtml).
CNR1 is predicted a theoretical target gene of miR-1273g-3p
(Table I). To obtain direct
evidence that CNR1 is a direct target of miR-1273g-3p, we
characterized the binding site of miR-1273g-3p in the 3′ UTR of
CNR1 mRNA (Fig. 2A). In a dual
luciferase reporter assay, miR-1273g-3p mimic was transfected in
LoVo cells causing the CNR1-3′-UTR-dependent luciferase activity
significantly decreased but did not affect the luciferase activity
of the mutant reporter compared with the mimic control (Fig. 2B). Meanwhile, qRT-PCR and western
blot analyses also confirmed this prediction. The results showed
that post LoVo cells transfection with miR-1273g-3p inhibitor, the
CNR1 mRNA (Fig. 2C) and protein
expression (Fig. 2D) level were
significantly upregulated, higher than that before transfection.
Collectively, these data confirmed that CNR1 as a direct target
gene of miR-1273g-3p in LoVo cells.
 | Table I.Potential targets mRNA of
miR-1273g-3p. |
Table I.
Potential targets mRNA of
miR-1273g-3p.
| miRNA | Target gene | Gene ID |
|---|
| miR-1273g-3p | CNR1 | NM_001160226 |
|
| PTEN | NM_000314 |
|
| MDM4 | NM_002393 |
|
| NOL9 | NM_024654 |
|
| PLCXD1 | NM_018390 |
|
| SAR1B | NM_016103 |
|
| ZNF850 | NM_001193552 |
|
| CYP20A1 | NM_020674 |
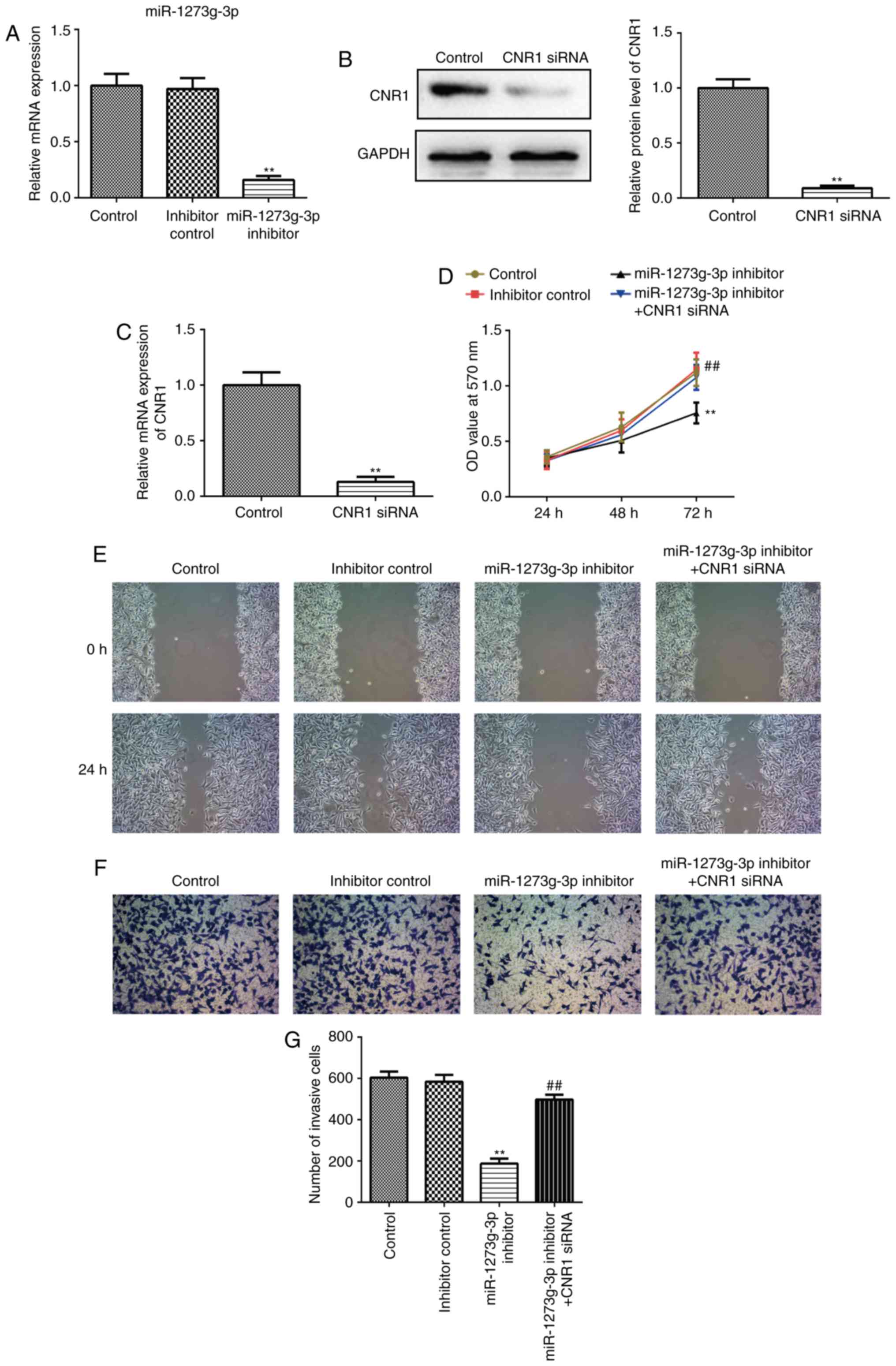
miR-1273g-3p promotes the
proliferation, migration and invasion of LoVo cells via directly
targeting CNR1
To further investigate the effect of miR-1273g-3p on
colon cancer cells, and whether this effect is mediated by CNR1
gene. LoVo cells were transfected with miR-1273g-3p inhibitor,
inhibitor control or co-transfected with miR-1273g-3p inhibitor and
CNR1 siRNA. Cells were harvested after 24 h and successful
transfection and subsequent downregulation of the expression of
miR-1273g-3p was confirmed by qRT-PCR (Fig. 3A).
Transfected with CNR1 small interfering RNA (siRNA)
in LoVo cells which resulted in a >90% reduction in the levels
of CNR1 protein (Fig. 3B) and mRNA
(Fig. 3C). MTT assay revealed that
miR-1273g-3p inhibitor significantly inhibited the growth of LoVo
cells compared with the control group (Fig. 3D). In addition, miR-1273g-3p
inhibitor significantly inhibited LoVo cells migration and invasion
compared with the control group in wound-healing assays (Fig. 3E) and cell invasion assay (Fig. 3F-G). Remarkably, the resulting
constitutively expressed CNR1siRNA abrogated the previously reduced
proliferation (Fig. 3D), migration
(Fig. 3E) and invasion (Fig. 3F-G) initiated by miR-1273g-3p
inhibitor. Taken together, these results proved that miR-1273g-3p
promotes LoVo cell proliferation, migration and invasion via
directly targeting CNR1.
miR-1273g-3p increases
ERBB4/PIK3R3/mTOR/S6K2 expression via directly targeting CNR1
It has been reported that CNR1 is coupled to several
signaling pathways directly involved in cell migration and tumor
growth, including cyclic AMP, and PI3K-Akt pathways (17,18).
Until now, ERBB4, S6K2, PIK3R3 and mTOR have been plays a critical
roles in cell movement, growth and development (6). To clarify the underlying molecular
mechanisms of miR-1273g-3p on LoVo cells proliferation, migration
and invasion. We then detected ERBB4, PIK3R3, mTOR, and S6K2
expression in LoVo cells were transfected with miR-1273g-3p
inhibitor, inhibitor control, or co-transfected with miR-1273g-3p
inhibitor and CNR1siRNA by western blot and qRT-PCR. As shown in
Fig. 4, the results revealed that
silencing of CNR1 could significantly upregulated the protein
(Fig. 4A) and mRNA (Fig. 4B-E) levels of ERBB4, PIK3R3, mTOR
and S6K2 in LoVo cells. Furthermore, inhibition of miR-1273g-3p
significantly decreased the protein (Fig. 4A) and mRNA (Fig. 4B-E) levels of ERBB4, PIK3R3, mTOR
and S6K2 compare with the control group in LoVo cells, which could
be revised by CNR1siRNA. Taken together, these data indicated that
miR-1273g-3p promotes proliferation, migration and invasion of LoVo
cells via CNR1 may be through the activation of ERBB4, PIK3R3,
mTOR, and S6K2 pathway.
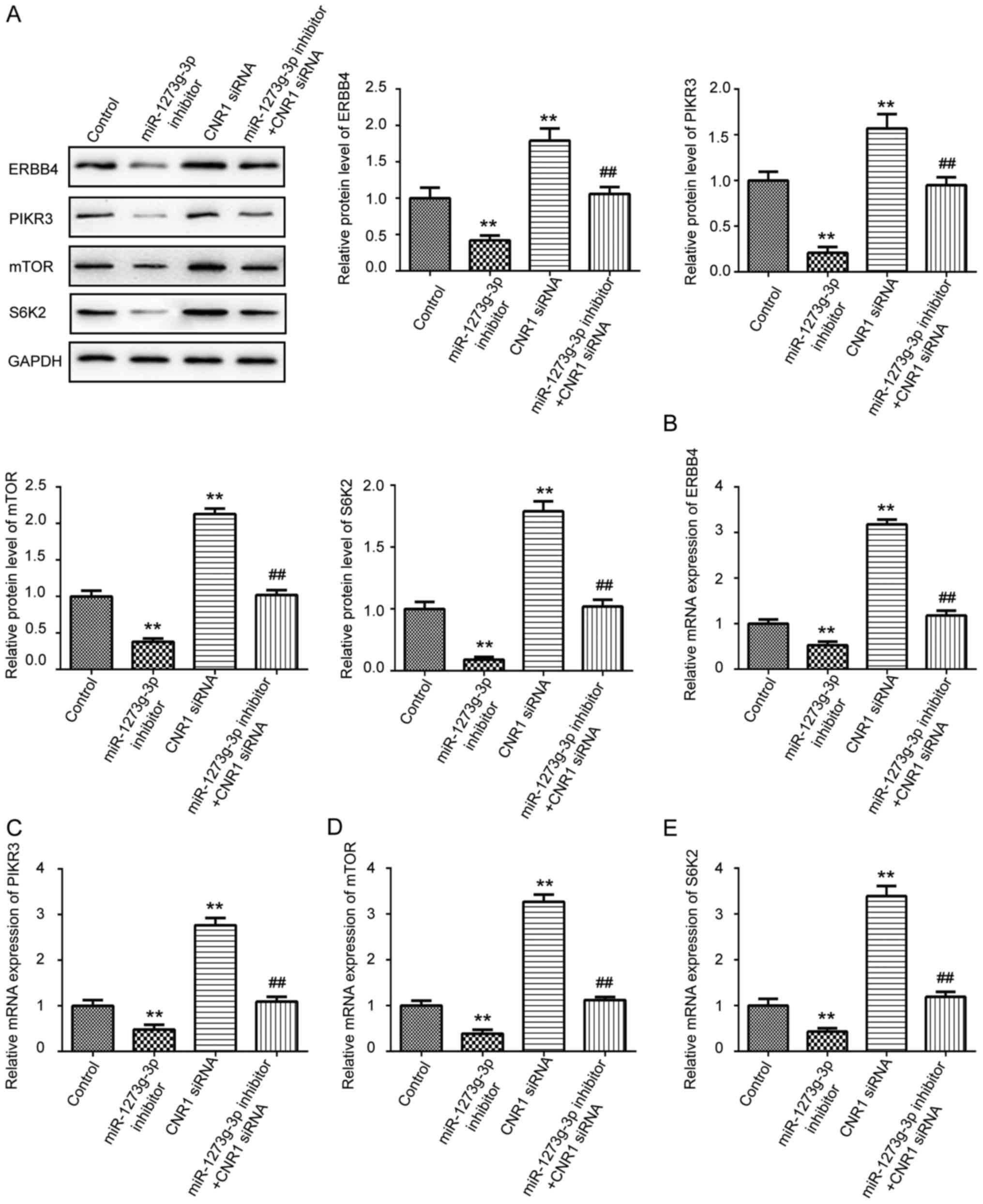 | Figure 4.miR-1273g-3p increases
ERBB4/PIK3R3/mTOR/S6K2 expression via directly targeting CNR1. (A)
Western blot was used to examine the protein expression levels of
ERBB4, PIK3R3, mTOR and S6K2 in LoVo cells transfected with
miR-1273g-3p inhibitor, CNR1siRNA, or co-transfected with
miR-1273g-3p inhibitor and CNR1siRNA. qRT-PCR was conducted to
examine the mRNA expression of ERBB4 (B), PIK3R3 (C), mTOR (D) and
S6K2 (E) in LoVo cells transfected with miR-1273g-3p inhibitor,
CNR1siRNA, or co-transfected with miR-1273g-3p inhibitor and
CNR1siRNA. Data are expressed as mean ± standard error. **P<0.01
vs. control. ##P<0.01 vs. miR-1273g-3p inhibitor.
CNR1, cannabinoid receptor 1; miR, microRNA. |
Discussion
Increasing evidence suggests that miRNAs have
important roles in carcinogenesis and metastasis (19). Dysregulation of miRNAs is
critically involved in the development and progression of
colorectal cancer (20). The
present study first validated that the expression of miR-1273g-3p
was significantly upregulated in LoVo cells by RT-PCR. In addition,
miR-1273g-3p promoted LoVo cells proliferation, migration and
invasion. The results indicated that miR-1273g-3p may act as a
tumor initiator and play an accelerating role in the progression of
CRC. However, the potential mechanisms of miR-1273g-3p in human
colorectal cancer LoVo cells remains unclear.
It has been reported that miR-1273g-3p has 1,330
binding sites on 1,074 mRNAs (7).
According to the prediction software, we list some potential
targets mRNA of miR-1273g-3p in Table
I. Previous study has indicated that miR-1273g-3p modulates
activation and apoptosis of hepatic stellate cells by directly
targeting PTEN in HCV-related liver fibrosis (9). In addition, recent study has
identified that miR-1273g-3p is upregulated in lung cancer A549
cells and promotes A549 cells migration by targeting CNR1 (10). CNR1, the gene encoding cannabinoid
receptors 1 (CB1), is highly expressed in the central and
peripheral nervous systems (PNS) in mammalians and is involved in
neuromodulatory functions (21).
CNR1 expression outside the brain has been associated with cell
migration and tumor growth (12).
Previous studies demonstrated that CNR1 may provide a therapeutic
target in tumor intervention and its anti-tumor actions by
proliferation arrest or induction of apoptosis, suppression of
angiogenesis and tumor metastatic spreading (18,22).
Recently, several studies have also suggested that CNR1 plays a
role in cell migration using cell lines, although the results
remain controversial (15,23). It has been reported that blocking
of the CB1 receptor, rather than its stimulation could inhibited
lung metastasis formation (13).
By contrast, loss of the CB1 receptor in a genetic model of colon
cancer induced tumor growth (24).
Our findings demonstrated that as a target gene of miR-1273g-3p,
CNR1 may be negatively regulated by miR-1273g-3p at the
transcriptional level via binding of the 3′UTR of CNR1 mRNA in LoVo
cells. Furthermore, our results revealed that CNR1 downregulation
was necessary to mediate the effects of miR-1273g-3p on cellular
proliferation, migration and invasion. Taken together, our data
clearly indicated that CNR1 was a direct target of miR-1273g-3p in
LoVo cells.
Recent studies have suggested that cell migratory
responses were often mediated by Gi protein-coupled receptors
(25,26). It has been reported that CNR1 is
coupled to pertussis toxin-sensitive G proteins and regulated
several signaling pathways directly involved in cell survival,
migration, proliferation and apoptosis, including MAPK, cyclic AMP,
and PI3K-Akt pathways (27,28).
Recent study has indicated that miR-1273g-3p suppresses PTEN
expression and negatively regulates the AKT pathway in LX-2 cells
(9). However, the potential
molecular mechanism of miR-1273g-3p promoting LoVo cells migration
via CNR1 was poorly understood. Until now, ERBB4, S6K2, PIK3R3 and
mTOR have been plays critical roles in cell movement, growth and
development. ERBB4, S6K2 (P70S6Kb), PIK3R3 and mTOR, potential
metastasis promoters, were identified as direct and functional
targets of miR-193a-3p/5p. ERBB4 and S6K2 were the direct targets
of miR-193a-3p and that PIK3R3 and mTOR were the direct targets of
miR-193a-5p in NSCLC. Therefore, miR-193a-3p/5p could inhibit the
metastasis of NSCLC by interacting with the ERBB signaling pathway
(6). ERBB4 activated the PI3K-AKT
cascade contribute to the progression of pancreatic cancer
(29). mTOR and its downstream
effector S6K2 were frequently upregulated in breast cancer, and
assumed to be driving forces in tumourigenesis (30). PIK3R3/AKT pathway was involved in
the tumorigenesis and progression of gastric cancer (31). Inhibition of PI3K/mTOR pathway
impaired breast tumor invasion and metastasis (32). The present study indicated that
cannabinoid receptor 1 (CNR1) was identified as a direct target
gene of miR-1273g-3p. Whether ERBB4/PIK3R3/mTOR/S6K2 signaling
pathway was involved in the effect of miR-1273g-3p on LoVo cells
remains unknown. In the present study, our results showed that
transfection of miR-1273g-3p inhibitor significantly downregulated
the proteins and mRNA levels of ERBB4, PIK3R3, mTOR and S6K2 in
LoVo cells, which could be reversed by CNR1siRNA. These results
suggested that the dysregulation of the ERBB4/PIK3R3/mTOR/S6K2
signaling pathway may be a potential mechanisms of miR-1273g-3p
promoting the proliferation, migration and invasion of LoVo
cells.
In conclusion, our results showed that miR-1273g-3p
was frequently upregulated in LoVo cells. The overexpression of
miR-1273g-3p promoted LoVo cells proliferation, migration and
invasion via directly targeting CNR1, and the potential mechanisms
may be through the activation of ERBB4/PIK3R3/mTOR/S6K2 signaling
pathway. Importantly, these findings indicated that miR-1273g-3p
may represent a novel target for therapeutic intervention to CRC.
However, a limitation of this study is normal cells should always
be included in these studies as manipulation of expression always
has an effect and would like to keep it to the minimum for control
cells. Further studies to investigate this possibility are ongoing
in our laboratory.
Acknowledgements
The present study was supported by the department of
Health projects of Lianyungang city (grant no: 201715) and clinic
science and technology projects of Jiangsu collage (grant no:
JLY20140133).
References
|
1
|
Xie H, Sun X, Piao Y, Jegga AG, Handwerger
S, Ko MS and Dey SK: Silencing or amplification of endocannabinoid
signaling in blastocysts via CB1 compromises trophoblast cell
migration. J Biol Chem. 287:32288–32297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wong MC, Ching JY, Chan VC, Lam TY, Luk
AK, Wong SH, Ng SC, Ng SS, Wu JC, Chan FK and Sung JJ: Colorectal
cancer screening based on age and gender: A cost-effectiveness
analysis. Medicine (Baltimore). 95:e27392016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang R, Liu R, Liu C, Niu Y, Zhang J, Guo
B, Zhang CY, Li J, Yang J and Chen X: A novel role for MiR-520a-3p
in regulating EGFR expression in colorectal cancer. Cell Physiol
Biochem. 42:1559–1574. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song B, Long Y, Liu D, Zhang W and Liu C:
MicroRNA-582 promotes tumorigenesis by targeting phosphatase and
tensin homologue in colorectal cancer. Int J Mol Med. 40:867–874.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and −5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ivashchenko A, Berillo O, Pyrkova A and
Niyazova R: Binding sites of miR-1273 family on the mRNA of target
genes. Biomed Res Int. 2014:6205302014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo J, Sang Y, Yin T, Wang B, Yang W, Li
X, Li H and Kang Y: miR-1273g-3p participates in acute glucose
fluctuation-induced autophagy, dysfunction, and proliferation
attenuation in human umbilical vein endothelial cells. Am J Physiol
Endocrinol Metab. 310:E734–E743. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Niu X, Fu N, Du J, Wang R, Wang Y, Zhao S,
Du H, Wang B, Zhang Y, Sun D and Nan Y: miR-1273g-3p modulates
activation and apoptosis of hepatic stellate cells by directly
targeting PTEN in HCV-related liver fibrosis. FEBS Lett.
590:2709–2724. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hou L, Su X, Qin X, et al: MiR-1273g-3p
regulates A549 cell migration by targeting CNR1. J Med Mol Biol.
11:125–131. 2014.
|
|
11
|
Pisanti S, Picardi P, D'Alessandro A,
Laezza C and Bifulco M: The endocannabinoid signaling system in
cancer. Trends Pharmacol Sci. 34:273–282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Freundt-Revilla J, Kegler K, Baumgärtner W
and Tipold A: Spatial distribution of cannabinoid receptor type 1
(CB1) in normal canine central and peripheral nervous system. PLoS
One. 12:e01810642017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marshall AD, Lagutina I and Grosveld GC:
PAX3-FOXO1 induces cannabinoid receptor 1 to enhance cell invasion
and metastasis. Cancer Res. 71:7471–7480. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guzmán M, Sánchez C and Galve-Roperh I:
Control of the cell survival/death decision by cannabinoids. J Mol
Med (Berl). 78:613–625. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sánchez MG, Ruiz-Llorente L, Sánchez AM
and Díaz-Laviada I: Activation of phosphoinositide 3-kinase/PKB
pathway by CB(1) and CB(2) cannabinoid receptors expressed in
prostate PC-3 cells. Involvement in Raf-1 stimulation and NGF
induction. Cell Signal. 15:851–859. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guindon J and Hohmann AG: The
endocannabinoid system and cancer: Therapeutic implication. Br J
Pharmacol. 163:1447–1463. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Greenhough A, Patsos HA, Williams AC and
Paraskeva C: The cannabinoid delta(9)-tetrahydrocannabinol inhibits
RAS-MAPK and PI3K-AKT survival signalling and induces BAD-mediated
apoptosis in colorectal cancer cells. Int J Cancer. 121:2172–2180.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jafri MA, Al-Qahtani MH and Shay JW: Role
of miRNAs in human cancer metastasis: Implications for therapeutic
intervention. Semin Cancer Biol. 44:117–131. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang LX, Hu CY, Jing L, Wang MC, Xu M,
Wang J, Wang Y, Nan KJ and Wang SH: microRNA-219-5p inhibits
epithelial-mesenchymal transition and metastasis of colorectal
cancer by targeting lymphoid enhancer-binding factor 1. Cancer Sci.
108:1985–1995. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pisanti S, Malfitano AM, Grimaldi C,
Santoro A, Gazzerro P, Laezza C and Bifulco M: Use of cannabinoid
receptor agonists in cancer therapy as palliative and curative
agents. Best Pract Res Clin Endocrinol Metab. 23:117–131. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Proto MC, Gazzerro P, Di Croce L, Santoro
A, Malfitano AM, Pisanti S, Laezza C and Bifulco M: Interaction of
endocannabinoid system and steroid hormones in the control of colon
cancer cell growth. J Cell Physiol. 227:250–258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song ZH and Zhong M: CB1 cannabinoid
receptor-mediated cell migration. J Pharmacol Exp Ther.
294:204–209. 2000.PubMed/NCBI
|
|
24
|
Refolo MG, D'Alessandro R, Malerba N,
Laezza C, Bifulco M, Messa C, Caruso MG, Notarnicola M and Tutino
V: Anti proliferative and pro apoptotic effects of flavonoid
quercetin are mediated by CB1 receptor in human colon cancer cell
lines. J Cell Physiol. 230:2973–2980. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Neptune ER and Bourne HR: Receptors induce
chemotaxis by releasing the betagamma subunit of Gi, not by
activating Gq or Gs. Proc Natl Acad Sci USA. 94:pp. 14489–14494.
1997; View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arai H, Tsou CL and Charo IF: Chemotaxis
in a lymphocyte cell line transfected with C-C chemokine receptor
2B: Evidence that directed migration is mediated by betagamma
dimers released by activation of Galphai-coupled receptors. Proc
Natl Acad Sci USA. 94:pp. 14495–14499. 1997; View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Howlett AC, Qualy JM and Khachatrian LL:
Involvement of Gi in the inhibition of adenylate cyclase by
cannabimimetic drugs. Mol Pharmacol. 29:307–313. 1986.PubMed/NCBI
|
|
28
|
Felder CC, Veluz JS, Williams HL, Briley
EM and Matsuda LA: Cannabinoid agonists stimulate both receptor-
and non-receptor-mediated signal transduction pathways in cells
transfected with and expressing cannabinoid receptor clones. Mol
Pharmacol. 42:838–845. 1992.PubMed/NCBI
|
|
29
|
Chen Y, Bai X, Zhang Q, Wen L, Su W, Fu Q,
Sun X, Lou Y, Yang J, Zhang J, et al: The hepatitis B virus X
protein promotes pancreatic cancer through modulation of the
PI3K/AKT signaling pathway. Cancer Lett. 380:98–105. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Karlsson E, Pérez-Tenorio G, Amin R,
Bostner J, Skoog L, Fornander T, Sgroi DC, Nordenskjöld B, Hallbeck
AL and Stål O: The mTOR effectors 4EBP1 and S6K2 are frequently
coexpressed, and associated with a poor prognosis and endocrine
resistance in breast cancer: A retrospective study including
patients from the randomised Stockholm tamoxifen trials. Breast
Cancer Res. 15:R962013. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cai JQ, Xu XW, Mou YP, Chen K, Pan Y and
Wu D: Upregulation of HOXB7 promotes the tumorigenesis and
progression of gastric cancer and correlates with clinical
characteristics. Tumour Biol. 37:1641–1650. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wander SA, Zhao D, Besser AH, Hong F, Wei
J, Ince TA, Milikowski C, Bishopric NH, Minn AJ, Creighton CJ and
Slingerland JM: PI3K/mTOR inhibition can impair tumor invasion and
metastasis in vivo despite a lack of antiproliferative action in
vitro: Implications for targeted therapy. Breast Cancer Res Treat.
138:369–381. 2013. View Article : Google Scholar : PubMed/NCBI
|