Introduction
The hair follicles of mammals have been reported to
be the only organ that continually cycles in growth, which includes
the growth period (anagen), the return period (catagen) and the
quiescence period (telogen) (1).
Alpaca hair has a high economic value; therefore, a number of
studies have investigated the mechanisms underlying the growth and
color formation of alpaca hair (2–4).
Previous studies have determined that microRNAs (miRNAs) serve
important roles in the development and differentiation of skin and
that miRNAs are associated with various skin diseases and skin
cancers (5); however, miRNAs
function in animal hair growth by regulating the expression of
target genes. A previous study identified 39 significantly
differentially expressed miRNAs in the ear and back skin of young
alpaca, including let-7b. Through three target prediction software
TargetScan (http://targetscan.org/), Pictar
(http://pictar.bio.nyu.edu/) and miRbase
targets (http:///microrna.sanger.ac
uk/targets/v1/), Ectodysplasin A (EDA) was predicted to be a target
gene of let-7b (6).
The EDA gene is located on chromosome Xq12-13 and
belongs to the tumor necrosis factor superfamily, as revealed by
the genetic analysis of EDA from syngeneic mice (7). EDA mutations may lead to a syndrome
that causes ectodermal dysplasia without sweating, called
hypohidrotic ectodermal dysplasia (HED). HED comprises >180
genetic syndromes and affects at least two ectodermal structures,
including the hair, teeth, nails or exocrine glands (8). The most common type of ectodermal
dysplasia is X-linked HED, which is characterized by sparse or
absent hair, dental dysplasia or missing teeth, reduced sweating
ability and defects of various lipid- or mucus-secreting glands.
EDA is crucial for morphogenesis of skin appendages, including
hair, teeth, sweat glands and eyelid glands (9). The EDA signaling pathway serves an
important role in the formation of ectoderm, hair, teeth and
exocrine glands (10). Enhanced
EDA signals were reported to enlarge wool fibers and alter coat
shape in mammals (11,12). A previous study demonstrated that
EDA mRNA expression was significantly higher in hair follicles in
goat skin tissue during the catagen phase compared with expression
during the telogen and anagen phases, and that expression was
higher during the anagen phase compared with the telogen phase
(13). This result indicated that
EDA may affect the regulation of the hair follicle growth
cycle.
Alpaca ear hair differs in characteristics from back
hair: Ear hair is thick, straight and has a long growth cycle,
whereas back hair is elongated, bent and has a short growth cycle
(14). Gene chip analyses
indicated that let-7b expression was significantly higher in the
back skin compared with the ear skin of the alpaca; boinformatics
analysis predicted EDA as a target gene of let-7b (6). In the present study, the differential
expression of EDA in alpaca ear and back skin tissues was detected
to analyze the effects of EDA on alpaca hair growth. In addition, a
dual-luciferase reporter assay was used to verify that EDA was a
target of let-7b, and the regulatory relationship between let-7b
and EDA was verified at the cellular level. The present study
provided additional information on the mechanisms underlying alpaca
hair growth and may serve as a theoretical basis to continue
exploring let-7b and its target genes. In addition, research into
the regulation of let-7b on alpaca hair growth through
downregulation of EDA may also inform further studies into HED.
Materials and methods
Animals, cells and plasmids
A total of 3 three-years-old male alpacas were
obtained from an alpaca breeding base in Shanxi, China. Animal care
and skin sample collection were in accordance with the
International Guiding Principles for Biomedical Research Involving
Animals (http://www.encyclopedia.com/science/encyclopedias-almanacs-transcripts-and-maps/international-guiding-principles-biomedical-research-involving-animals)
and approved by the Animal Experimentation Ethics Committee of
Shanxi Agricultural University (Taigu, China). Two skin samples
were collected from the ear and back (n=2/site) of each alpaca: One
biopsy sample was selected for RNA extraction, and the other biopsy
sample was selected for protein extraction; all samples were soaked
in liquid nitrogen.
Alpaca fibroblasts and 293T cells were stored in
liquid nitrogen at the Alpaca Biological Laboratory in Shanxi
Agricultural University (Taigu, China) and the primary alpaca
fibroblasts were cultured in Alpaca Biological Laboratory, Shanxi
Agricultural University. 293T cells were obtained from Shanghai
Bioleaf Biotech Co., Ltd. (Shanghai, China). Alpaca fibroblasts and
293T cells were cultured in 37°C, 5% CO2, in Alpaca
fibroblasts medium (Thermo Fisher Scientific, Inc., Waltham, MA,
USA) and Dulbecco's modified Eagle's medium (Thermo Fisher
Scientific, Inc.), respectively.
The pmirGLO dual-luciferase miRNA target expression
vector and the negative control (empty vector) were purchased from
Promega Corporation (Madison, WI, USA). The
pcDNA6.2-GW/EmGFP-let-7b overexpression vector based on the let-7b
mature sequence; the negative control (empty vector) did not
contain the let-7b sequence. The vectors were obtained from a
previous deep sequencing analysis on alpaca skin (15) and were purchased from Invitrogen
(Thermo Fisher Scientific, Inc., Waltham, MA, USA).
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the six skin samples
(diameter=8 mm; three samples each from the ear skin and the back
skin) and the transfected and untransfected alpaca fibroblast cells
by using TRIzol LS Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocols. The specific
transfection method is mentioned below in ‘Alpaca fibroblast
culture and transfection of the let-7b eukaryotic expression
vector’. Total RNA concentration was determined with a NanoDrop
1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.), and the integrity of total RNA was confirmed by
1% agarose gel electrophoresis and ethidium bromide staining for 5
min at room temperature, then rinse with water at room temperature.
cDNA was synthesized using a PrimeScript II First-Strand cDNA
Synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China)
according to the manufacturer's protocols. The PCR product of EDA
was 202 bp, as confirmed by 1% agarose gel and 2000 DNA Marker; the
EDA gene was sequenced by the Beijing Genomics Institute (Shenzhen,
China). Gene sequencing analysis revealed that the alpaca EDA gene
was homologous with other species [published on National Center for
Biotechnology Information (NCBI), Bethesda, MD, USA] by NCBI-BLAST
https://blast.ncbi.nlm.nih.gov/Blast.cgi; accession
nos. XM_015250693.1 XM_014566996.1, XM_010978765.1, XM_010973676.1,
XM_008155957.1, XM_008155955.1, XM_004871764.2, XM_004871761.2,
XM_012643114.1 and XM_012643097.1. Then qPCR was conducted with
SYBR Premix Ex Taq II (Perfect Real Time; Takara Biotechnology Co.,
Ltd.) according to the manufacturer's protocols. The primer
sequences used were: EDA, forward, 5′-GTGGACGGCACCTACTTCATCTAT-3′,
reverse: 5′-GCACCATCTTCACAGCGATCTTCT-3′; EDA expression was
normalized to 18S rRNA. The reaction conditions constituted
pre-denaturation: 95°C for 10 min; denaturation: 95°C for 15 sec;
annealing: 63°C for 30 sec; extension 72°C 30 sec; 40 cycles; 95°C
15 sec, 60°C 1 min and 95°C, 15 sec. and the relative expression
level was calculated using the 2−ΔΔCq method (16).
Western blot analysis
Total protein content from the six skin samples
(diameter=8 mm; three samples each from the ear skin and the back
skin), the transfected and untransfected alpaca fibroblasts were
extracted with a protein extraction kit (radioimmunoprecipitation
lysis buffer, Beyotime Institute of Biotechnology, Haimen, China)
according to the manufacturer's protocols. The specific
transfection method is mentioned below in ‘Alpaca fibroblast
culture and transfection of the let-7b eukaryotic expression
vector’. The protein concentrations were determined using a
NanoDrop 1000 spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.). Equal amounts (300 µg/lane) of protein
from alpaca skin samples and equal amounts (500 µg/lane) of protein
from cells were separated by 10% SDS-PAGE and transferred onto
polyvinylidene fluoride membranes (constant pressure 100 V, 1 h).
The membranes were blocked with 5% nonfat milk (w/v) in TBST (TBS
with 0.1% Tween 20) at room temperature for 1 h. Membranes were
incubated with primary antibodies against EDA (1:200, ab125233,
Abcam, Cambridge, UK) and anti β-actin antibody (1:1,500; CW0096 M,
CWBIO, Beijing, China) at 4°C overnight. Following washing in TBST,
membranes were incubated with the goat anti-rabbit
horseradish-peroxidase (HRP) secondary antibody (1:8,000 in 3%
TBST; bs-0295G-HRP, BIOSS, Beijing, China) and the goat anti-mouse
(HRP) secondary antibody (1:5,000 in 3% TBST; bs-0296G-HRP, BIOSS)
for 1 h at 37°C. Membranes were washed with TBST and protein bands
were visualized with an Enhanced Chemiluminescence Western Blot kit
(CWBIO). EDA signal intensity was quantified using ImageJ (ImageJ
Launcher 1.4.3.67, National Institutes of Health, Bethesda, MD,
USA) and Quantity One software (Quantity One 4.6.2.70, Bio-Rad
Laboratories Inc, Hercules, CA, USA); relative expression levels
were normalized to β-actin.
Dual-luciferase reporter assay
The 3′-untranslated region (UTR) of EDA from Alpaca
back and ear skin samples was amplified by PCR using the following
primers to construct the dual-luciferase vector pmirGLO-EDA-3′-UTR:
Forward (SacI restriction site underlined)
5′-CGAGCTCGACCCTGAATCCGTACTTGG-3′, reverse (XbaI restriction
site underlined) 5′-GCTCTAGAGCAAATCCCTCTGTCCTATCCTC-3′. The
reaction conditions were: Pre-denaturation: 94°C, 2 min;
denaturation: 94°C, 30 sec; annealing 53°C, 30 sec; extension 72°C,
30 sec; 35 cycles; end extension: 72°C, 2 min. A 2×Taq MasterMix
(CWBIO) was used, and composed of Es Taq DNA Polymerase, Mg2 +,
dNTPs and PCR stabilizer and enhancer. The amplified product (469
bp) was connected to the T-Vector pMD19 (Takara Biotechnology Co.,
Ltd.), and then the EDA-3′-UTR-T-Vector pMD19 and the pmirGLO
vector (Promega Corporation) were double digested by the
SacI and XbaI. Finally, the two digested products
were ligated with T4 ligase. The dual-luciferase recombinant vector
was constructed. The empty pmirGLO vector (368.7 µg/µl) and let-7b
negative control (349.6 ng/ul) served as negative controls.
Untransduced 293T cells were plated in 24-well plates to ~60%
confluence and transfected with the pmirGLO-EDA-3′-UTR (422.3
ng/µl; pmirGLO-EDA) vector and the pcDNA6.2-GW/EmGFP-let-7b (348.4
ng/µl) overexpression vector using the Attractene Transfection
Reagent (Qiagen, Shanghai, China). Dual-luciferase assays (Promega
Corporation) were performed 48 h post-transfection according to the
manufacturer's protocols, and luciferase activity was determined
using a GloMax96 Microplate Luminometer (Promega Corporation).
Firefly and Renilla luciferase served as the experimental
and control, respectively. The firefly/Renilla ratios were
normalized to a negative control and that of an empty plasmid
control.
Alpaca fibroblast culture and
transfection of the let-7b eukaryotic expression vector
Alpaca fibroblasts were cultured in six-well plates
at 37°C and 5% CO2 to ~60% confluence; the cells were
transfected with 1.2 µg of the let-7b expression vector (treatment
group) by using the Attractene Transfection Reagent (Qiagen)
according to the manufacturer's protocols. Untransfected cells
(cell control) incubated in Attractene Transfection Reagent
(Qiagen) and cells transfected with let-7b negative control
expression vectors served as negative controls. Following 48 h
incubation, fluorescence from the GFP was observed under a
fluorescence microscope (Leica Microsystems GmbH). The cells were
collected for total RNA and protein extraction for the analysis of
EDA expression by RT-qPCR and western blotting, respectively as
aforementioned.
Immunocytochemical analysis of EDA in
fibroblasts
Alpaca fibroblasts were seeded onto slides in
24-well plates at 37°C and 5% CO2 to ~60% confluence.
The cells were transfected with 0.4 µg of the let-7b expression
vector (treatment group) by using the Attractene Transfection
Reagent (Qiagen) according to the manufacturer's protocols, and
cells incubated in Attractene Transfection Reagent (Qiagen) served
as an untransfected control. Following 48 h incubation (37°C, 5%
CO2), the fibroblasts were processed for
immunocytochemistry as previously described (17). Cells were fixed with 4%
paraformaldehyde for 30 min at room temperature, incubated in 3%
hydrogen peroxide at 37°C for 10 min, blocked with 5% bovine serum
albumin (Beijing Solarbio Science & Technology, Beijing, China)
at 37°C for 30 min and incubated with anti-EDA rabbit polyclonal
antibody (1:30, ab198022, Abcam Cambridge, UK) at 4°C for 14 h;
cells only for the negative staining control were incubated in PBS
in similar conditions. HRP-conjugated goat anti-rabbit
immunoglobulin G (SP Rabbit HRP kit, CWBIO) was used as a secondary
antibody and cells were incubated at 37°C for 30 min.
3–3′diaminobenzidine (DAB) was used with the SP Rabbit HRP kit
(CWBIO), which served as a positive control, containing DAB-A (20×)
and DAB-B (1×) reagents; DAB produces a brown precipitate following
catalysis by HRP. Cells of the -positive control group were
incubated in DAB ~3 min in the dark at room temperature and
subsequently counterstained with hematoxylin for ~5 min at room
temperature. The slides were mounted, cover-slipped and evaluated
under a Leitz DMRB microscope (Leica Microsystems GmbH), five
fields were examined per slide.
Statistical analysis
Data are presented as the mean ± standard deviation
of three replicates. Data were analyzed using SPSS v17.0 software
(SPSS, Inc., Chicago, IL, USA). Multiple comparison analyses were
performed by one-way analysis of variance followed by a
Newman-Keuls post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
RT-qPCR analysis of EDA in alpaca ear
and back skin
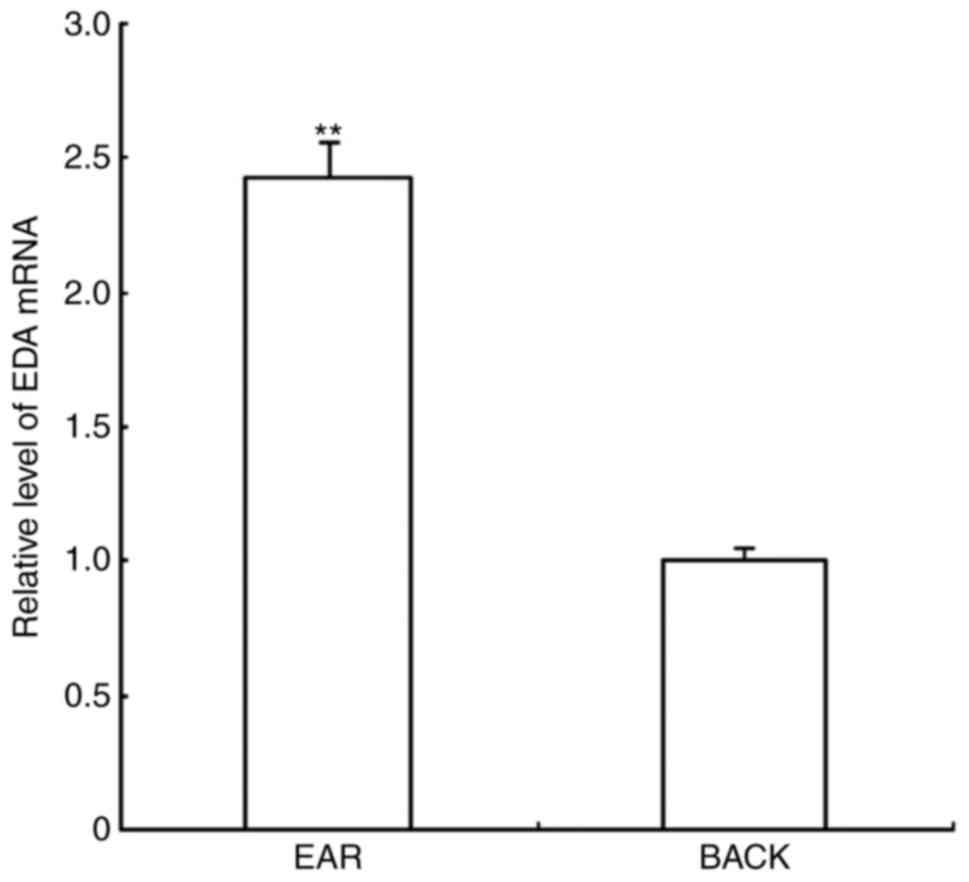
RT-qPCR results demonstrated that the amplification
curves of EDA and 18S rRNA were smooth; the inflection points were
clear with marked repeatability, and the melting curves revealed a
single peak, indicating that there was no nonspecific amplification
during the amplification process. EDA mRNA expression levels in ear
skin were relatively 2.425 higher compared with expression in the
back skin (Fig. 1).
Western blot analysis of EDA in alpaca
ear and back skin
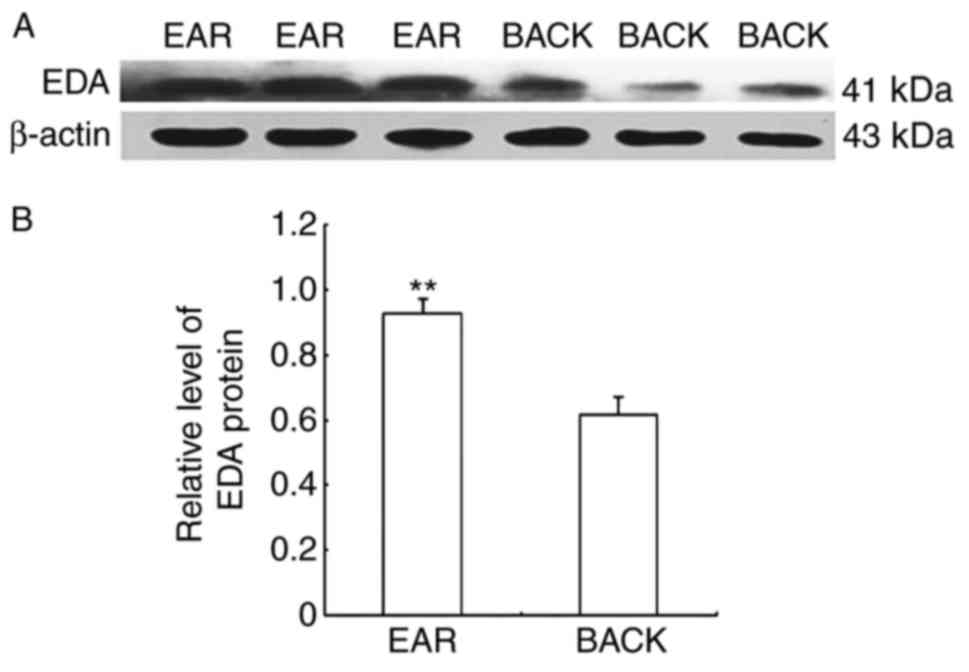
EDA protein expression levels were detected in
alpaca ear and back skin by western blot. The molecular size of the
EDA protein was 41 kDa, whereas that of the β-actin protein was 43
kDa (Fig. 2A). The relative
protein expression levels in the ear and back skin were
0.9254±0.0473 and 0.6145±0.0582, respectively. EDA protein
expression in alpaca ear skin was 1.506-fold higher compared with
expression in back skin (Fig.
2B).
Dual-luciferase reporter assay
Our previous study reported that let-7b may be
involved in regulating hair growth and was differentially expressed
in alpaca skin from various parts of the body with different fiber
qualities (6). Previous
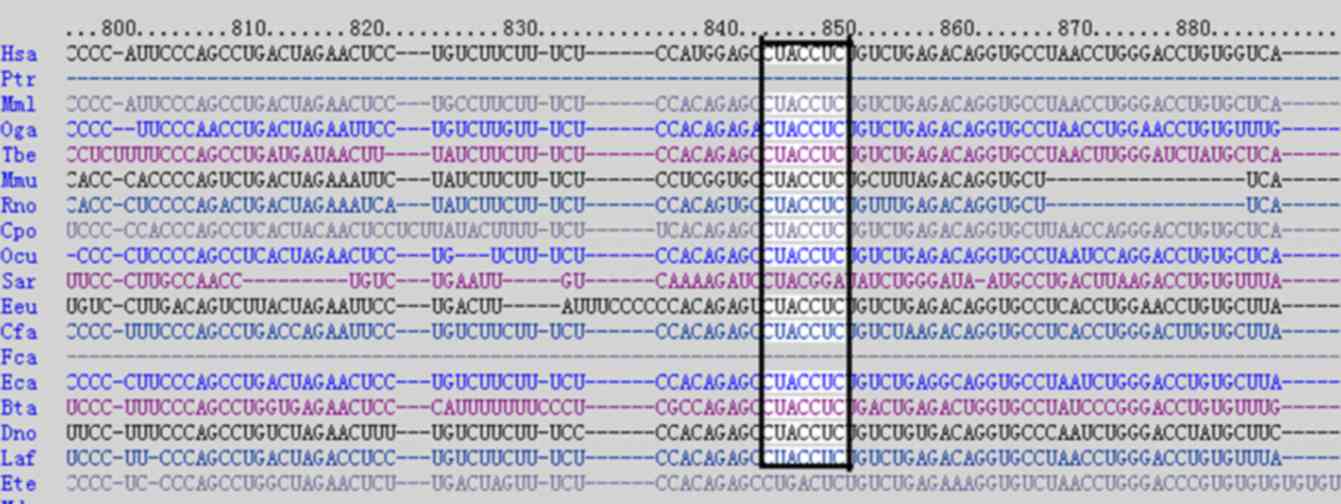
bioinformatics predictions using the publicly available algorithm
from TargetScan release 4.1 (www.targetscan.org) indicated that EDA may be a target
of let-7b (Fig. 3) (6). In the present study, a
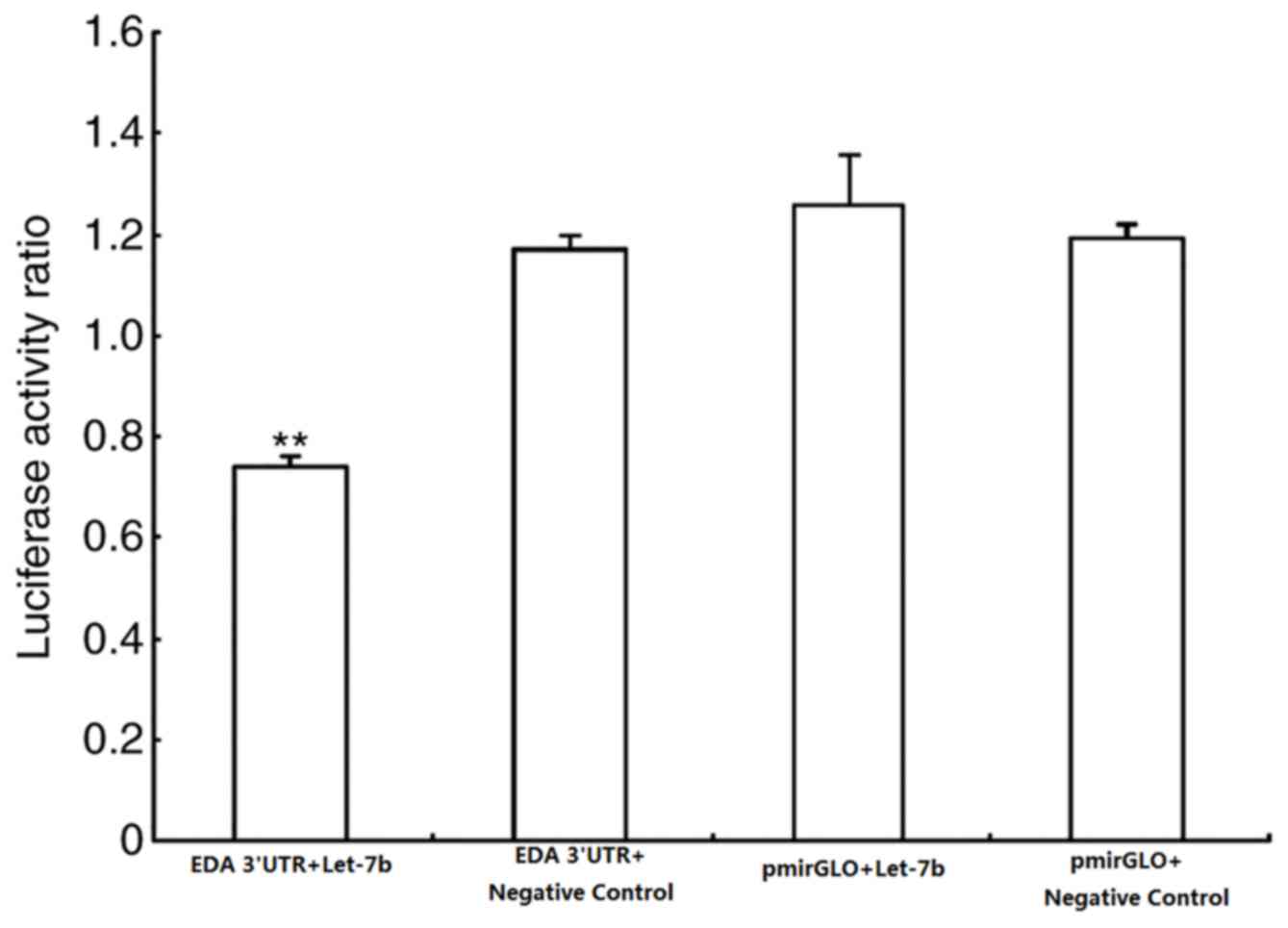
dual-luciferase recombinant vector that contained the 3′-UTR of EDA
was constructed by combining target prediction algorithms. The
firefly/Renilla ratio collected from the experimental group
(cells transfected with pmirGLO-EDA-3′-UTR and let-7b
overexpression vector) significantly decreased, whereas of the
control groups (cells transfected with pmirGLO and let-7b negative
controls) did not change (Fig.
4).
EDA expression in alpaca fibroblasts
overexpressing let-7b
Alpaca fibroblasts were transfected with the let-7b
overexpression and negative control vectors, and cultured under
standard conditions, to further evaluate let-7b regulation of EDA.
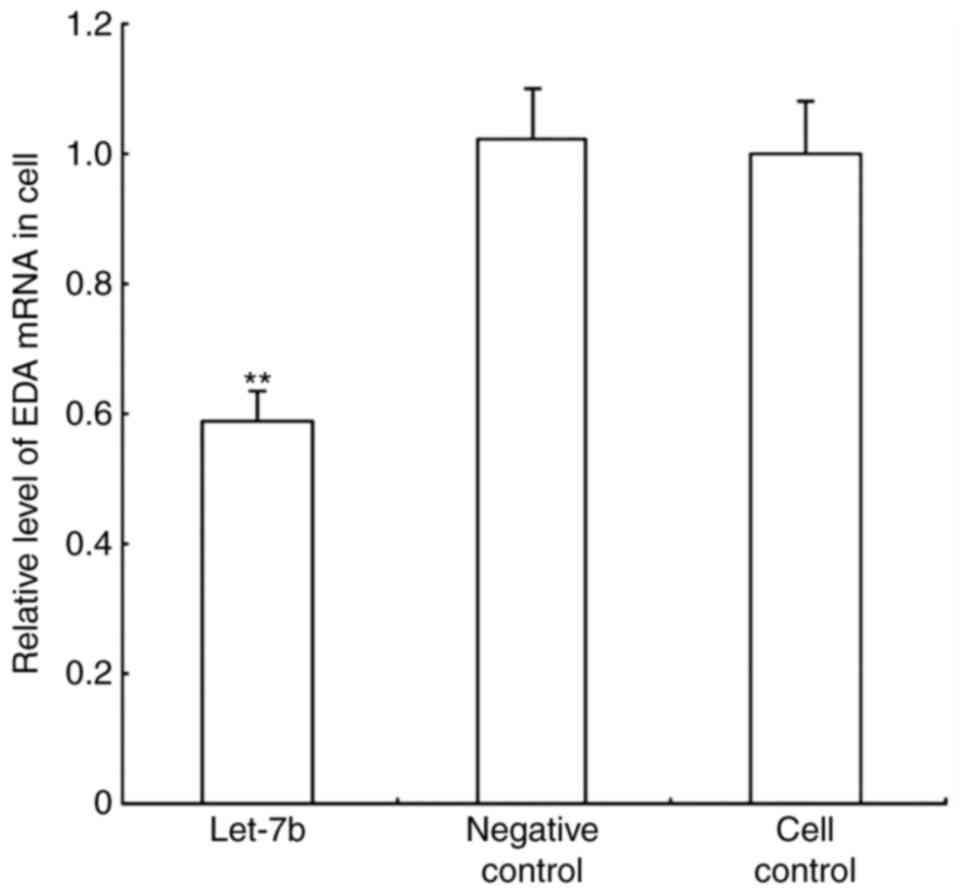
Relative EDA mRNA expression levels were determined by RT-qPCR
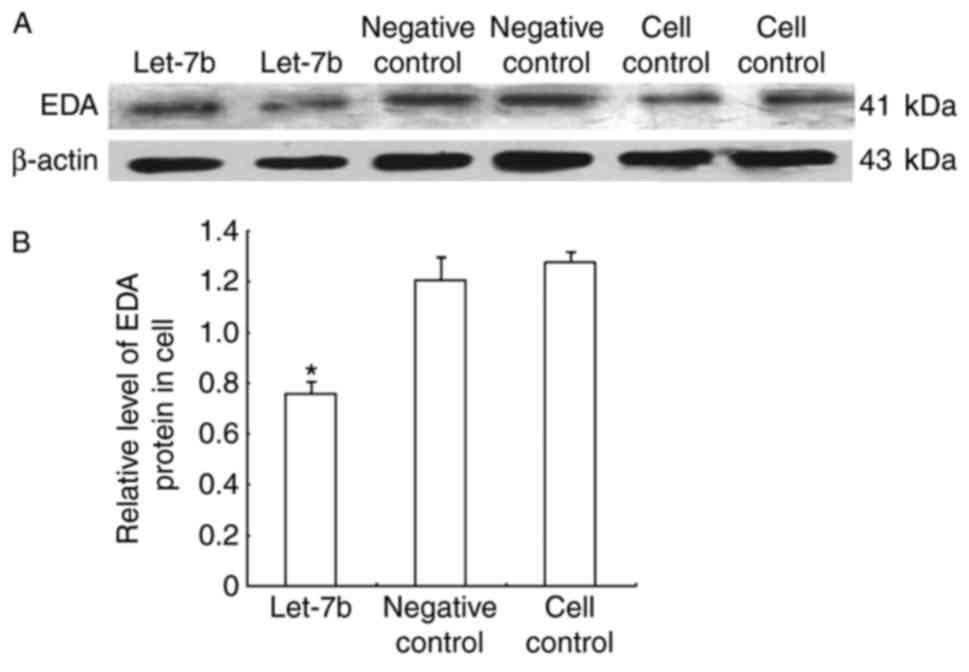
(Fig. 5), and protein expression
levels were detected through western blot analysis (Fig. 6). The results demonstrated that
let-7b overexpression resulted in decreased EDA expression at both
the mRNA and protein level. let-7b transfected cells exhibited a
1.914-fold reduction in EDA mRNA quantity compared with the control
groups, whereas the EDA protein expression level was 1.589-fold
lower in the let-7 overexpressing cells, compared with the control
groups.
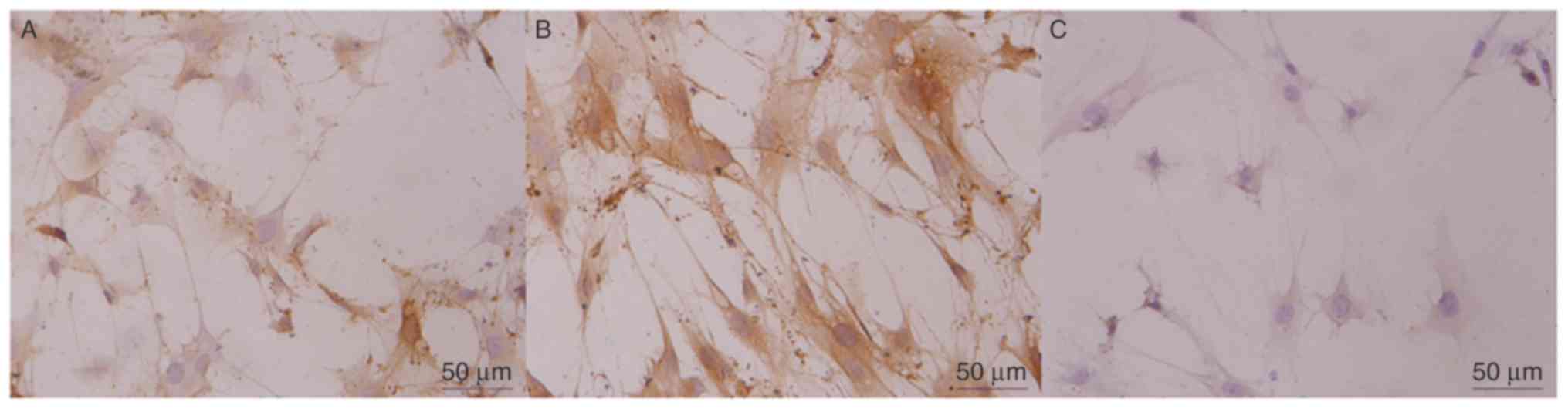
EDA expression in fibroblast cells. Alpaca
fibroblasts were transfected with the let-7b overexpression vector.
Positive reactivity was noted as predominantly fibroblastic
cytoplasmic staining, and the negative staining control cells
exhibited no specific immunostaining of fibroblasts as EDA was not
expressed (Fig. 7).
Immunoreactivity staining was barely detected in the let-7b
overexpression vector-transfected group, which was consistent with
the results of RT-qPCR and western blotting.
Discussion
The first miRNA was discovered in a nematode in
1993, and numerous miRNAs have subsequently been identified in a
variety of species (18–27); an increasing number of reports on
miRNA represent a new approach of gene regulation. miRNAs regulate
the expression of target genes in two ways, degradation and
inhibition of expression. miRNA (miR)-17, let-7, miR-30 and miR-8
are expressed in mouse, goat and sheep skin; these miRNAs may be
related to skin development (28).
Fibroblast growth factor (FGF) 5 and transforming growth factor β
receptor type I (TGFβRI) have been previously identified as target
genes of let-7b. Let-7b negatively regulates its target genes by
binding to the 3′ end of the mRNA (29,30).
FGF5 serves an important role in the transformation process during
the early-mid growth period of hair follicles. FGF5 overexpression
inhibits the growth of hair length. In the hair follicle cycle
period, TGFβRI is highly expressed in the hair follicle root
sheath, which aids in starting and maintaining the hair follicle
catagen (29,30). EDA was predicted as a target gene
of let-7b (6).
Previous genetic studies demonstrated that the
EDA/EDA receptor signaling pathway serves an important role in the
development of skin appendages (31). Regulation of the EDA signaling
pathway is complex, and previous studies have reported that sonic
hedgehog, Wnt/Dickkopf, bone morphogenetic protein and lymphotoxin
β are located downstream of EDA signaling (32,33).
Some of these downstream genes were revealed to inhibit the
development of skin appendages, whereas other genes only inhibited
the development of specific organs, and yet others are only
expressed at a given period; these data indicated that EDA had a
wide range of time and organ specificity during skin development
(34). For example, EDA signaling
may regulate the hair follicle transition from the growth phase to
the catagen phase; in addition, EDA signaling serves a role in hair
follicle morphogenesis and regulates the apoptosis of hair follicle
keratinocytes in the catagen phase. EDA knockout or the
pharmacological inhibition of EDA signalling within wild type mice
may accelerate the growth of the hair follicle catagen (35). Alpaca ear hair is thick and
straight, and has a long growth cycle, whereas back hair is
elongated, bent and has a short growth cycle (14). To study the role of EDA in the
growth of alpaca hair, differential expression of EDA in alpaca ear
and back skin was examined, and the results demonstrated that EDA
expression in alpaca ear skin was significantly higher compared
with expression in back skin. These results suggested that the
growth cycle of the ear and back hair of alpaca may be associated
with the differential expression of EDA in the ear and back skin of
alpaca.
Results from the present study verified that EDA
expression was higher in the ear compared with expression in the
back skin, however the expression of let-7b was reported to be
significantly higher in the back compared with expression in the
ear skin and EDA was predicted to be a target gene of let-7b
(6). Let-7b probably regulates EDA
expression by binding with the EDA 3′-UTR. The present study
successfully amplified the EDA 3′-UTR and used this sequence to
construct the EDA dual-luciferase reporter vector, the results of
which confirmed that EDA was the target gene of let-7b. Following
transfection of the let-7b eukaryotic overexpression vector into
alpaca skin fibroblasts, the mRNA and protein expression levels of
EDA decreased. Immunocytochemical analysis of EDA expression in the
let-7b-transfected and untransfected groups demonstrated that the
EDA protein was abundantly expressed in the fibroblast cytoplasm
and that expression in the transfected cells was weaker compared
with expression in the untransfected cells. These results
demonstrated that let-7b negatively regulated EDA at the mRNA and
protein levels.
miRNAs serve important roles in the incidence of
disease, as well as cell differentiation, cell growth, apoptosis
and immune response (36–38). EDA is a target gene of let-7b, and
EDA mutations may lead to HED (8),
which indicated that let-7b may be associated with HED; however,
further research is required. In conclusion, EDA was demonstrated
to be differentially expressed in alpaca ear and back skin, which
suggested that EDA may serve role in the growth cycle of alpaca
hair. In addition, EDA is the target gene of let-7b, and let-7b
negatively regulates EDA mRNA and protein.
Acknowledgements
The present study was supported by The National
Natural Science Foundation of China (grant nos. 31172283 and
31302049).
References
|
1
|
Wang N, Rong EG and Yan XH: Research
progress of hair follicle development and hair production. J
Northeast Agric Univ. 43:6–11. 2012.
|
|
2
|
Zhu Z, He J, Dong C, Jiang J, Bai R, Yu X,
Lv L, Fan R, He X, Geng J, et al: MicroRNA-25 functions in
regulation of pigmentation by targeting the transcription factor
MITF in alpaca (Lama pacos) skin melanocytes. Domest Anim
Endocrinol. 38:200–209. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dong Y, Cao J, Wang H, Zhang J, Zhu Z, Bai
R, Hao H, He X, Fan R and Dong C: Nitric oxide enhances the
sensitivity of alpaca melanocytes to respond to
alpha-melanocyte-stimulating hormone by up-regulation
melanocortin-1 receptor. Biochem Biophys Res Commun. 396:849–853.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geng JJ, Mu XL, Suan LT, Jiang JB, Zhang
J, Li HQ, Zhang Y and Dong CS: Expression and immunolocalization of
endothelin receptor B in Alpaca skin of different colors. Acta
Veterinaria et Zootechnica Sinica. 41:1478–1484. 2010.
|
|
5
|
Sand M, Gambichler T, Sand D, Skrygan M,
Altmeyer P and Bechara FG: MicroRNAs and the skin: Tiny players in
the body's largest organ. J Dermatol Sci. 53:169–175. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He XY, Hao HQ, Liu DD, Fan RW, Cao J, Zhu
ZW, Dong YJ and Xing HQ: Difference of microRNA expression in the
ear and back skin of Young Alpaca (Lama pacos). Chin J Biochem Mol
Biol. 26:1016–1022. 2010.
|
|
7
|
Kere J, Srivastava AK, Montonen O, Zonana
J, Thomas N, Ferguson B, Munoz F, Morgan D, Clarke A, Baybayan P,
et al: X-linked anhidrotic (hypohidrotic) ectodermal dysplasia is
caused by mutation in a novel transmembrane protein. Nat Genet.
13:409–416. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Visinoni AF, Lisboa-Costa T, Pagnan NA and
Chautard-Freire-Maia EA: Ectodermal dysplasias: Clinical and
molecular review. Am J Med Genet A. 149A:1–2002. 2009. View Article : Google Scholar
|
|
9
|
Pinheiro M and Freire-Maia N: Ectodermal
dysplasias: A clinical classification and a causal review. Am J Med
Genet. 53:153–162. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Clarke A, Phillips DI, Brown R and Harper
PS: Clinical aspects of X-linked hypohidrotic ectodermal dysplasia.
Arch Dis Child. 62:989–996. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mustonen T, Pispa J, Mikkola ML, Pummila
M, Kangas AT, Pakkasjärvi L, Jaatinen R and Thesleff I: Stimulation
of ectodermal organ development by ectodyplasin-A1. Dev Biol.
259:123–136. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang M, Brancaccio A, Weiner L, Missero C
and Brissette J: Ectodysplasin regulates pattern formation in the
mammalian hair coat. Genesis. 37:30–37. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang W, Xue P, He YX and Chen YL:
Molecular cloning, sequence analysis and expression of goat Eda
gene. J Northwest A F Univ (Nat. Sci. Ed). 40:7–12. 2012.
|
|
14
|
Liu DD, Zhang ZY, He XY, Dong YJ, Zhu ZW,
Bai R, Zhang J, Hao HQ and Xing HQ: FGF5 expression and
immunolocalization in back and ear skin of young alpaca (Lama
pacos). Chin J Bioch Mol Biol. 5:473–479. 2011.
|
|
15
|
Tian X, Jiang J, Fan R, Wang H, Meng X, He
X, He J, Li H, Geng J, Yu X, et al: Identification and
characterization of microRNAs in white and brown alpaca skin. BMC
Genomics. 13:5552012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cekanova M, Uddin MJ, Bartges JW, Callens
A, Legendre AM, Rathore K, Wright L, Carter A and Marnett LJ:
Molecular imaging of cyclooxygenase-2 in canine transitional cell
carcinoma as in vitro and in vivo. Cancer Prev Res (Phila).
6:466–476. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee RC, Feinbaum RL and Ambros V: The C.
Elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lagos-Quintata M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dostie J, Mourelatos Z, Yang M, Sharma A
and Dreyfuss G: Numerous microRNPs in neuronal cells containing
novel microRNAs. RNA. 9:180–186. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lim LP, Glasner ME, Yekta S, Burge CB and
Bartel DP: Vertebrate microRNA genes. Science. 299:15402003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lim LP, Lau NC, Weinsstein EG, Abdelhakim
A, Yekta S, Rhoades MW, Burge CB and Bartel DP: The microRNAs of
Caenorhabditis elegans. Genes Dev. 17:991–1008. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Grad Y, Aach J, Hayes GD, Reinhart BJ,
Church GM, Ruvkun G and Kim J: Computational and experimental
identification of C. Elegans microRNAs. Mol Cell. 11:1253–1263.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Reinhart BJ, Weinsstein EG, Rhoades MW,
Bartel B and Bartel DP: MicroRNAs in plants. Genes Dev.
16:1616–1626. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lau NC, Lim LP, Weinsstein EG and Bartel
DP: An abundant class of tiny RNAs with probable regulatory roles
in Caenorhabditis elegans. Science. 294:858–862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim J, Krichevsky A, Grad Y, Hayes GD,
Kosik KS, Church G and Ruvkun G: Identification of many microRNAs
that copurify with polyribosomes in mammalian neurons. Pro Natl
Acad Sci USA. 101:360–365. 2004. View Article : Google Scholar
|
|
28
|
Liu DD, He XY and Hao HQ: The regulatory
role of microRNAs in the development of animal skin and hair. Chin
J Biochem Mol Biol. 26:802–808. 2010.
|
|
29
|
Wang T, Zhang Y, Wang HD, Shen Y, Liu N,
Cao J, Yu XJ, Dong CS and He XY: Alpaca fiber growth is mediated by
microRNA let-7b via down-regulation of target gene FGF5. Genet Mol
Res. 14:13754–13763. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yan S, Yu Z, Ning L, Hai-Dong W, Jian-Shan
X, Shu-Yuan G, Jia-Qi C, Xiu-Ju Y, Ting W, Chang-Sheng D and
Xiao-Yan H: Let-7b promotes alpaca hair growth via transcriptional
repression of TGFβR I. Gene. 577:32–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Weih F and Caamaño J: Regulation of
secondary lymphoid organ development by the nuclear factor-kappaB
signal transduction pathway. Immunol Rev. 195:91–105. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Headon DJ, Emmal SA, Ferguson BM, Tucker
AS, Justice MJ, Sharpe PT, Zonana J and Overbeek PA: Gene defect in
ectodermal dysplasia implicates a death domain adapter in
development. Nature. 414:913–916. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cui CY and Schlessinger D: EDA signaling
and skin appendage development. Cell Cycle. 5:2477–2483. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Botchkarev VA and Fessing MY: Edar
signaling in the control of hair follicle development. J Investig
Dermatol Symp Proc. 10:pp. 247–251. 2005; View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Naguibneva I, Ameyar-Zazoua M, Polesskaya
A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S and Harel-Bellan
A: The microRNA miR-181 targets the homeobox protein Hox-A11 during
mammalian myoblast differentiation. Nat Cell Biol. 8:278–284. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jakymiw A, Lian SL, Eystathioy T, Li S,
Satoh M, Hamel JC, Fritzler MJ and Chan KL: Disruption of GW bodies
impairs mammalian RNA interference. Nat Cell Biol. 7:1267–1274.
2005. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Naguibneva I, Ameyar-Zazoua M, Nonne N,
Polesskaya A, Ait-Si-Ali S, Groisman R, Souidi M, Pritchard LL and
Harel-Bellan A: An LNA-based loss-of-function assay for micro-RNAs.
Biomed Pharmacother. 60:633–638. 2006. View Article : Google Scholar : PubMed/NCBI
|