Introduction
Bone marrow-derived mesenchymal stem cells (BMSCs)
have been reported to have the potential ability to differentiate
into bone, cartilage, muscle and adipose tissue (1), which may be used for the clinical
treatment of associated diseases (2,3).
Osteogenic differentiation of BMSCs is an important step (4). Previous studies have indicated that
certain signaling pathways have vital roles in regulating
osteoblastic differentiation of BMSCs, including transforming
growth factor-β (TGF-β)/bone morphogenetic protein (BMP), Notch and
Hedgehog (5–7). In addition, altering phosphorylation
of the p38 mitogen-activated protein kinase (MAPK) signaling
pathway has been verified to be involved in osteogenic
differentiation of BMSCs (8).
However, the molecular mechanisms underlying osteogenic
differentiation of BMSCs remain to be fully elucidated.
MicroRNAs (miRNAs/miRs) are a type of small
non-coding RNA with 18 to 22 nucleotides, and are involved in the
post-transcriptional regulation of gene expression by binding to
the translation section, lead to either mRNA degradation or
translational inhibition (9,10).
In addition, previous studies have demonstrated that microRNAs
regulate the process of osteogenic differentiation of BMSCs. For
example, miR-26a conversely regulates osteogenic differentiation of
BMSCs depending on distinct activation and roles of the Wnt and BMP
signaling pathways (11), and
miR-30e regulates osteogenic differentiation of BMSCs by directly
targeting insulin-like growth factor II (12). miR-124 negatively regulates
osteogenic differentiation of BMSCs (13). Therefore, miRNAs have vital roles
in the process of osteogenic differentiation in BMSCs.
Previous studies have demonstrated that miR-23a is
upregulated in dexamethasone-induced human (h)BMSCs during
osteogenic differentiation (14,15).
In addition, in our previous study, it was revealed miR-23a-5p was
upregulated in dexamethasone-induced hBMSCs during osteogenic
differentiation, and target prediction analysis also suggested that
MAPK13 may be a target gene of miR-23a-5p, which is involved in the
p38 MAPK signaling pathway. However, whether MAPK13 may be
regulated by miR-23a-5p, and its involvement in osteoblast
differentiation of hBMSCs remains unclear. Therefore, the present
study aimed to investigate the role of miR-23a-5p and MAPK13 in
osteogenic differentiation of hBMSCs, and to identify whether
MAPK13 is a direct target of miR-23a-5p. The results indicated that
miR-23a-5p serves a vital role in osteogenic differentiation of
hBMSCs, which may act by targeting MAPK13.
Materials and methods
Tissue acquisition and cell
culture
The present study was approved by the Ethics
Committee of Tianjin First Central Hospital (Tianjin, China), and
written consent was obtained from the donor. Bone marrow tissue was
obtained from the iliac crest of three disease-free control donors
(male, n=2; female, n=1; age, 29.4±5.9 years) who were recruited
from Tianjin First Central Hospital (Tianjin, China), and hBMSCs
were isolated from the human bone marrow tissue according to a
previously described protocol (16). hBMSCs were resuspended
(2×106 cells) in growth medium [high glucose Dulbecco's
modified Eagle's medium (DMEM), 0.29 mg/ml Glutamax, 10%
inactivated fetal bovine serum (FBS), 100 mg/ml streptomycin and
100 U/ml penicillin] (all from Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), cultured in a 75
cm2 flask and maintained at 3°C in 5% CO2.
Passage 3 cells were used for following experiments.
Osteogenic differentiation and
staining in vitro
Following RNA and protein isolation and staining,
cells at 70–80% confluence had regular growth medium replaced with
osteogenic differentiation medium (high glucose DMEM supplemented
with 10% FBS, 10 mM β-glycerophosphate, 0.2 mM l-ascorbic acid and
1 nM dexamethasone; all Guangzhou Saiguo Biotech Co., Ltd.,
Guangzhou, China). The medium was replaced every 3 days for 15
days. The osteoblast phenotype was evaluated by determining
alkaline phosphatase (ALP) activity. An alkaline phosphatase
detection kit (Jiancheng Bioengineering, Nanjing, China) and an ALP
staining kit (Blood Institute, Chinese Academy of Medical Sciences,
Shanghai, China) were used for ALP activity and ALP staining,
according to the manufacturer's protocol. 2% Alizarin Red S (ARS;
2%; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to
detect matrix mineralization using an inverted microscope (Olympus
IX73; Olympus Corporation, Tokyo, Japan). Individual experiments
were repeated at least three times.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction
Total RNA was extracted using the RNeasy Plus Mini
kit (Qiagen China Co., Ltd., Shanghai, China). Total RNA
concentration was determined using a NanoVue Plus spectrophotometer
(GE Healthcare, Chicago, IL, USA). cDNA was synthesized with a
PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian,
China). miR-23a-5p was detected with TaqMan miRNA assays (Applied
Biosystems; Thermo Fisher Scientific, Inc.), and U6 served as an
internal control. The human osteogenic gene ALP, osteopontin (OPN),
runt-related transcription factor 2 (RUNX2) and MAPK13 mRNA
expression levels were detected by qPCR with a SYBR Premix Ex Taq
II kit (Takara Biotechnology Co., Ltd.) and the Applied Biosystems
ABI Prism 7500 HT sequence detection system, with β-actin as an
internal control. Primer sequences were as follows: Forward,
5′-ACGTGGCTAAGAATGTCATC-3′ and reverse, 5′-CTGGTAGGCGATGTCCTTA-3′
for ALP; forward, 5′-ACTCGAACGACTCTGATGA-TGT-3′ and reverse,
5′-GTCAGGTCTGCGAAACTTCTTA-3′ for OPN; forward,
5′-TCTTCACAAATCCTCCCC-3′ and reverse, 5′-TGGATTAAAAGGACTTGG-3′ for
RUNX2; forward, 5′-AGGTCTCTGGGGGTTGAGT-TGGG-3′ and reverse,
5′-AGGGGCAGCAACGTCTCATTGC-3′ for MAPK13; forward,
5′-CCACCATGTACCCAGGCATT-3′ and reverse, 5′-CAGCTCAGTAACAGTCCGCC-3′
for β-actin. The thermocycling conditions were as follows: 95°C for
2 min, followed by 40 cycles of denaturation at 95°C for 20 sec,
annealing at 60°C for 30 sec and an extension at 72°C for 45 sec.
The expression of mRNA or microRNA was evaluated based on the
quantitation cycle (Cq) as n = 2−ΔΔCq (17), where ΔCq = Cq related mRNA-Cq
β-actin and ΔΔCq = ΔCq experimental-ΔCq control. Individual
experiments were repeated at least three times.
Transfection of relative miRNAs
Synthetic miR-23a-5p mimics, an miR-23a-5p inhibitor
and the relative controls were purchased from BioVectra
(Charlottetown, PE, Canada). Synthetic microRNAs were transfected
into hBMSCs (at a final concentration of 200 nM) with Lipofectamine
2000™ (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. This process was performed with the
non-serum medium Opti-Minimum Essential medium (Opti-MEM; Gibco;
Thermo Fisher Scientific, Inc.) at 37°C in 5% CO2. After
6 h, Opti-MEM was replaced with induction medium of differentiation
(high glucose DMEM supplemented with 10% FBS, 10 mM
β-glycerophosphate, 0.2 mM l-ascorbic acid and 1 nM dexamethasone)
(all from Guangzhou Saiguo Biotech Co., Ltd.). Cells were collected
72 h after transfection.
Western blot analysis
Cells were lysed with radioimmunoprecipitation lysis
buffer (Beyotime Institute of Biotechnology, Nanjing, China) for 15
min on ice, and then centrifuged at 14,000 × g and 4°C for 10 min.
Protein concentrations were determined using the Bicinchoninic Acid
assay (Wuhan Booute Biotechnology Co., Ltd., Wuhan, China). A total
of 50 µg protein/lane was loaded and separated by 10% SDS-PAGE and
subsequently transferred onto polyvinylidene difluoride membranes.
Membranes were blocked with 5% non-fat milk in TBST buffer (TBS
containing 0.5% Tween-20) at room temperature for 1 h, washed with
TBST, then incubated with the following primary antibodies:
Anti-ALP (1:1,000; cat no. sc-365765; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), anti-OPN (1:1,000; cat no. ab128284; Abcam,
Cambridge, MA, USA), anti-RUNX2 (1:500; cat no. 12556S; Cell
Signaling Technology, Inc., Danvers, MA, USA), anti-MAPK13 (1:200;
cat no. ab188324; Abcam) and anti-β-actin (1:1,000; cat no. 4970S;
Cell Signaling Technology, Inc.) at 4°C overnight. Membranes were
then incubated with anti-rabbit or anti-mouse horseradish
peroxidase-conjugated secondary antibodies (1:5,000; cat no.
TA140003; OriGene Technologies, Inc., Beijing, China) at room
temperature for 1 h following washing with triple TBST buffer (TBS
containing 0.5% Tween-20). Antibody and antigen complexes were
detected using an Enhanced Chemiluminescence reagent (EMD
Millipore, Billerica, MA, USA). For relative quantification, the
integrated optical density (IOD) was estimated using ImageJ
(version 2.1.4.7; National Institutes of Health, Bethesda, MD,
USA). The relative protein expression level was calculated as
IODExperimental/IODControl.
miRNA target prediction
Prediction of hsa-miR-23a-5p target genes was
performed using the TargetScan (version 7.1; www.targetscan.org/), PicTar (2007 release; www.pictar.org) and miRanda (2010 release; www.microrna.org/) databases.
Dual-luciferase reporter assay
To detect MAPK13 3′-untranslated region (UTR)
luciferase reporter activity, stable miR-23a-5p-overexpressing
HEK293T and hBMSCs were cultured and transfected with 100 ng
luciferase reporter plasmid and 5 ng pRL-TK vector expressing
Renilla luciferase (Promega Corporation, Madison, WI, USA)
using Lipofectamine 2000. To detect T-cell factor/lymphoid enhancer
factor transcriptional activity, miR-23a-5p-overexpressing hBMSCs
were transfected with 5 ng pRL-TK and 200 ng TOPflash or FOPflash
(EMD Millipore) using Lipofectamine 2000. After 48 h, cells were
harvested and lysed, and luciferase activity was measured using the
Dual-Luciferase Reporter Assay system (Promega Corporation).
Individual experiments were repeated at least three times.
Construction of the MAPK13 short
hairpin (sh)RNA expression vector
Lentiviral plasmids were used to construct the
MAPK13 shRNA expression vector. Three pairs of shRNA coding
sequences were used to direct against different sites of MAPK13
mRNA, and then ligated into the lentiviral vector plasmid pLB to
construct pLB-MAPK13/shRNA1, pLB-MAPK13/shRNA2, pLB-MAPK13/shRNA3
and pLB-null/shRNA-negative control (NC) constructs. Recombinant
lentiviruses were obtained from Shanghai Genepharma Co., Ltd.
(Shanghai, China).
Statistical analysis
Data are presented as the mean ± standard deviation.
Independent-samples t-test or one-way analysis of variance followed
by the Tukey post hoc test were used to identify differences
between groups. SPSS v19.0 software (IBM Corp., Armonk, NY, USA)
was used for data analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression level of miR-23a-5p and
MAPK13 during osteogenic differentiation of hBMSCs
After hBMSCs were cultured with osteogenic
differentiation medium, the expression of osteoblastic marker genes
including RUNX2, ALP and OPN was detected by RT-qPCR. The results
indicated that RUNX2, ALP and OPN were significantly increased
after induction of differentiation (Fig. 1A). The results of the ALP and
Alizarin Red S staining experiments revealed that the osteogenic
differentiation of hBMSCs, ALP activity and calcium deposition were
all markedly increased when compared with the NC group (indicated
by white arrows; Fig. 1B).
Following this, the expression levels of miR-23a-5p and MAPK13 were
detected by RT-qPCR and western blotting at different time points.
The results demonstrated that the expression of miR-23a-5p was
significantly decreased during osteogenic differentiation of hBMSCs
(Fig. 1C), the mRNA (Fig. 1D) and protein (Fig. 1E) expression levels of MAPK13 were
significantly increased during osteogenic differentiation of
hBMSCs.
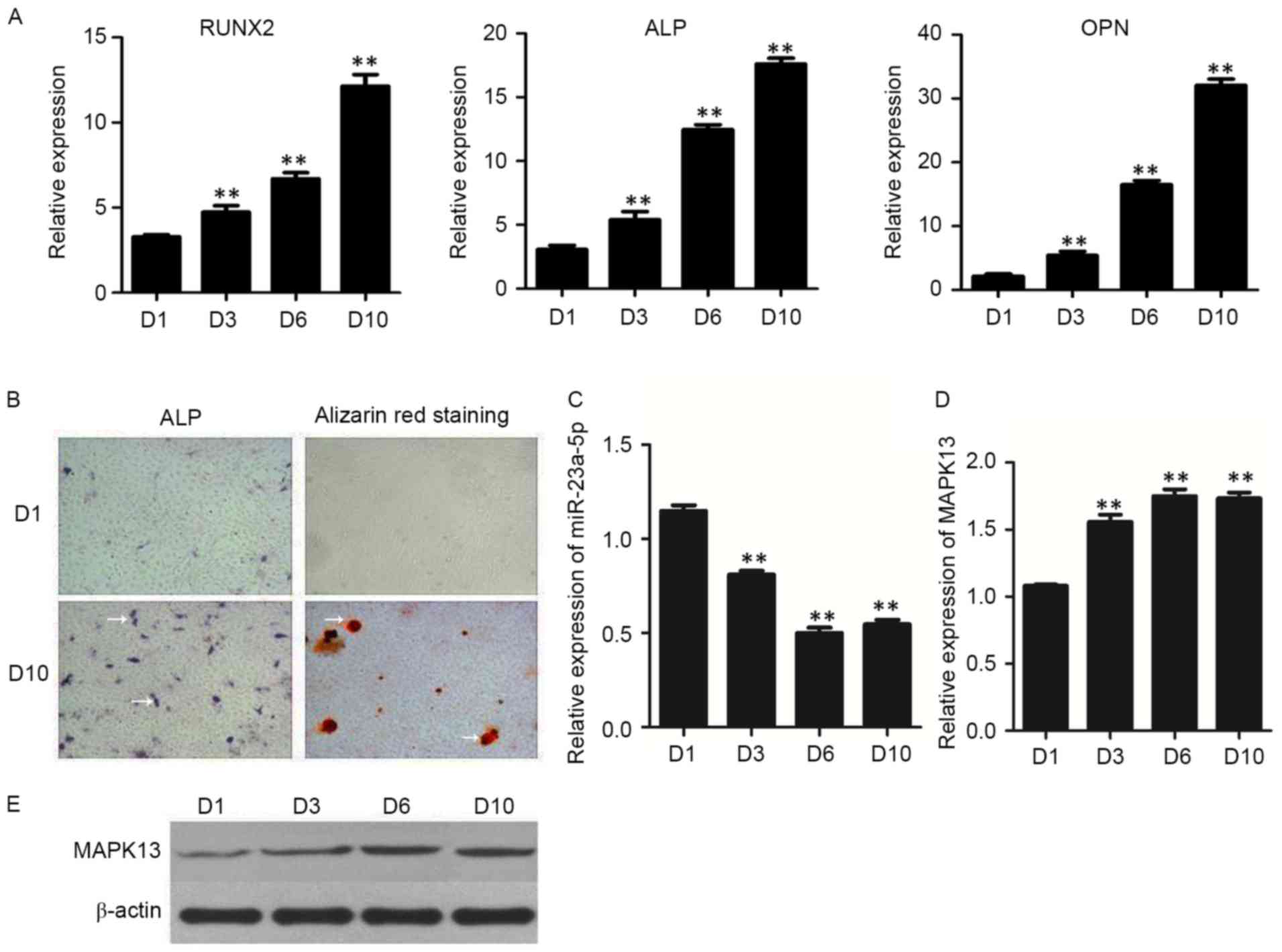 | Figure 1.Expression of osteoblastic marker
genes in hBMSCs. (A) mRNA expression levels of RUNX2, ALP and OPN,
as detected by RT-qPCR. (B) ALP staining and Alizarin Red S
staining were performed at days 1 and 10, respectively
(magnification, ×100). White arrows indicate areas of ALP activity
and calcium deposition. Expression levels of (C) miR-23a-5p and (D)
MAPK13, as detected by RT-qPCR. (D) Quantification and (E)
representative western blot images of MAPK13 protein expression
levels during osteogenic differentiation of hBMSCs, as assessed by
western blotting. Data are presented as the mean ± standard
deviation. **P<0.01 vs. D1. RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; D, day;
MAPK13, mitogen-activated protein kinase-13; RUNX2, runt-related
transcription factor 2; ALP, alkaline phosphatase; OPN,
osteopontin; hBMSCs, human bone marrow-derived mesenchymal stem
cells; miR, microRNA. |
miR-23a-5p inhibits osteogenic
differentiation of hBMSCs
To further investigate whether miR-23a-5p is
involved in osteogenic differentiation, synthetic mimics and
inhibitors of miR-23a-5p were transfected into hBMSCs. The
expression of miR-23a-5p and osteoblast markers were detected by
RT-qPCR and western blotting. The results demonstrated that the
expression of intracellular miR-23a-5p was significantly increased
by miR-23a-5p mimics, and substantially decreased by miR-23a-5p
inhibitors (Fig. 2A). In addition,
upregulation of miR-23a-5p inhibited osteogenic differentiation,
and the mRNA (Fig. 2B) and protein
(Fig. 2C) expression levels of the
osteoblastic marker genes RUNX2, ALP and OPN were decreased.
Furthermore, downregulation expression of miR-23a-5p promoted
osteogenic differentiation; the protein (Fig. 2C) and mRNA (Fig. 2D) expression levels of RUNX2, ALP
and OPN were increased. Osteogenic differentiation levels were
confirmed by ALP and Alizarin Red S staining experiments. When
compared with the NC, ALP activity and calcium deposition were not
markedly increased in miR-23a-5p mimics, however, a marked increase
was observed in the miR-23a-5p inhibitors (Fig. 2E).
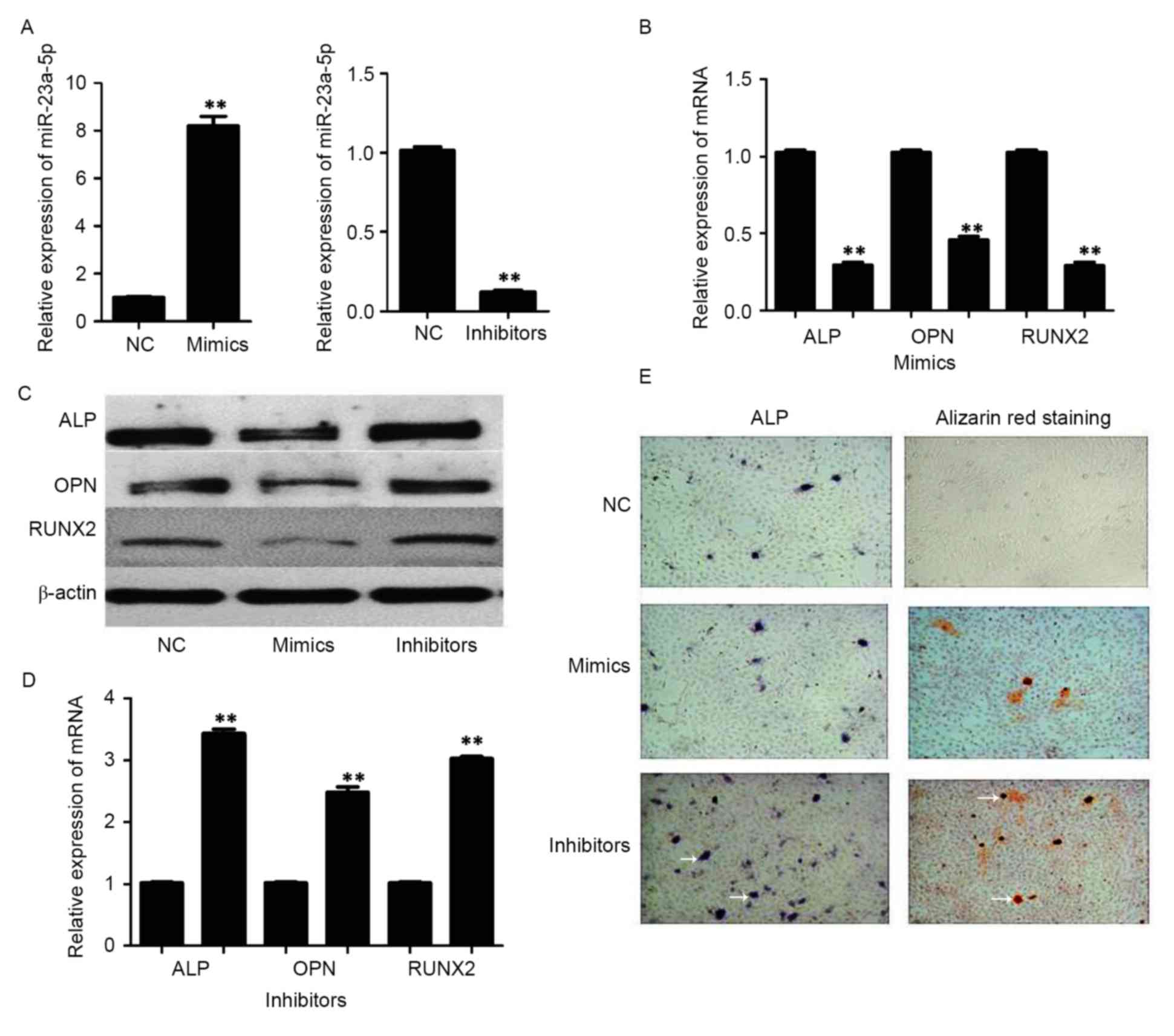 | Figure 2.Expression of osteoblastic marker
genes in bone marrow-derived mesenchymal stem cells following
transfection with mimics and inhibitors of miR-23a-5p. (A)
miR-23a-5p expression levels 72 h after transfection. (B) mRNA
expression levels of RUNX2, ALP and OPN following transfection with
miR-23a-5p mimics. (C) Protein expression levels of RUNX2, ALP and
OPN after transfection with miR-23a-5p mimics and inhibitors, as
assessed by western blotting. (D) mRNA expression levels of RUNX2,
ALP and OPN following transfection with miR-23a-5p inhibitors. (E)
Representative images of ALP and Alizarin Red S staining
(magnification, ×100). White arrows indicate areas of ALP activity
and calcium deposition. Data are presented as the mean ± standard
deviation. **P<0.01 vs. NC. RUNX2, runt-related transcription
factor 2; ALP, alkaline phosphatase; OPN, osteopontin; miR,
microRNA; NC, negative control. |
miR-23a-5p directly targets
MAPK13
According to TargetScan, PicTar and miRanda
prediction analyses, MAPK13 has a putative miR-23a-5p-binding site
mapped to the 3′-UTR, and was identified as one of the high-scoring
candidate genes for miR-152 targeting (Fig. 3A). To further investigate whether
miR-23a-5p directly targets MAPK13, luciferase reporters that
contained either a wild-type MAPK13 3′-UTR or a mutant MAPK13
3′-UTR containing mutant sequences of the miR-23a-5p binding site
were used (Fig. 3A). The results
showed that miR-23a-5p inhibited luciferase activity through the
3′-UTR of MAPK13 compared with the control group (Fig. 3B). In addition, the relative
luciferase activity of miR-23a-5p containing the mutated 3′-UTR
site or the control group lacking an MAPK13 3′-UTR sequence had no
statistically significant difference (Fig. 3B).
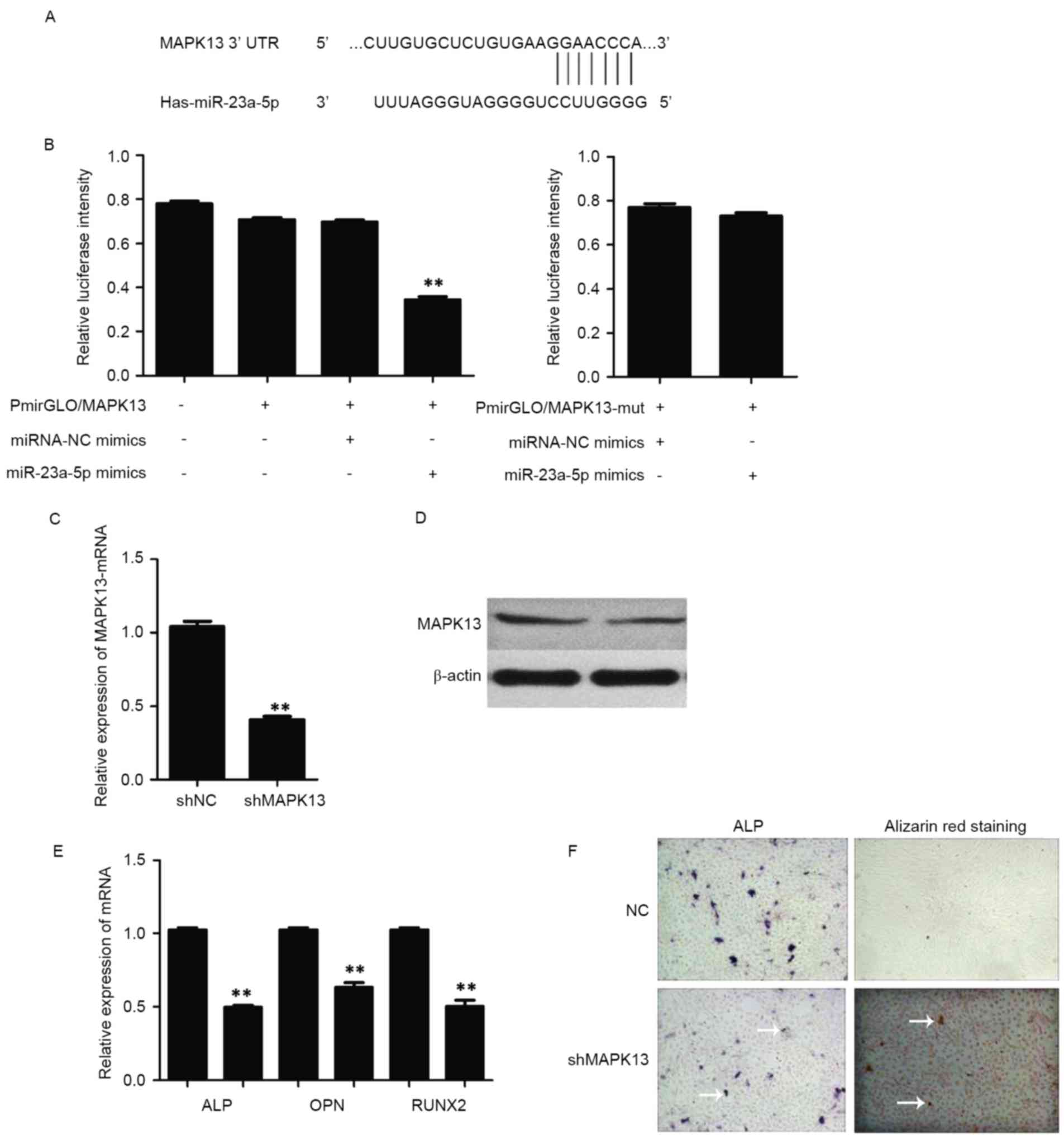 | Figure 3.Assessment of miR-23a-5p activity. (A)
Putative miR-23a-5p binding sequence in the MAPK13 3′-UTR. (B)
Wild-type or mut reporter plasmids and miRNA-NC or miR-23a-5p
mimics were co-transfected into hBMSCs. **P<0.01 vs.
pmirGLO/MAPK13+miRNA-NC mimics. (C) mRNA and (D) protein expression
levels of MAPK13 following transfection with shRNA-NC or shRNA
against MAPK13, as assessed by RT-qPCR and western blotting,
respectively. (E) mRNA expression levels of RUNX2, ALP and OPN were
detected by RT-qPCR, following transfection of hBMSCs with shRNA
against MAPK13. **P<0.01 vs. shNC. (F) Representative images of
ALP and Alizarin Red S staining (magnification, ×100). White arrows
indicate areas of ALP activity and calcium deposition. Data are
presented as the mean ± standard deviation. RUNX2, runt-related
transcription factor 2; ALP, alkaline phosphatase; OPN,
osteopontin; miR, microRNA; NC, negative control; mut, mutant;
MAPK13, mitogen activated protein kinase 13; UTR, untranslated
region; shRNA, short hairpin RNA; hBMSCs, human bone marrow-derived
mesenchymal stem cells; RT-qPCR, reverse transcription-quantitative
polymerase chain reaction. |
Knockdown of MAPK13 inhibits
osteogenic differentiation of hBMSCs
To further investigate the role of MAPK13 on
osteogenic differentiation, the expression of MAPK13 was inhibited
by transfecting hBMSCs with shRNA against MAPK13. The results
demonstrated that the mRNA (Fig.
3C) and protein (Fig. 3D)
expression levels of MAPK13 were significantly decreased by the
shRNA. The results also indicated that knockdown of MAPK13
inhibited osteogenic differentiation of hBMSCs, as was confirmed by
the reduced mRNA expression levels of RUNX2, ALP, OPN (Fig. 3E) and ALP and Alizarin Red S
staining experiments. ALP activity and calcium deposition was not
markedly altered in the knockdown MAPK13 group when compared with
the NC (Fig. 3F).
MAPK13 knockdown inhibits the effect
of miR-23a-5p in osteogenic differentiation of hBMSCs
To further investigate whether MAPK13 knockdown
inhibits the effect of miR-23a-5p in osteogenic differentiation of
hBMSCs, after MAPK13 knockdown, an miR-23a-5p inhibitor was
transfected into hBMSCs. Osteoblast markers were detected by
RT-qPCR and western blotting. The results demonstrated that
miR-23a-5p inhibitors could promote osteogenic differentiation in
the NC-MAPK13 knockdown group, which was confirmed by expression of
RUNX2, ALP, OPN (Fig. 4A and B).
However, osteogenic differentiation levels of hBMSCs in the
presence of miR-23a-5p inhibitors were blocked following MAPK13
knockdown (Fig. 4A and B). ALP and
Alizarin Red S staining experiments confirmed these results. ALP
activity and calcium deposition were not altered in miR-23a-5p
inhibitors with MAPK13 knockdown when compared with NCs, however,
ALP activity and calcium deposition were markedly increased in
miR-23a-5p inhibitors without MAPK13 knockdown (Fig. 4C).
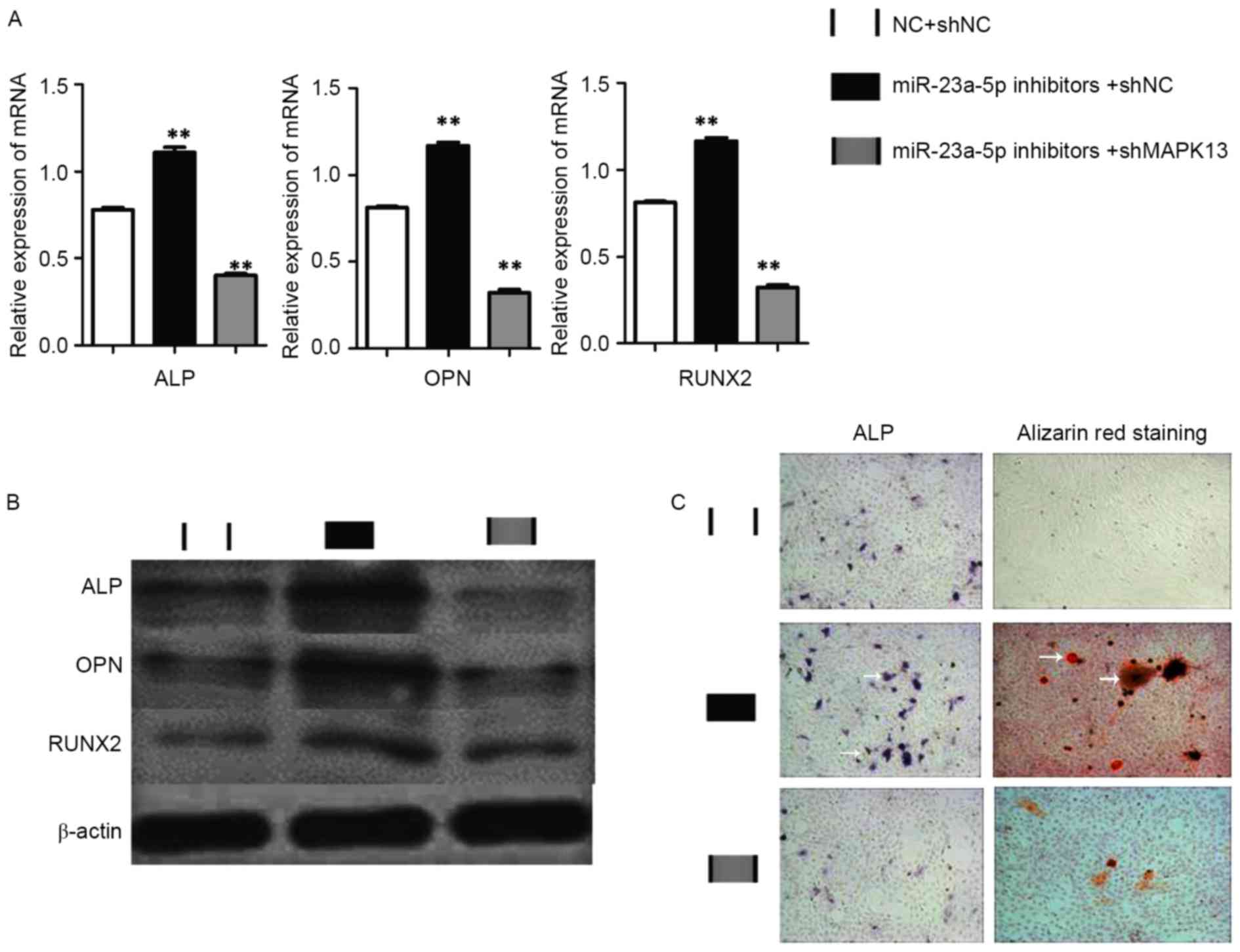 | Figure 4.Investigating MAPK13 knockdown. hBMSCs
were transfected with NC+shNC, miR-23a-5p inhibitors+shNC or
miR-23a-5p inhibitors+shMAPK13. (A) mRNA and (B) protein expression
levels of RUNX2, ALP and OPN, as detected by reverse
transcription-quantitative polymerase chain reaction and western
blotting, respectively. (C) Representative images of ALP and
Alizarin Red S staining (magnification, ×100). White arrows
indicate areas of ALP activity and calcium deposition. Data are
presented as the mean ± standard deviation. **P<0.01 vs.
NC+shNC. RUNX2, runt-related transcription factor 2; ALP, alkaline
phosphatase; OPN, osteopontin; miR, microRNA; NC, negative control;
sh, short hairpin; MAPK13, mitogen-activated protein kinase-13. |
Discussion
The present study identified that miR-23a-5p has a
potentially regulative role in the osteogenic differentiation of
hBMSCs. The results demonstrated that miR-23a-5p was significantly
decreased during the process of osteogenic differentiation.
Downregulation of miR-23a-5p in hBMSCs enhanced osteogenic
differentiation; however, upregulation of miR-23a-5p in hBMSCs
inhibited their osteogenic potential, which was associated with
downstream regulation of MAPK13.
miRNAs are a type of small non-coding RNA, as serve
as post-transcriptional regulators of target gene expression.
Previous studies have demonstrated that microRNAs serve vital roles
in regulation of stem cell differentiation into osteoblasts. For
example, miR-138 is reduced during the process of osteoblast
differentiation of hBMSCs, which targets focal adhesion kinase
(FAK) and subsequently suppresses the FAK-extracellular
signal-regulated kinase 1/2 signaling pathway (18). miR-20a enhances the osteogenesis
level of hBMSCs, which targets peroxisome proliferator-activated
receptor-γ, BMP and activing membrane-bound inhibitor homolog and
cysteine-rich motor neuron 1 protein, and then suppresses BMP
signaling (19). For miR-23a-5p, a
study revealed that miR-23a is increased in the bone tissue of
osteoporotic patients compared with healthy controls (20). In addition, another study
demonstrated that overexpression of miR-23a inhibits osteogenic
differentiation of hBMSCs at the cellular, mRNA and protein levels
(14), and acts by targeting
low-density lipoprotein receptor-related protein 5 (15). These findings support the
hypothesis that miR-23a-5p may inhibit osteogenesis.
To further investigate the downstream molecular
mechanism of miR-23a-5p during osteogenic differentiation of
hBMSCs, the prediction analysis method was used, and the results
demonstrated that MAPK13 had a putative miR-23a-5p-binding site
mapped to the 3′-UTR. Furthermore, a dual luciferase reporter assay
identified MAPK13 as a direct target of miR-23a-5p. In addition,
knockdown of MAPK13 blocked the effect of miR-23a-5p in osteogenic
differentiation of hBMSCs. Therefore, it was hypothesized that
MAPK13 is regulated by miR-23a-5p during osteogenic
differentiation.
MAPK13 can encode a member of the MAPK family. MAPKs
serve as an integration point of multiple biochemical signals, and
are involved in a wide variety of cellular processes such as
proliferation, differentiation, transcription regulation and
development (21,22). The encoded protein is a p38 MAPK,
and has the ability of activating the p38 MAPK signaling pathway
(23). Notably, previous studies
have reported that altering phosphorylation of the p38 MAPK
signaling pathway is involved in osteogenic differentiation of
hBMSCs (8,24). The present study demonstrated that
MAPK13 is increased in the process of osteogenic differentiation,
and knockdown of hBMSCs inhibits osteogenic differentiation of
hBMSCs. Therefore, MAPK13 may be promote osteogenic differentiation
of hBMSCs.
In conclusion, the present study demonstrated that
downregulation of endogenous MAPK13 inhibits the osteogenesis level
of hBMSCs, which was similar to the effect of upregulation of
miR-23a-5p. In addition, the effects of miR-23a-5p on osteogenic
differentiation of hBMSCs could be blocked by MAPK13 shRNA. These
results provide evidence that miR-23a-5p inhibits osteogenic
differentiation of hBMSCs directly by negatively regulating MAPK13.
Therefore, miR-23a-5p serves an important role in the osteogenic
differentiation of hBMSCs by targeting MAPK13, which may provide a
basis for the treatment of bone injuries, however, further studies
are required.
Acknowledgements
The present study was supported by the Health Bureau
of Science and Technology Fund Project of Tianjin (grant no. 2013
KZ 032).
References
|
1
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao D, Cui D, Wang B, Tian F, Guo L, Yang
L, Liu B and Yu X: Treatment of early stage osteonecrosis of the
femoral head with autologous implantation of bone marrow-derived
and cultured mesenchymal stem cells. Bone. 50:325–330. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hare JM, Fishman JE, Gerstenblith G,
DiFede Velazquez DL, Zambrano JP, Suncion VY, Tracy M, Ghersin E,
Johnston PV, Brinker JA, et al: Comparison of allogeneic vs.
autologous bone marrow-derived mesenchymal stem cells delivered by
transendocardial injection in patients with ischemic
cardiomyopathy: The POSEIDON randomized trial. JAMA. 308:2369–2379.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rodrigues MT, Lee SJ, Gomes ME, Reis RL,
Atala A and Yoo JJ: Amniotic fluid-derived stem cells as a cell
source for bone tissue engineering. Tissue Eng Part A.
18:2518–2527. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bandyopadhyay A, Tsuji K, Cox K, Harfe BD,
Rosen V and Tabin CJ: Genetic analysis of the roles of BMP2, BMP4,
and BMP7 in limb patterning and skeletogenesis. PLoS Genet.
2:e2162006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hilton MJ, Tu X, Wu X, Bai S, Zhao H,
Kobayashi T, Kronenberg HM, Teitelbaum SL, Ross FP, Kopan R and
Long F: Notch signaling maintains bone marrow mesenchymal
progenitors by suppressing osteoblast differentiation. Nat Med.
14:306–314. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Long F, Chung UI, Ohba S, McMahon J,
Kronenberg HM and McMahon AP: Ihh signaling is directly required
for the osteoblast lineage in the endochondral skeleton.
Development. 131:1309–1318. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou Y, Wu Y, Jiang X, Zhang X, Xia L, Lin
K and Xu Y: The effect of quercetin on the osteogenesic
differentiation and angiogenic factor expression of bone
marrow-derived mesenchymal stem cells. PLoS One. 10:e01296052015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Thomas M, Lieberman J and Lal A:
Desperately seeking microRNA targets. Nat Struct Mol Biol.
17:1169–1174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Su X, Liao L, Shuai Y, Jing H, Liu S, Zhou
H, Liu Y and Jin Y: MiR-26a functions oppositely in osteogenic
differentiation of BMSCs and ADSCs depending on distinct activation
and roles of Wnt and BMP signaling pathway. Cell Death Dis.
6:e18512015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding W, Li J, Singh J, Alif R,
Vazquez-Padron RI, Gomes SA, Hare JM and Shehadeh LA: miR-30e
targets IGF2-regulated osteogenesis in bone marrow-derived
mesenchymal stem cells, aortic smooth muscle cells, and
ApoE−/−mice. Cardiovasc Res. 106:131–142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qadir AS, Um S, Lee H, Baek K, Seo BM, Lee
G, Kim GS, Woo KM, Ryoo HM and Baek JH: miR-124 negatively
regulates osteogenic differentiation and in vivo bone formation of
mesenchymal stem cells. J Cell Biochem. 116:730–742. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li T, Li H, Li T, Fan J, Zhao RC and Weng
X: MicroRNA expression profile of dexamethasone-induced human bone
marrow-derived mesenchymal stem cells during osteogenic
differentiation. J Cell Biochem. 115:1683–1691. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li T, Li H, Wang Y, Li T, Fan J, Xiao K,
Zhao RC and Weng X: microRNA-23a inhibits osteogenic
differentiation of human bone marrow-derived mesenchymal stem cells
by targeting LRP5. Int J Biochem Cell Biol. 72:55–62. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Oskowitz AZ, Lu J, Penfornis P, Ylostalo
J, McBride J, Flemington EK, Prockop DJ and Pochampally R: Human
multipotent stromal cells from bone marrow and microRNA: Regulation
of differentiation and leukemia inhibitory factor expression. Proc
Natl Acad Sci USA. 105:pp. 18372–18377. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qu B, Xia X, Wu HH, Tu CQ and Pan XM:
PDGF-regulated miRNA-138 inhibits the osteogenic differentiation of
mesenchymal stem cells. Biochem Biophys Res Commun. 448:241–247.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang JF, Fu WM, He ML, Xie WD, Lv Q, Wan
G, Li G, Wang H, Lu G, Hu X, et al: MiRNA-20a promotes osteogenic
differentiation of human mesenchymal stem cells by co-regulating
BMP signaling. RNA Biol. 8:829–838. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Seeliger C, Karpinski K, Haug AT, Vester
H, Schmitt A, Bauer JS and van Griensven M: Five freely circulating
miRNAs and bone tissue miRNAs are associated with osteoporotic
fractures. J Bone Miner Res. 29:1718–1728. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei C, Ren H, Xu L, Li L, Liu R, Zhang L,
Zhao F, Lu J, Zhang X and Du L: Signals of Ezh2, Src, and Akt
involve in myostatin-Pax7 pathways regulating the myogenic fate
determination during the sheep myoblast proliferation and
differentiation. PLoS One. 10:e01209562015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tan FE and Elowitz MB: Brf1
posttranscriptionally regulates pluripotency and differentiation
responses downstream of Erk MAP kinase. Proc Natl Acad Sci USA.
111:pp. E1740–E1748. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yurtsever Z, Scheaffer SM, Romero AG,
Holtzman MJ and Brett TJ: The crystal structure of phosphorylated
MAPK13 reveals common structural features and differences in p38
MAPK family activation. Acta Crystallogr D Biol Crystallogr.
71:790–799. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hu N, Feng C, Jiang Y, Miao Q and Liu H:
Regulative Effect of Mir-205 on Osteogenic Differentiation of Bone
Mesenchymal Stem Cells (BMSCs): Possible role of SATB2/Runx2 and
ERK/MAPK pathway. Int J Mol Sci. 16:10491–10506. 2015. View Article : Google Scholar : PubMed/NCBI
|


















