Introduction
Endometriosis is an estrogen-dependent chronic
disorder that affects between 5 and 10% of women at reproductive
age (1,2). It is characterized by chronic pelvic
pain and subfertility or infertility, and affects the quality life
of the affected individuals (3–5).
Despite the progress made in elucidating the molecular etiology of
endometriosis, due to heterogeneity of this disorder, there are
individuals with unknown molecular/genetic alterations which are
affected by endometriosis (6–9).
Endometriosis has long been considered to be closely
associated with the development of ovarian endometrioid and clear
cell carcinoma, and has been previously proposed to be a
precancerous lesion (10–12). Previous studies further supported
this hypothesis as an increasing number of genetic alterations in
oncogenes and tumor suppressor genes, such as KRAS proto-oncogene
GTPase, tumor protein p53, phosphatase and tensin homolog, breast
cancer type 2 susceptibility protein and protein phosphatase 2
scaffold subunit Aα, have been identified in patients with
endometriosis (13–15).
Transcriptional regulating factor 1 (TRERF1) acts as
a zinc-finger transcriptional regulatory protein and modifies the
expression of cholesterol side-chain cleavage enzyme (P450scc)
(16,17). Endometriosis is frequently
characterized by perturbed levels of estrogen (18,19)
and diverse somatic mutations in multiple genes (14). TRERF1 may regulate the expression
of P450scc, thus affecting the conversion of cholesterol to
pregnenolone, which is the first and rate-limiting step in the
synthesis of the steroid hormones, such as estrogen (20). The authors of the present study
hypothesize that certain TRERF1 aberrations, including gene
mutations, may contribute to the development of endometriosis. To
verify this hypothesis, samples were collected from 92 patients
with ovarian endometriosis and the entire coding region and
corresponding intron-exon boundaries of TRERF1 were sequenced to
analyze the potential presence of TRERF1 mutations.
Materials and methods
Samples
The ectopic endometria and paired blood samples were
obtained from a total of 92 patients with ovarian endometriosis who
underwent surgical resection in Jiangxi Provincial Maternal and
Child Health Hospital between June 2013 and July 2014. Tissue and
blood samples were stored at −80°C immediately following
collection.
Clinical data
Clinical data was determined for individuals with
ovarian endometriosis, including the following information: Age of
diagnosis, age at the time of menarche, the serum levels of
estrogen (E2), progesterone (P), cancer antigen 125 (CA125),
thyroid stimulating hormone (TSH), free triiodothyronine (FT3),
free thyroxine (FT4), carcinoembryonic antigen (CEA), α-fetoprotein
(AFP) and squamous cell carcinoma antigen (SCCA) is presented in
Table I. The levels for the
aforementioned factors were determined according to the previously
described protocols (21).
 | Table I.Association of
transcriptional-regulating factor 1 mutations with clinical
characteristics in the 92 individuals with ovarian
endometriosis. |
Table I.
Association of
transcriptional-regulating factor 1 mutations with clinical
characteristics in the 92 individuals with ovarian
endometriosis.
| Feature | Wild type (n=90) | Mutant type
(n=2) | P-value |
|---|
| Age (years) |
33.54±7.45 | 39.00±4.24 | 0.31 |
| Age of menarche
(years) |
13.68±1.38 |
14±2.83 | 0.75 |
| E2 (pg/ml) | 126.65±99.15 | 84.61±61.51 | 0.54 |
| P (ng/ml) |
1.54±3.68 |
0.91±0.49 | 0.81 |
| CA125 (µ/ml) | 103.38±191.13 | 57.03±14.60 | 0.73 |
| TSH (mIU/ml) |
2.60±1.25 |
1.90±0.43 | 0.31 |
| FT3 (pg/ml) |
3.05±0.37 |
3.00±0.04 | 0.87 |
| FT4 (ng/dl) |
1.29±0.13 |
1.30±0.06 | 0.56 |
| CEA (ng/ml) |
1.16±0.42 |
0.96±0.11 | 0.36 |
| AFP (ng/ml) |
2.53±1.62 |
3.47±0.77 | 0.43 |
| SCC (ng/ml) |
1.48±1.01 |
1.68±1.28 | 0.65 |
Compliance with ethical standards
The present study was approved by the Ethics
Committee at the Jiangxi Provincial Maternal and Child Health
Hospital (Nanchang, China). All procedures were performed according
to the tenets of the Declaration of Helsinki. Written informed
consent was obtained from each patient prior to this study.
DNA isolation, polymerase chain
reaction (PCR) amplification and DNA sequencing
Genomic DNA (gDNA) was isolated from tissues and
paired blood samples using a TIANamp Genomic DNA kit (cat no.
DP304; Tiangen Biotech Co., Ltd., Beijing, China). The quantity and
quality of the isolated gDNA was assessed by SmartSpec Plus
Spectrophotometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
at a wavelength of 260 nm and 1.5% agarose gel electrophoresis with
ethidium bromide staining for visualization, respectively. The
entire coding sequence of the TRERF1 gene was amplified and
sequenced to analyze potential somatic mutations in 92 ovarian
endometriosis samples. A total of 50 ng DNA from each sample was
amplified using PCR in a final volume of 30 µl containing 1.0 U of
rTaq DNA polymerase (Takara Biotechnology Co., Ltd., Dalian,
China), 3 µl 10X PCR buffer (Takara Biotechnology Co., Ltd. Dalian,
China), 1.0 mM MgCl2, 200 µM dNTPs (Takara Biotechnology
Co., Ltd., Dalian, China), 1.5 µM each primer in a Thermal Cycler
2720 (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The
following thermocycling conditions were used for the PCR: Initial
denaturation for 5 min at 94°C, 35 cycles of 94°C for 30 sec,
52–62°C for 30 sec, and 72°C for 30 sec, and a final extension at
72°C for 8 min. The purification of the amplified PCR products was
performed using TIANgel Midi Purification kit (Tiangen Biotech Co.,
Ltd. Beijing, China). The purified PCR products were subjected to a
sequencing reaction using ABI PRISM® BigDye®
Terminator v3.1 cycle Sequencing kit (Thermo Fisher Scientific,
Inc.) with ABI Prism 3730 DNA sequencer (Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. The sequencing
electropherograms were analyzed using DNASTAR software version 4.0
(DNASTAR, Inc., Madison, WI, USA). Somatic mutations were confirmed
by sequencing DNA from paired blood samples. The PCR primer
sequences for the entire coding sequence of the TRERF1 gene are
listed in Table II.
 | Table II.Polymerase chain reaction primer
sequences for the mutation analysis of transcriptional-regulating
factor 1 gene. |
Table II.
Polymerase chain reaction primer
sequences for the mutation analysis of transcriptional-regulating
factor 1 gene.
|
| Primer sequence
(5′-3′) |
|
|
|---|
|
|
|
|
|
|---|
| Exon | Forward | Reverse | Annealing temperature
(°C) | Amplicon length
(bp) |
|---|
| 5–1 |
GACGTCTCCTCACCACAGTG |
CCAGTGAAACCAGGGTGAGG | 55 | 891 |
| 5–2 |
TCAGCAGTGATGGATGGAGC |
CTGACCCTGTAGCACACTGG | 60 | 981 |
| 6 |
GGTGGTCCCAAGTCAAGGAG |
CACCCCAGAAAATCCTCCCC | 57 | 312 |
| 7 |
CTCTAAAGGGCACTGGGGTG |
CAAGCAGCACACGACCTAGA | 50 | 355 |
| 8 |
AAGTGCATCCCCCTTGTGAG |
GGGTAGGGTTCCCAATGTGG | 52 | 521 |
| 9 |
CAACCAGAACTCGCTTTGCC |
GTCCCAGGACTTTACCCAGC | 52 | 620 |
| 10 |
CACCATACTCCACCCAGCTC |
AGGGCTTCATGCTTTGACCA | 52 | 446 |
| 11, 12 |
CAGTGAAAAGGCCACGTGTG |
CCTACCCACCGAGAGAAGGA | 55 | 697 |
| 13 |
TCCCTCTGGGTTTCCTTCCA |
CACAACCGAACATGCAAGCA | 55 | 279 |
| 14 |
GAACCCAGGTGTCAGAGCTC |
CCAGCGAGTGTGGAAGACAT | 50 | 393 |
| 15 |
GGTAAGGACAGGCGTGTGAA |
GGCTATCTTGGCAGCAAAGC | 52 | 382 |
| 16 |
TCCTAAGCATCCGGAGACCA |
CCCTCTGCCAAACTGTGACT | 57 | 519 |
| 17 |
ACAGGATCTGTGGTTGTGGT |
TCCCATAGAGCGACTACCCA | 62 | 457 |
| 18 |
AGGAGGTCCTAGAAGCCGAG |
TTATTTATTCCCCCAACCCCC | 57 | 663 |
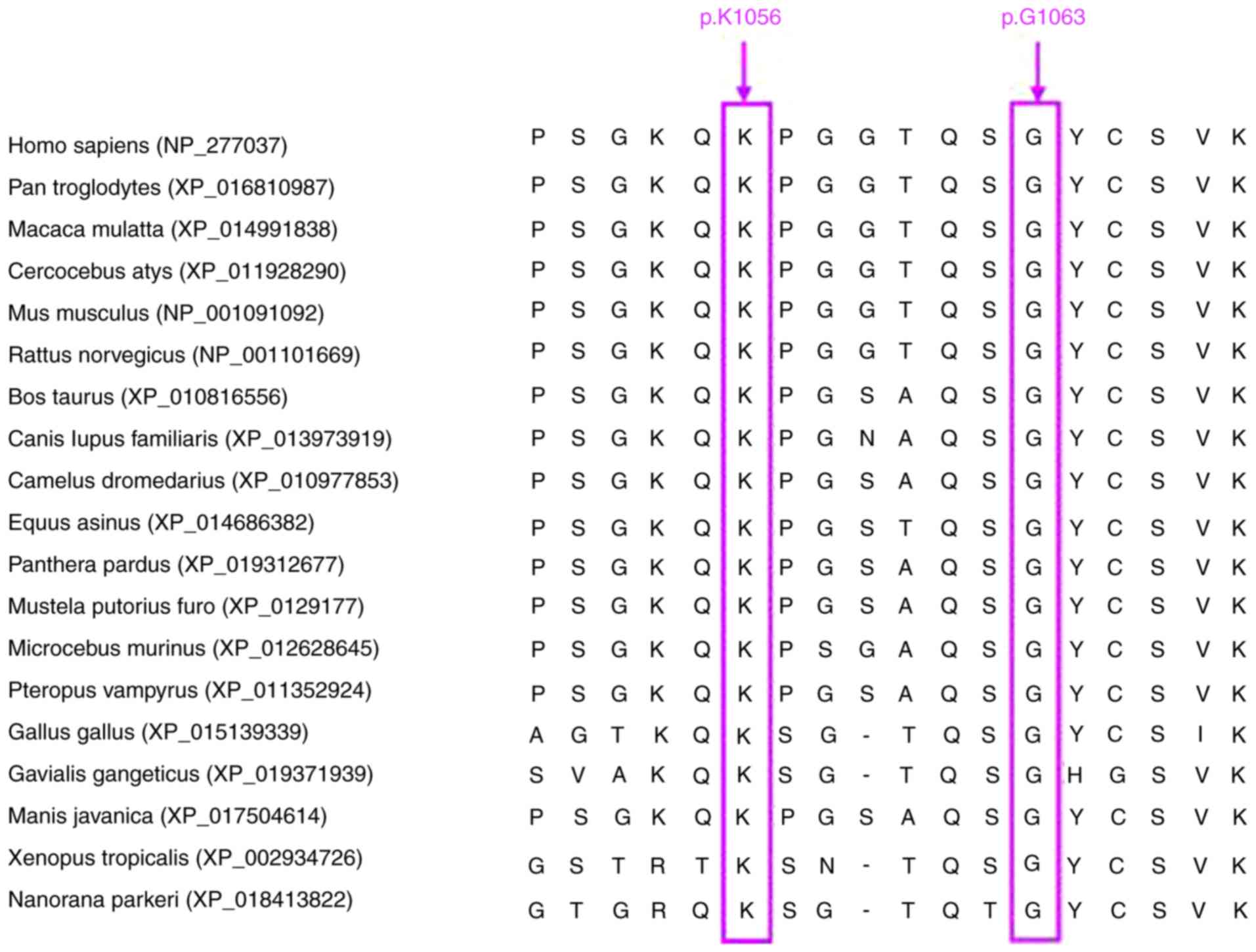
Evolutionary conservation
analysis
To evaluate the role of the identified TRERF1
somatic mutations, evolutionary conservation analysis of the TRERF1
protein sequence was performed. The amino acid sequences of 19
vertebrate species were obtained from GenBank database and
subjected to the evolutionary conservation analysis of TRERF1,
including: Homo sapiens (accession no. NP_277037.1), Pan
troglodytes (accession no. XP_016810987), Macaca mulatta
(accession no. XP_014991838), Cercocebus atys (accession no.
XP_011928290), Mus musculus (accession no. NP_001091092),
Rattus norvegicus (accession no. NP_001101669), Bos
taurus (accession no. XP_010816556), Canis lupus
familiaris (accession no. XP_013973919), Camelus
dromedarius (accession no. XP_010977853), Equus asinus
(accession no. XP_014686382), Panthera pardus (accession no.
XP_019312677), Mustela putorius furo (accession no.
XP_012917709), Microcebus murinus (accession no.
XP_012628645), Pteropus vampyrus (accession no.
XP_011352924), Gallus gallus (accession no. XP_015139339),
Gavialis gangeticus (accession no. XP_019371939), Manis
javanica (accession no. XP_017504614), Xenopus
tropicalis (accession no. XP_002934726) and Nanorana
parkeri (accession no. XP_018413822). The alignment of
sequences was carried out by ClusterW method using MEGA version 4.0
(22).
Bioinformatics prediction of TRERF1
mutations
The online bioinformatics programs, PolyPhen-2
(23,24) and MutationTaster version 2
(25) were used to predict the
potential disease-causing roles for the identified TRERF1
mutations, using the instructions of the programs.
Statistical analysis
Student's t-test was used to compare the potential
association between nominal variables referring to TRERF1
mutations, and continuous variables were compared using the
Mann-Whitney method. P-values were 2-tailed and P<0.05 was
considered to indicate a statistically significant difference. All
statistical analyses were performed using SPSS version 18.0 (SPSS,
Inc., Chicago, IL, USA).
Results
TRERF1 mutations in ovarian
endometriosis
In the present study, the coding sequence and the
corresponding intron-exon boundaries of the TRERF1 gene in the
ectopic endometria from 92 individuals with ovarian endometriosis
were sequenced. Two heterozygous missense somatic mutations in
TRERF1 [NM_033502; c.3166A>C (p.K1056Q) and c.3187 G>A
(p.G1063R)] located in exon 17 were identified in 2 out of 92
ectopic endometria (2.2%) samples. The somatic status of these
mutations was confirmed by sequencing of the respective paired
blood samples (Fig. 1). From the
two samples with TRERF1 mutations, one sample was from a
42-year-old diagnosed with uterine leiomyoma, and the other
mutation carrier was a 36-year-old woman exhibiting no other
apparent gynecological conditions. No TRERF1 mutations were
identified in the remaining 90 samples of ovarian endometriosis.
Furthermore, the two novel mutations were not identified in the
Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php) (26) and dbSNP database (https://www.ncbi.nlm.nih.gov/snp).
Association between TRERF1 mutations
and clinical data
The potential association between TRERF1 mutations
and the available clinical data was analyzed using SPSS software.
Therefore, no association between TRERF1 mutations and clinical
characteristics was identified (Table
I).
Evolutionary conservation analysis of
TRERF1 mutations
Evolutionary conservation analysis revealed that the
mutated amino acids p.K1056 and p.G1063 led to alterations of
highly conserved amino acid sequences in vertebrate species
(Fig. 2).
Pathogenic potential of TRERF1
mutations
The TRERF1 mutations were predicted by
MutationTaster online program and the two TRERF1 mutations
(p.K1056Q and p.G1063R) were predicted to be ‘disease-causing’,
with a score of 125 (p.K1056Q) and 53 (p.G1063R), where the
mutations were considered as ‘disease-causing’ when the mutation
frequency was >0.01 and the mutation was not present in the
known single-nucleotide polymorphisms (SNPs) database in the
National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/snp/). Additionally, the two
mutations were also predicted to be ‘probably damaging’ by
PolyPhen-2, with a score of 0.999 (sensitivity, 0.14; specificity,
0.99) for p.K1056Q mutation and a score of 1.000 (sensitivity:
0.00; specificity: 1.00) for p.G1063R mutation, where the mutations
were considered as ‘probably damaging’ when the prediction score
value was >0.05.
Discussion
Previous studies have suggested that the balance
between estrogen and progesterone production is frequently
perturbed in endometriosis (18,19)
and that endometriosis is a potential precancerous lesion harboring
multiple somatic mutations in certain cancer-associated genes
(10,11,13–15).
It remains to be determined whether endometriosis harbors mutations
in genes involved in the regulation of production of steroid
hormones. Considering that TRERF1 has a role in the production of
steroid hormones, including estrogen and progesterone (16,20),
the authors of the present study hypothesized that TRERF1 may
harbor mutations in endometriosis.
In the present study, two heterozygous somatic
mutations in the TRERF1 gene were identified in two out of 92
tissue samples with ovarian endometriosis. To the best of our
knowledge, both mutations were not previously reported and are
located in exon 17. One individual carrying the somatic mutation of
TRERF1 gene was diagnosed with uterine leiomyoma while the other
exhibited no other apparent gynecological conditions. In addition,
no association between TRERF1 mutation and the collected clinical
data was observed in the samples included in the present study,
including age at diagnosis, age of menarche, the levels of serum
E2, P, CA125, TSH, FT3, FT4, CEA, AFP and SCCA. The results of the
statistical analysis should be treated with caution due to small
sample size in the TRERF1 mutation group (n=2). The evolutionary
conservation analysis suggested that both the p.K1056 and p.G1063
residues were evolutionarily highly conserved in a number of
vertebrate species. Furthermore, the two TRERF1 mutations were
predicted to be ‘disease-causing’ and ‘probably damaging’ according
to MutationTaster and PolyPhen-2 prediction programs, respectively.
However, whether these TRERF1 mutations have a role in the
development of ovarian endometriosis remains to be confirmed.
The mutation frequency of TRERF1 among patients with
ovarian endometriosis was 2.2% (2/92). Previous studies aiming to
determine the somatic mutation profiles in ovarian endometriosis
did not identify any somatic mutations in the TRERF1 gene in
endometriotic lesions from 16 patients with ovarian endometriosis
(14) and 27 patients with deep
infiltrating endometriosis (15).
The authors of the present study hypothesize that the relatively
small sample sizes analyzed in the prior studies may have prevented
identification of the potential rare somatic mutations (14,15).
In conclusion, the present study identified somatic
mutations in TRERF1 (p.K1056Q and p.G1063R) in ovarian
endometriosis and the mutation frequency was 2.2% (2/92). In
silico prediction suggested that the two somatic mutations may
be ‘disease-causing’. Future functional assays should be performed
to confirm the pathogenic roles of the TRERF1 mutations, which may
elucidate the underlying mechanism of ovarian endometriosis.
Acknowledgements
The present study was supported by a grant from the
Natural Science Foundation of Jiangxi Province (grant no.
20151BAB205012).
References
|
1
|
Arosh JA, Lee J, Balasubbramanian D,
Stanley JA, Long CR, Meagher MW, Osteen KG, Bruner-Tran KL,
Burghardt RC, Starzinski-Powitz A and Banu SK: Molecular and
preclinical basis to inhibit PGE2 receptors EP2 and EP4 as a novel
nonsteroidal therapy for endometriosis. Proc Natl Acad Sci USA.
112:pp. 9716–9721. 2015; View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Samartzis EP, Noske A, Dedes KJ, Fink D
and Imesch P: ARID1A mutations and PI3K/AKT pathway alterations in
endometriosis and endometriosis-associated ovarian carcinomas. Int
J Mol Sci. 14:18824–18849. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leone Roberti Maggiore U, Ferrero S,
Mangili G, Bergamini A, Inversetti A, Giorgione V, Viganò P and
Candiani M: A systematic review on endometriosis during pregnancy:
Diagnosis, misdiagnosis, complications and outcomes. Hum Reprod
Update. 22:70–103. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vigano P, Corti L and Berlanda N: Beyond
infertility: Obstetrical and postpartum complications associated
with endometriosis and adenomyosis. Fertil Steril. 104:802–812.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fadhlaoui A, Bouquet de la Jolinière J and
Feki A: Endometriosis and infertility: How and when to treat? Front
Surg. 1:242014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tosti C, Pinzauti S, Santulli P, Chapron C
and Petraglia F: Pathogenetic mechanisms of deep infiltrating
endometriosis. Reprod Sci. 22:1053–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Richards EG, Zheng Y, Shenoy CC, Ainsworth
AJ, Delaney AA, Jones TL, Khan Z and Daftary GS: KLF11 is an
epigenetic mediator of DRD2/dopaminergic signaling in
endometriosis. Reprod Sci. 24:1129–1138. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Uimari O, Rahmioglu N, Nyholt DR, Vincent
K, Missmer SA, Becker C, Morris AP, Montgomery GW and Zondervan KT:
Genome-wide genetic analyses highlight mitogen-activated protein
kinase (MAPK) signaling in the pathogenesis of endometriosis. Hum
Reprod. 32:780–793. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yotova I, Hsu E, Do C, Gaba A, Sczabolcs
M, Dekan S, Kenner L, Wenzl R and Tycko B: Epigenetic alterations
affecting transcription factors and signaling pathways in stromal
cells of endometriosis. PLoS One. 12:e01708592017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matsumoto T, Yamazaki M, Takahashi H,
Kajita S, Suzuki E, Tsuruta T and Saegusa M: Distinct β-catenin and
PIK3CA mutation profiles in endometriosis-associated ovarian
endometrioid and clear cell carcinomas. Am J Clin Pathol.
144:452–463. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pavlidou A and Vlahos NF: Endometriosis
and ovarian cancer: Clinical and molecular aspects. Minerva
Endocrinol. 39:155–165. 2014.PubMed/NCBI
|
|
12
|
Burghaus S, Fasching PA, Häberle L, Rübner
M, Büchner K, Blum S, Engel A, Ekici AB, Hartmann A, Hein A, et al:
Genetic risk factors for ovarian cancer and their role for
endometriosis risk. Gynecol Oncol. 145:142–147. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vestergaard AL, Thorup K, Knudsen UB, Munk
T, Rosbach H, Poulsen JB, Guldberg P and Martensen PM: Oncogenic
events associated with endometrial and ovarian cancers are rare in
endometriosis. Mol Hum Reprod. 17:758–761. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li X, Zhang Y, Zhao L, Wang L, Wu Z, Mei
Q, Nie J, Li X, Li Y, Fu X, et al: Whole-exome sequencing of
endometriosis identifies frequent alterations in genes involved in
cell adhesion and chromatin-remodeling complexes. Hum Mol Genet.
23:6008–6021. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Anglesio MS, Papadopoulos N, Ayhan A,
Nazeran TM, Noë M, Horlings HM, Lum A, Jones S, Senz J, Seckin T,
et al: Cancer-associated mutations in endometriosis without cancer.
N Engl J Med. 376:1835–1848. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gizard F, El-Alfy M, Duguay Y, Lavallée B,
DeWitte F, Staels B, Beatty BG and Hum DW: Function of the
transcriptional regulating protein of 132 kDa (TReP-132) on human
P450scc gene expression. Endocr Res. 28:559–574. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gizard F, Teissier E, Dufort I, Luc G,
Luu-The V, Staels B and Hum DW: The transcriptional regulating
protein of 132 kDa (TReP-132) differentially influences
steroidogenic pathways in human adrenal NCI-H295 cells. J Mol
Endocrinol. 32:557–569. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Reis FM, Petraglia F and Taylor RN:
Endometriosis: Hormone regulation and clinical consequences of
chemotaxis and apoptosis. Hum Reprod Update. 19:406–418. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vercellini P, Viganò P, Somigliana E and
Fedele L: Endometriosis: Pathogenesis and treatment. Nat Rev
Endocrinol. 10:261–275. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gizard F, Lavallee B, DeWitte F, Teissier
E, Staels B and Hum DW: The transcriptional regulating protein of
132 kDa (TReP-132) enhances P450scc gene transcription through
interaction with steroidogenic factor-1 in human adrenal cells. J
Biol Chem. 277:39144–39155. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu J, Zou Y, Luo Y, Guo JB, Liu FY, Zhou
JY, Zhang ZY, Wan L and Huang OP: Prevalence and clinical
significance of mediator complex subunit 12 mutations in 362 Han
Chinese samples with uterine leiomyoma. Oncol Lett. 14:47–54. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tamura K, Dudley J, Nei M and Kumar S:
MEGA4: Molecular evolutionary genetics analysis (MEGA) software
version 4.0. Mol Biol Evol. 24:1596–1599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Adzhubei I, Jordan DM and Sunyaev SR:
Predicting functional effect of human missense mutations using
PolyPhen-2. Curr Protoc Hum Genet. Jan;2013.doi:
10.1002/0471142905.hg0720s76. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stenson PD, Mort M, Ball EV, Evans K,
Hayden M, Heywood S, Hussain M, Phillips AD and Cooper DN: The
human gene mutation database: Towards a comprehensive repository of
inherited mutation data for medical research, genetic diagnosis and
next-generation sequencing studies. Hum Genet. 136:665–677. 2017.
View Article : Google Scholar : PubMed/NCBI
|