Introduction
Esophageal neoplasm is one of most common human
cancers in Africa, South America, China, Europe and North America,
and includes esophageal squamous cell carcinoma (ESCC) and
esophageal adenocarcinoma (EA), small cell undifferentiated
carcinoma and sarcoma divided by histological types (1). Previous reports have indicated that
the globalincidence of esophageal neoplasm has increased ~500% over
the past 30 years (2–4). Statistics indicate that ESCC and EA
account for 95 and 5% of esophageal cancer diagnoses, respectively
(5,6). As the number of patients diagnosed
with esophageal cancer is increasing, there is a focus on the
development of novel strategies for early esophageal neoplasm
diagnosis and treatments using novel and efficient anticancer
treatments, including surgical treatment, radiotherapy, chemical
therapy, biological therapy and comprehensive therapy for patients
(7–9).
The expression of oncogenic and oncolytic proteinsin
human tumors has previously been investigated to analyze gene
expression patterns during cellular transcription and translation,
in addition to tumor cell growth, migration and invasion (10–12).
Gene associated with retinoid-interferon (IFN)-induced mortality-19
(Grim-19) is reported to be a cell death activator that is used to
define mechanisms involved in IFN-β- and retinoic acid-induced cell
death and apoptosis in various tumor cell lines (13). Oncolytic proteins have been
demonstrated to inhibited tumor cell growth by activating specific
sets of genes and initiating the apoptotic program of cells
(14). In a previous study,
Grim-19 upregulation exhibited antitumor effects via induction of
IFN-β and retinoic acid in human tumor cells (15). Li et al (15) reported that Grim-19 upregulation
inhibited signal transducer and activator of transcription 3
(STAT3) transcriptional activity by constitutive inactivation of
the signal transducer, and this regulatory pathway contributed to
the inhibition of progression and metastasis in several different
tumor types. In addition, Grim-19 bound to the transcription factor
STAT3 and led to the ablation of pro-oncogenes Fas cell surface
death receptor and Jun proto-oncogene, and inhibition of the
pro-oncogenic effects of v-Src independently of STAT3 (16). Furthermore, reduced Grim-19
expression was reported to beassociated with high-risk human
papilloma virus infection in cervical squamous intraepithelial
neoplasia and cancer (17).
Adenovirus vectors are the most widely used vectors
and adenovirus-mediated delivery of antitumor genes or polypeptides
into tumor cells has been described extensively (18,19).
In addition, gene transfer strategies have presented increased
clinical potential for clinicians, which may be applied
asimmunotherapy for patients with cancer using oncolytic proteins
delivered by recombinant adenovirus (rAd) to exert various effects,
including inhibition of oncogenes and restoration of tumor
suppressor genes, immunotherapy, anti-angiogenesis and virotherapy
(20).
The present study investigated the efficacy of rAd
expressing Grim-19 (rAd-Grim-19) on esophageal tumor growth and
investigated the potential for rAd-Grim-19 as a therapeutic
intervention for patients with esophageal cancer. The results of
the current study indicated that esophageal tumor growth was
suppressed by rAd-Grim-19 and survival was prolonged during a
120-day observation period, indicating that rAd-Grim-19 may be an
efficient antitumor agent for patients with esophageal
neoplasm.
Materials and methods
Ethics statement
The investigation was conducted according to the
Guide for the Care and Use of Laboratory Animals (21). All experimental protocols and
surgery on animals were performed in accordance with the First
Affiliated Hospital of Soochow University on the Ethics of Animal
Experiments Defense Research (Suzhou, China). All surgical
operations and euthanasia were performed in a manner that minimized
suffering.
Patients
A total of 5 patients with esophageal tumor (2
female and 3 male; mean age, 54.2 years old) were admitted to the
First Affiliated Hospital of Soochow University (Suzhou, China)
between May 2013 and January 2014. Both human and animal
experiments were approved by Ethics Committee of the First
Affiliated Hospital of Soochow University.
Construction of recombinant adenovirus
by genetic engineering
Adeno-X expression system (cat. no. 632269; Takara
Biotechnology Co., Ltd., Dalian, China) was used for the
construction of recombinant adenovirus. Human Grim-19 (the First
Affiliated Hospital of Soochow University, Suzhou, China) linked
with cell-penetrating peptide was extracted as previously described
(22) and cloned into the pAd-X
expression system plasmid. The recombinant adenoviral plasmid
expressing Grim-19 was selected and confirmed by polymerase chain
reaction (PCR) and sequencing (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and is termed rAd-Grim-19. The
recombinant adenoviruses (5 MOI) were generated by transfecting
into 293 cells (1×106) for 144 h at 37°C and followed
10th generation propagation to purify viruses. rAd-Grim-19 was
purified as described in a previous report (23). The titers of rAd-Grim-19 and rAd
were determined by TCID50 as plaque-forming units
(pfu)/ml. The EC-109 cells (1×104) divided into three
treatment groups: i) The control group (PBS); ii) the rAd group
(0.25 mg/ml at 37°C for 24 h); and iii) the rAd-Grim-19 group (0.25
mg/ml at 37°C for 24 h).
Cell culture
EC-109 and Het-1A cells were purchased from American
Type Culture Collection (Manassas, VA, USA). All cells were
cultured in EMEM supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) at 37°C, 5% CO2
and reasonable humidity.
MTT assay
EC-109 cells (1×103) were cultured and
then inoculated with rAd-Grim-19 [0.5 multiplicity of infection
(MOI)] or rAd-EGFP 0.5 (MOI) or PBS in 96-well plates for 48 h at
37°C in triplicate for each condition. Following culture, 20 µl MTT
(5 mg/ml) in PBS solution was added to each well, the plate was
further incubated for 4 h at 37°C. The medium was entirely removed
and 100 µl dimethyl sulfoxide was added to the wells to solubilize
the crystals. The optical density (OD) was measured using an ELISA
microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
reader at a wave length of 450 nm.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was obtained from EC-109 cells and Het-1A
normal esophageal epithelial cells by using an RNA easy Mini kit
(Qiagen Sciences, Inc., Gaithersburg, MD). The expression of
Grim-19 in EC-109 and Het-1A cells was measured using a RT-qPCR kit
(Qiagen Sciences, Inc.) with β-actin expression as an endogenous
control, according to the manufacturer's protocol. All the primers
(Grim-19 forward, 5′-TCGGGGACTGTCGGGGTAC-3′ reverse,
5′-AGGGTCCTCCGGTCCTTCT-3′; β-actin forward,
5′-CGGAGTCAACGGATTTGGTC-3′, reverse, 5′-AGCCTTCTCCATGGTCGTGA-3′)
were synthesized by Invitrogen (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). cDNA (10 ng) was used for qPCR with the
SYBR-Green Master Mix system (Bio-Rad Laboratories, Inc.; 50 ng of
genomic DNA, 200 µM dNTP, 2.5 units of Taq DNA polymerase, and 200
µM primers) performed followed by initial denaturation at 94°C for
2 min, followed by 45 cycles of 94°C for 30 sec, annealing
temperature reduced to 58°C for 30 sec and 72°C for 10 min.
Relative Grim-19 expression levels were calculated by the
2−ΔΔCq method (24).
The results are presented relative to β-actin.
Animal study
A total of 60 male BALB/c (SPF) nude mice
(6-week-old; 26–35 g body weight) were purchased from SLAC
Laboratory Animal Co., Ltd. (Shanghai, China). All animals were fed
under pathogen-free conditions. All mice were housed under
controlled temperature (23±1°C, 50–60% humidity, 0.02–0.03%
CO2) in a 12 h light/dark cycle with free access to food
and water. A total volume of 200 µl EC-109 cells (1×106)
were injected into the left flank of male BALB/c nude mice.
EC-109-bearing mice were treated with PBS, rAd and rAd-Grim-19 when
tumor diameters reached 5–8 mm on day 6 after tumor inoculation.
Tumor-bearing mice were randomly divided into 3 groups (n=20) and
intratumorally treated with 2×108 pfurAd or rAd-Grim-19,
and PBS was used as a control. Treatments were performed 10 times
at 2 day intervals. Tumor diameters were recorded every 2 days and
tumor volume was calculated by using the following formula: 0.52 ×
smallest diameter2 × largest diameter. Furthermore, a
120-day observation was employed to evaluate the long-term efficacy
of rAd-Grim-19. The mice were euthanized when the tumor reached 10
mm. All experimental mice were housed for a total of 120 days.
Splenocyte collection and cytotoxic T
lymphocyte (CTL) responses
Splenocytes were isolated from the spleens of the
therapeuticmice with esophageal neoplasm on day 30 as previously
described (25). The splenocytes
(1×106) were incubated with the mitomycin (10 µg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany)-inactivated EC-109
cells (1×106) for 6 h at 37°C after washing with PBS.
Supernatants were obtained using centrifugation (2,000 × g) for 10
min at 37°C. Release of IFN-γ was determined by ELISA (cat. no
ab46025; Abcam, Cambridge, UK) in the supernatants after culture
for 72 h. In addition, T cells (1×106) were purified
from the splenocytesas previously described (26) and cultured with EC-109 cells for 4
h at the effector: Target ratios of 6:1, 12:1 and 24:1 (27). Specific CTL activity of T cells for
tumor cells was performed by MTT cytotoxicity assays (28).
Flow cytometry analysis
EC-109 cells were obtained from the American Type
Culture Collection (Manassas, VA, USA) and cultured in minimum
essential medium supplemented with 10% fetal calf serum. EC-109
cells (1×106) were treated with PBS, rAd and rAd-Grim-19
for 72 h at 37°C. Subsequently, apoptosis in suspended cells was
analyzed by flow cytometry using Annexin V-FITC and PI (Annexin
V-FITC kit; BD Biosciences, Franklin Lakes, NJ, USA). In addition,
on day 30 cells (1×106) from tumors in mice treated with
PBS, rAd and rAd-Grim-19 were prepared for PE-CD4+ and
PI-CD8+ analysis by fluorescence-activated cell sorting.
Tumors were removed from euthanized animals and tumor cells were
isolated by passing through 100 µm nylon mesh filters and then
cells washed with PBS and resuspended. Tumor cells were labeled by
CD3 and CD45, followed by PE-CD4+ (1:2,000, cat no.
88-8999-40; Thermo Fisher Scientific, Inc.) and PI-CD8+
(1:2,000, cat no. 8804-6825-74; Thermo Fisher Scientific, Inc.) for
4 h at 4°C, staining to determine the frequency of CD4 and CD8 cell
subsets in the total infiltrated immune cells. Cells were washed
with PBST three times and the stained cells were analyzed by using
a FACScan flow cytometer (BD Biosciences) and analyzed using BD
FACSDiva™ software (version 1.2; BD Biosciences).
Western blot analysis and histological
immunostaining
EC-109 and Het-1A cells were lysed and used to
analyzed Grim-19 protein expression, according to a previous study
(29). For immunostaining, EC-109
cells (1×106) pre-treated with PBS, rAd or rAd-Grim-19
for 24 h, or tumors from esophageal carcinoma xenograph mice in
PBS, rAd and rAd-Grim-19 groups on day 30 were fixed using 10% for
maldehyde for 15 min at room temperature and embedded in paraffin.
Protein concentration was measured by a Bicinchoninic acid protein
assay kit (Thermo Scientific, Pittsburgh PA, USA). Protein samples
(20 µg/lane) wereseparated by 15% SDS-PAGE and then transferred to
polyvinylidene fluoride membrane. After 1 h blocking at 37°C
temperature using 10% blocking reagent (Roche Applied Science,
Penzberg, Germany), membrane was incubated with primary antibodies:
Grim-19 (1:1,000; cat. no. ab134325; Abcam) and β-actin (1:1,000,
cat. no. ab8226; Abcam) for 12 h at 4°C. Following the incubation,
membrane was washed three times in TBST and incubated with
horseradish peroxidase-conjugated goat anti-rabbit IgG mAb
(1:1,000; cat. no. PV-6001; OriGene Technologies, Inc., Beijing,
China) for 1 h at 37°C. After three-time washing in TBST, membrane
was developed using a chemiluminescence assay system (Roche Applied
Science) and exposed to Kodak exposure films. In addition, tumor
samples were cut in to tumor sections (4 µm) and antigen retrieval
was also performed. EC-109 cells and tumor sections were incubated
with TUNEL reaction mixture (Sigma-Aldrich; Merck KGaA) at 37°C for
2 h. Cells were three-time washing in TBST and stained with DAPI
(Sigma-Aldrich; Merck KGaA) at 37°C for 2 h. TUNEL assays were
conducted using a TUNEL fluorescence FITC kit (Roche, Indianapolis,
IN, USA) according to the manufacturer's instructions. All sections
were stained with hematoxylin and eosin (H&E) for 2 h at 37°C.
Images were captured on MicroChemi 4.2 (Eastwin, Shenzen,
China).
Immunohistochemistry
Esophageal tumors from patients were fixed with 10%
for maldehyde, then embedded in paraffin and cut into tumor
sections for 2 h at 37°C. Antigen retrieval was performed on the
tumor sections using eBioscience™ IHC Antigen Retrieval Solution
(cat. no 00-4955-58; Invitrogen), and the sections were
subsequently incubated with rabbit anti-mouse Grim-19 (1:1,000,
ab134325; Abcam). Following antibody incubation, Alexa Fluor
488-labeled secondary antibodies (1:500; Beyotime Institute of
Biotechnology) for 2 h at 37°C and the specimens were visualized. A
Ventana Benchmark automated Diaminobenzidine staining system
(Ventana Medical Systems, Inc., Tucson, AZ, USA) was used to detect
Grim-19 protein expression.
Statistical analysis
Data are presented as the mean ± standard error of
the mean of triplicate experiments. All data were analyzed using
SPSS software (version 19.0; IBM Corp., Armonk, NY, USA). Unpaired
data were analyzed by Student's t-test. Comparisons between
multiple groups were analyzed by one-way analysis of variance
followed by Bonferroni post hoc tests. The Kaplan-Meier test was
used to estimate survival during the 120-day observation period.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of Grim-19 and its function
in esophageal tumor cells
In order to analyze the function of Grim-19 in tumor
cells, the present study first investigated Grim-19 expression in
esophageal tumor cells. EC-109 cells were cultured and cells were
harvested to isolate total RNA. Grim-19 expression was analyzed by
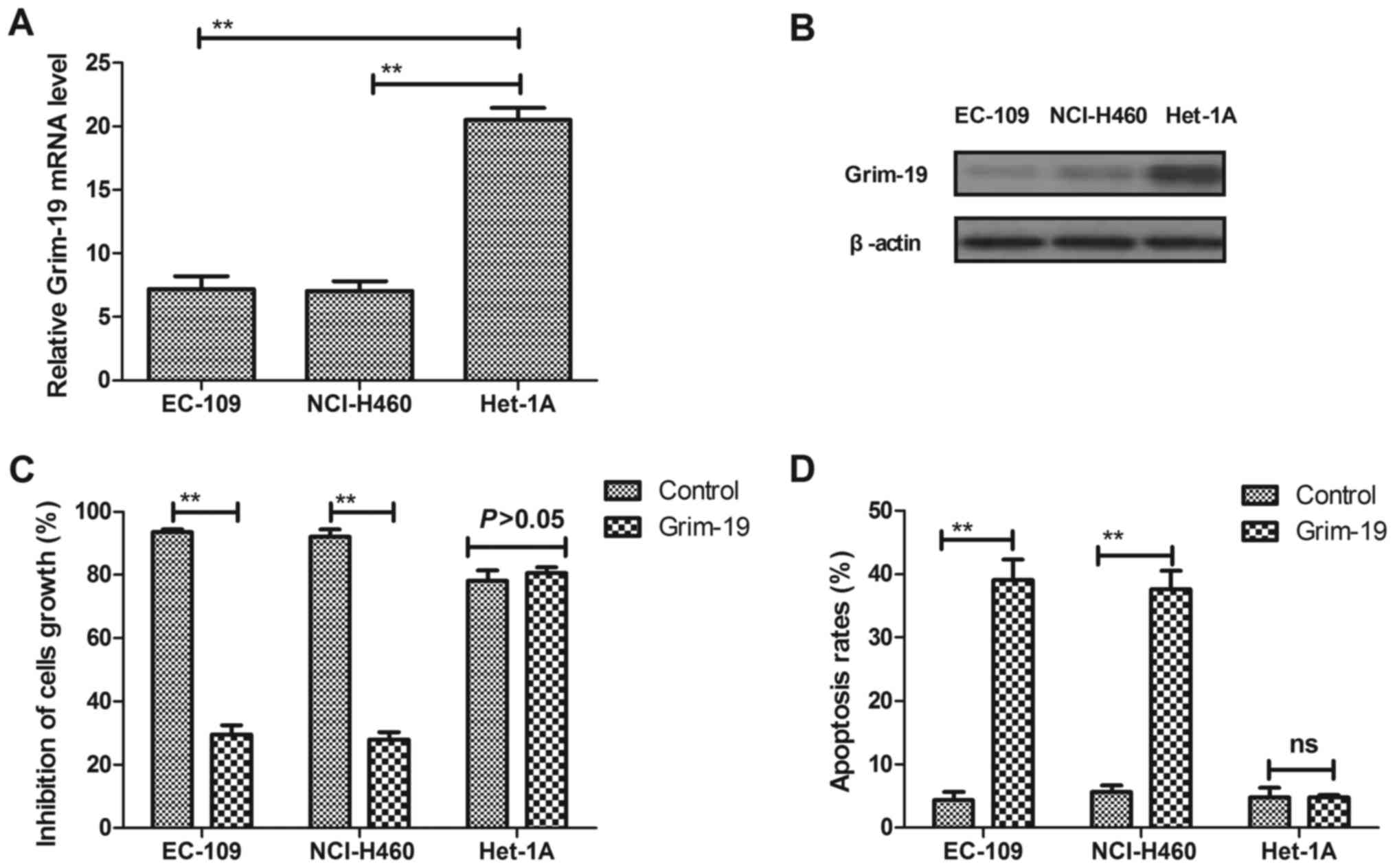
RT-qPCR and western blot analysis. The results in Fig. 1A and B demonstrate that Grim-19
expression was downregulated in EC-109 cells at mRNA and protein
levels compared with HET-1A normal esophageal epithelial cells. In
addition, the function of Grim-19 was studied in EC-109 cells.
Furthermore, the results of MTT assays indicated that Grim-19
exhibited significant inhibitory effects on EC-109 cell growth
(Fig. 1C), andthe results in
Fig. 1D indicate that Grim-19
significantly induced apoptosis in EC-109 cells after 72 h
treatment. These results indicated that Grim-19 expression was
downregulated in EC-109 cells, and Grim-19 treatment led to growth
inhibitory effects and enhanced apoptosis in EC-109 cells.
Detection and bioactivity of Grim-19
in rAd-Grim-19-infected cells in vitro
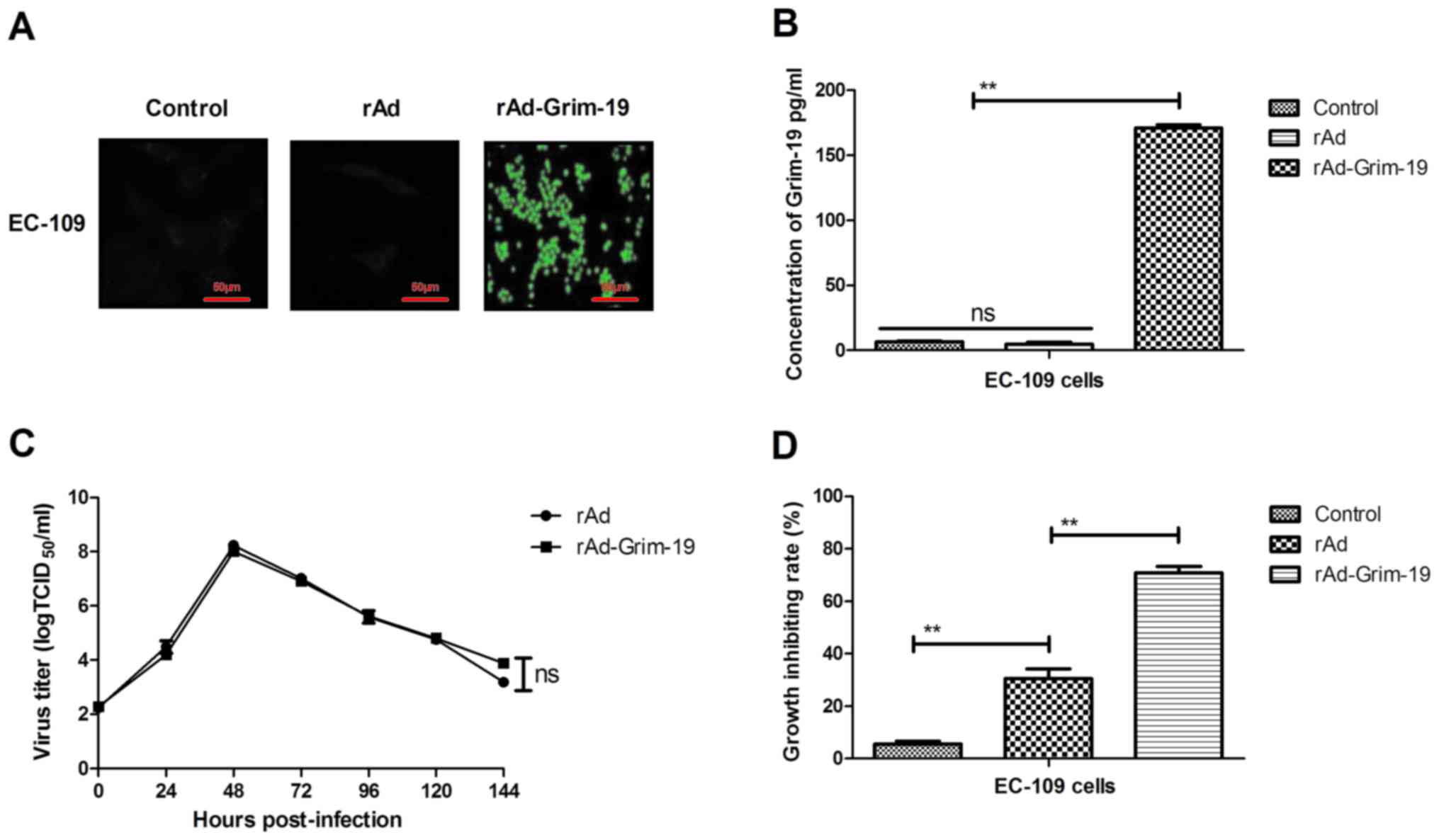
In order to investigate the effect of Grim-19 over
expression, the expression of Grim-19 proteins in
rAd-Grim-19-infected EC-109 cells was observed by fluorescent
microscopy following incubation with anti-Grim-19 antibody. The
results demonstrated that Grim-19 protein was observed in EC-109
cells infected with rAd-Grim-19, indicating that Grim-19 was
efficiently expressed in tumor cells (Fig. 2A). The results in Fig. 2B indicate that Grim-19 was
effectively expressed and secreted extracellularly in EC-109 cells
infected with rAd-Grim-19. In addition, the kinetic curve of
rAd-Grim-19 was analyzed, which demonstrated that Grim-19 gene
insertion did not affect the recombinant adenovirus (Fig. 2C). Furthermore, the results
demonstrated that EC-109 cell growth was significantly inhibited by
rAd-Grim-19 compared with rAd and PBS groups (Fig. 2D). These results indicated that
Grim-19 was efficiently upregulated by rAd-Grim-19 and rAd-Grim-19
led to tumor growth inhibitory effects on EC-109 cells.
In vivo effects of rAd-Grim-19 in
esophageal tumor-bearing mice
In order to analyze the in vivo effects of
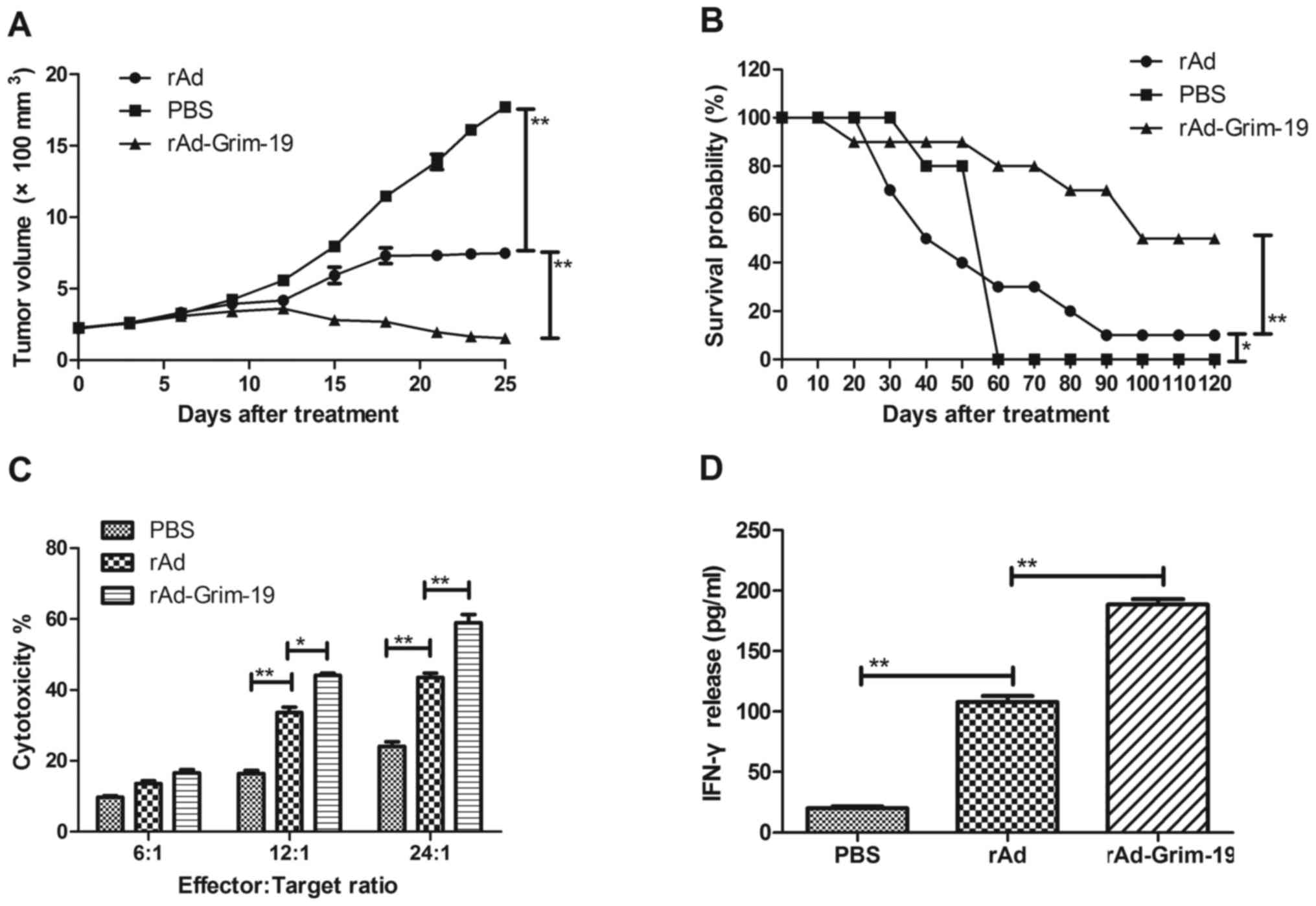
rAd-Grim-19 on tumor growth inhibition, its anticancer effect was
determined in an esophageal tumor-bearing mouse model. The results
in Fig. 3A demonstrate that
rAd-Grim-19 significantly suppressed tumor growth, while xenograft
mice treated with rAd-only exhibited reduced inhibitory effects,
compared with PBS treatment. The results indicate that tumors in
the rAd-Grim-19-treated group decreased by a volume of 32.18±8.38
mm3 by day 25, which is significant difference compared
with the rAd and PBS groups (P<0.01). During the period of
treatment, no side effects were observed other than swelling at the
injection site. In addition, a 120-day long-term survival
observation period was performed following treatment with PBS, rAd
and rAd-Grim-19. The results in Fig.
3B demonstrated that rAd-Grim-19 (n=20) prolonged the survival
of mice compared with rAd and PBS groups. In addition, CTL
responses and IFN-γ release were determined on day 25 after
treatments. The results in Fig. 3C and
D indicated that mice treated with rAd-Grim-19 developed a
strong CTL response for EC-109 cells and exhibited an increased
release of IFN-γ compared with rAd and PBS groups. These results
demonstrate therapeutic effects of rAd-Grim-19 against EC-109 in
tumor-bearing animals, as long-term survival was increased in mice
treated with rAd-Grim-19.
rAd-Grim-19 increases immune cell and
apoptotic body accumulation in EC-109 tumor-bearing mice
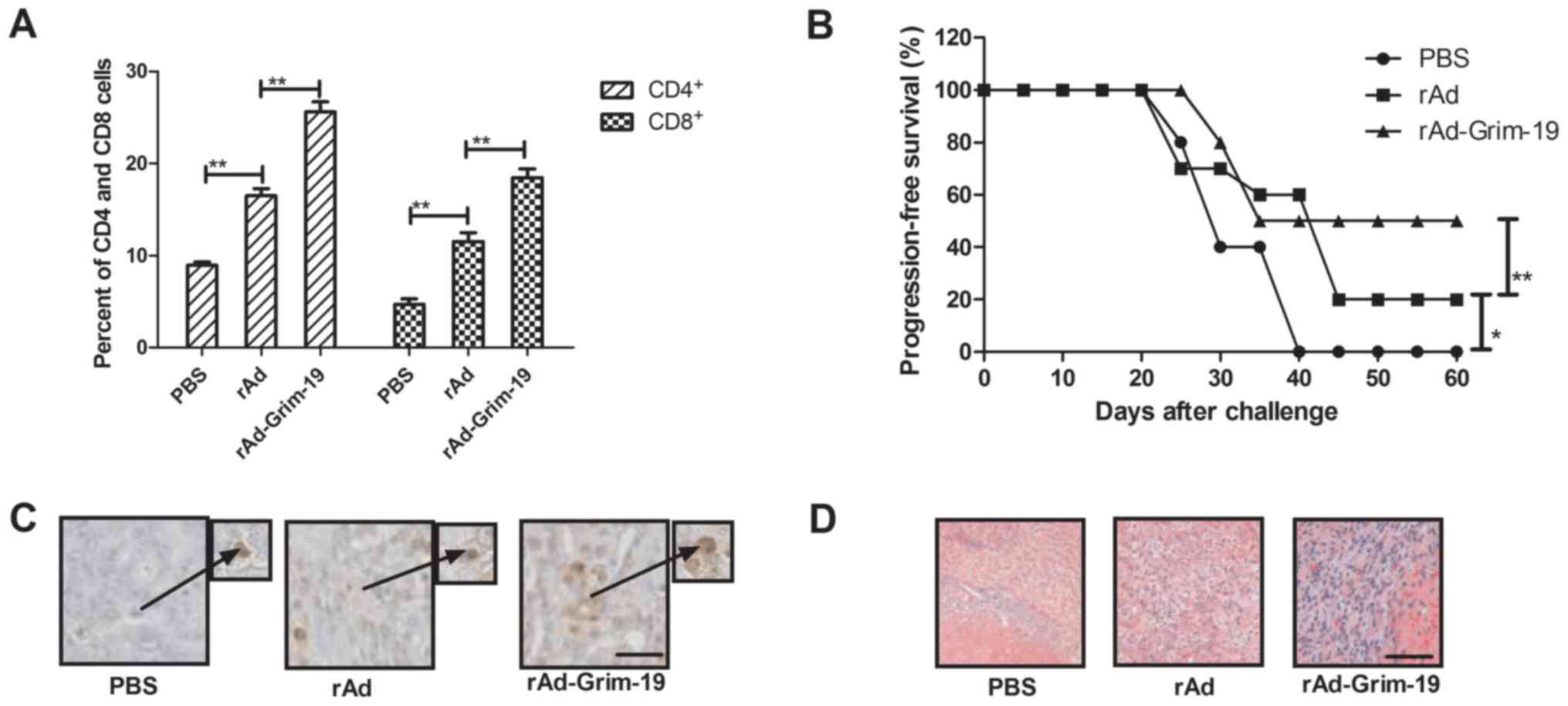
To further investigate the long-term survival of
tumor-bearing mice, immune responses were analyzed and tumor
challenge experiments were performed. The results in Fig. 4A demonstrate that CD4+
and CD8+ expression levels were increased in tumors from
rAd-Grim-19-treated mice compared with rAd and PBS groups. Tumor
challenge experiments were performed after a 120-day survival
period observation. As indicated in Fig. 4B, rAd-Grim-19-treated mice
demonstrated an immune memory for EC-109 tumor cells and their
survival was prolonged compared with rAd-treated mice. Furthermore,
apoptotic bodies were analyzed by immunohistochemistry staining in
tumors from experimental mice following treatment with rAd-Grim-19,
rAD and PBS. The results in Fig.
4C demonstrated that an increased number of apoptotic bodies on
tumors were observed in rAd-Grim-19-treated mice compared with rAd
and PBS groups. In addition, Grim-19 expression levels in tumors on
day 25 after treatment were also determined. The results
demonstrated that rAd-Grim-19 promoted Grim-19 expression following
infection with rAd-Grim-19 in tumors compared with rAd and
PBS-treated tumors (Fig. 4D). In
conclusion, these results indicate that rAd-Grim-19 led to
beneficial outcomes for mice with esophageal neoplasms potentially
through increased immune accumulation and immune memory against
homologous tumor cells, which may contribute to the formation of
apoptotic bodies, improve long-term survival in mice and reduce the
risk of recurrence.
Discussion
Patients with esophageal neoplasms frequently
receive surgery, chemotherapy, radiotherapy, immunotherapy and
comprehensive therapythatin majority of cases results in deaths and
represents a global health burden (30). Patients with esophageal neoplasms
are frequently diagnosed at an advancedstage, and combined
treatment with 5-fluorouracil and cisplatin with concurrent
irradiationisa standard treatment regimen (31,32).
In addition, statistics have indicated that ESCC and EA account for
95 and 5% of esophageal cancer diagnoses, respectively, with ESCC
accounting for the highest number of cases, and being associated
with poor survival outcome and a high mortality rate (33). Given the higher morbidity and
mortality, the identification and development of novel strategies
for the treatment of esophageal neoplasms are required to improve
the treatment and eradication of these tumors. In the present
study, a mouse model of esophageal neoplasms was established to
analyze the tumor growth and long-term survival following
treatments.
Grim-19 is a protein with a molecular weight of ~16
kDa that was isolated as a novel gene product by using a genetic
technique and was reported to enhance tumor cell apoptosis and
death induced by IFN/retinoic acid (RA) (34). It is established that the
Src-family of tyrosine kinases are important regulators of the cell
growth, migration and invasion of various tumor cell types.
Inactivation of Src by mutation caused cellular transformation
through alterations in transcription and cytoskeletal properties
(35). Kalakonda et al
(36) reported that the tumor
suppressive protein Grim-19 inhibited Src-induced oncogenic
transformation. In addition, previous studies have also
investigated the mechanisms of anticancer effects of IFN-β and RA
in human tumor cells. Grim-19 upregulation exhibited antitumor
effects that occurred via IFN-β and RA in human tumor cells
(15,34). In the current study, the results
demonstrated that Grim-19 expression was reduced in EC-109 tumor
cells compared with Het-1 A normal esophageal epithelial cells, and
overexpression of Grim-19 inhibited EC-109 growth and induced tumor
cell apoptosis.
Previous studies have demonstrated that recombinant
adenoviruses based on the Adeno-X expression system resulted in a
gene and oncolytic therapy vehicle for certain types of human
carcinoma (23,37). The recombinant adenovirusvehicle is
the most widely used gene therapy vector to deliver functional
protein in preclinical and clinical applications for the treatment
of various human diseases, including cancer (38). In the present study, the oncolytic
protein Grim-19 delivered by recombinant adenovirus was constructed
and the in vitro and in vivo rAd-GRIM-19 inhibitory
effects on esophageal cancer tumor growth were investigated. The
results indicated that rAd-GRIM-19 significantly suppressed tumor
growth in EC-109-bearing mice. However, there are few reports
concerning immunotherapy delivered by a recombinant adenovirus.
A previous study reported that dysregulation of
Grim-19 expression in human renal cell carcinomas and proapoptotic
inactivity were mediated by signal transducer and activator of
transcription 3 and JAK signaling pathways, indicating that Grim-19
may be a potential tumor suppressor that may be useful for cancer
therapy development (39). In
addition, Grim-19 overexpression in various human tumor cells led
to reduced cell growth and apoptosis induction via IFN-β and
RA-activated regulator of cell death (40). The results of the current study
indicated that esophageal tumor cells exhibited reduced Grim-19
mRNA and protein expression compared with normal esophageal
epithelial cells. Grim-19 has been associated with reactive oxygen
species induced by an IFN/RA pathway and has a conserved
evolutionary history in eukaryotic cells (41,42).
In addition, the present study also demonstrated
that CTL responses and IFN-γ release were significantly enhanced
following rAd-GRIM-19 treatment compared with rAd and PBS
treatments. The enhanced cytotoxic sensitivity of tumor cells
induced by rAd-Grim-19 treatment was also associated with
improvements in the long-term survival and immune memory of mice.
Furthermore, tumor cells from mice treated with rAd-GRIM-19
exhibited higher levels of CD4+ and CD8+ cella ccumulation, which
enhanced immunotherapy in esophageal neoplasm mice. Importantly,
the volume of tumors was significantly reduced in
rAd-GRIM-19-treated mice compared with rAd and PBS-treated tumors,
indicating that stronger immunotherapy may be stimulated by
rAd-GRIM-19 in vivo. Therefore, therapeutic effects against
esophageal neoplasm cells in xenograph mice were achieved through
treatment with rAd-GRIM-19. Based on the results of the present
study, rAd-GRIM-19 may have potential as an anticancer strategy for
future clinical trials for esophageal neoplasm and potentially
other types of cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
JS, WS and SZ analyzed, and interpreted the patient
data regarding the esophageal neoplasm and SZ was the major
contributor in writing the manuscript. WW and YZ performed the
animal experiments in the present study.
Ethics approval and consent to
participate
Both human and animal experiments were approved by
Ethics Committee of the First Affiliated Hospital of Soochow
University. Written informed consent was signed by all
participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Straatman J, Joosten PJ, Terwee CB, Cuesta
MA, Jansma EP and van der Peet DL: Systematic review of
patient-reported outcome measures in the surgical treatment of
patients with esophageal cancer. Dis Esophagus. 29:760–772. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fahey PP, Mallitt KA, Astell-Burt T, Stone
G and Whiteman DC: Impact of pre-diagnosis behavior on risk of
death from esophageal cancer: A systematic review and
meta-analysis. Cancer Causes Control. 26:1365–1373. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yodying H, Matsuda A, Miyashita M,
Matsumoto S, Sakurazawa N, Yamada M and Uchida E: Prognostic
significance of neutrophil-to-lymphocyte ratio and
platelet-to-lymphocyte ratio in oncologic outcomes of esophageal
cancer: A systematic review and meta-analysis. Ann Surg Oncol.
23:646–654. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Parry K, Ruurda JP, van der Sluis PC and
van Hillegersberg R: Current status of laparoscopic transhiatal
esophagectomy for esophageal cancer patients: A systematic review
of the literature. Dis Esophagus. 30:1–7. 2017.PubMed/NCBI
|
|
5
|
Gong J, Huang Z and Huo JR: Involvement of
F-box proteins in esophageal cancer (Review). Int J Oncol.
48:886–894. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tian X, Zhou JG, Zeng Z, Shuai T, Yi LJ,
Ma L, Wang Y, Cao H and Song GM: Cetuximab in patients with
esophageal cancer: A systematic review and meta-analysis of
randomized controlled trials. Med Oncol. 32:1272015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Du D, Song T, Liang X, Fang M and Wu S:
Concurrent chemoradiotherapy with elective lymph node irradiation
for esophageal cancer: A systemic review and pooled analysis of the
literature. Dis Esophagus. 30:1–9. 2017.
|
|
8
|
Goense L, van Rossum PS, Reitsma JB, Lam
MG, Meijer GJ, van Vulpen M, Ruurda JP and van Hillegersberg R:
Diagnostic performance of 18F-FDG PET and PET/CT for the
detection of recurrent esophageal cancer after treatment with
curative intent: A systematic review and meta-analysis. J Nucl Med.
56:995–1002. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chung CS, Lo WC, Lee YC, Wu MS, Wang HP
and Liao LJ: Image-enhanced endoscopy for detection of second
primary neoplasm in patients with esophageal and head and neck
cancer: A systematic review and meta-analysis. Head Neck. 38 Suppl
1:E2343–E2349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Koo J, Wang X, Owonikoko TK, Ramalingam
SS, Khuri FR and Sun SY: GSK3 is required for rapalogs to induce
degradation of some oncogenic proteins and to suppress cancer cell
growth. Oncotarget. 6:8974–8987. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bahlawane C, Eulenfeld R, Wiesinger MY,
Wang J, Muller A, Girod A, Nazarov PV, Felsch K, Vallar L, Sauter
T, et al: Constitutive activation of oncogenic PDGFRα-mutant
proteins occurring in GIST patients induces receptor
mislocalisation and alters PDGFRα signalling characteristics. Cell
Commun Signal. 13:212015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hakimi R: Treatment of a questionable
prostate carcinoma recurrence with oncolytic viruses, dendritic
cells and heat shock proteins in established naturopathy practice.
Versicherungsmedizin. 64:87–88. 2012.(In German). PubMed/NCBI
|
|
13
|
Kalakonda S, Nallar SC, Jaber S, Keay SK,
Rorke E, Munivenkatappa R, Lindner DJ, Fiskum GM and Kalvakolanu
DV: Monoallelic loss of tumor suppressor GRIM-19 promotes
tumorigenesis in mice. Proc Natl Acad Sci USA. 110:pp. E4213–E4222.
2013; View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wen LJ, Gao LF, Jin CS, Zhang HJ, Ji K,
Yang JP, Zhao XJ, Wen MJ and Guan GF: Small interfering RNA
survivin and GRIM-19 co-expression salmonella plasmid inhibited the
growth of laryngeal cancer cells in vitro and in
vivo. Int J Clin Exp Pathol. 6:2071–2081. 2013.PubMed/NCBI
|
|
15
|
Li M, Li Z, Liang C, Han C, Huang W and
Sun F: Upregulation of GRIM-19 suppresses the growth of oral
squamous cell carcinoma in vitro and in vivo. Oncol
Rep. 32:2183–2190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kalakonda S, Nallar SC, Lindner DJ, Sun P,
Lorenz RR, Lamarre E, Reddy SP and Kalvakolanu DV: GRIM-19
mutations fail to inhibit v-Src-induced oncogenesis. Oncogene.
33:3195–3204. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cheng Y, Zhang HY, Zhou Y, Tao F and Yu
YH: Decreased expression of GRIM-19 and its association with
high-risk HPV infection in cervical squamous intraepithelial
neoplasias and cancer. Clin Invest Med. 37:E77–E84. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou W, Luo C, Zhang Z, Liu J, Gu J, Pei Z,
Qian C and Liu X: A novel oncolytic adenovirus targeting to
telomerase activity in tumor cells with potent. Oncogene.
23:457–464. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li G, Jiang P, Li Y, Wang X, Huang J, Du Y
and Zeshan B: Effective suppression of replication of porcine
reproductive and respiratory syndrome virus by adenovirus-mediated
small interfering RNAs targeting ORF1b, 5 and 7 genes. J Virol
Methods. 157:40–46. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hernández-Alcoceba R, Sangro B and Prieto
J: Gene therapy of liver cancer. Ann Hepatol. 6:5–14.
2007.PubMed/NCBI
|
|
21
|
Woodger T: Restrainers in laboratory
animal research. Lab Anim (NY). 45:310–311. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tomioka R: Expression of EGFP by
adenovirus-mediated gene transfer in the central nervous system.
Methods Mol Biol. 515:97–106. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan F, Zheng Y and Huang L:
Adenovirus-mediated combined anti-angiogenic and pro-apoptotic gene
therapy enhances antitumor efficacy in hepatocellular carcinoma.
Oncol Lett. 5:348–354. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chan YS, Wong JH, Fang EF, Pan W and Ng
TB: Isolation of a glucosamine binding leguminous lectin with
mitogenic activity towards splenocytes and anti-proliferative
activity towards tumor cells. PLoS One. 7:e389612012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bustos-Valenzuela JC, Halcsik E, Bassi EJ,
Demasi MA, Granjeiro JM and Sogayar MC: Expression, purification,
bioactivity, and partial characterization of a recombinant human
bone morphogenetic protein-7 produced in human 293T cells. Mol
Biotechnol. 46:118–126. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Greaves MF and Brown G: Purification of
human T and B lymphocytes. J Immunol. 112:420–423. 1974.PubMed/NCBI
|
|
28
|
Zamarin D, Vigil A, Kelly K, Garcia-Sastre
A and Fong Y: Genetically engineered Newcastle disease virus for
malignant melanoma therapy. Gene Ther. 16:796–804. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jang BI and Hwang MJ: Do esophageal
squamous cell carcinoma patients have an increased risk of
coexisting colorectal neoplasms? Gut Liver. 10:6–7. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Law S and Wong J: Lymph node dissection in
surgical treatment of esophageal neoplasms. Surg Oncol Clin N Am.
16:115–131. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Almhanna K, Hoffe S, Strosberg J,
Dinwoodie W, Meredith K and Shridhar R: Concurrent
chemoradiotherapy with protracted infusion of 5-fluorouracil (5-FU)
and cisplatin for locally advanced resectable esophageal cancer. J
Gastrointest Oncol. 6:39–44. 2015.PubMed/NCBI
|
|
32
|
Cottreau J, Gruchy S, Kamionek M, Lauwers
GY and Arnason T: Prevalence of oesophageal epidermoid metaplasia
in 1048 consecutive patients and 58 patients with squamous
neoplasms. Histopathology. 68:988–995. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aksel' EM, Davydov MI and Ushakova TI:
Statistics of lung, stomach and esophageal cancer: Status of
oncological care, morbidity and mortality. Vestn Ross Akad Med
Nauk. 61–65. 2001.(In Russian).
|
|
34
|
Kalakonda S, Nallar SC, Lindner DJ, Hu J,
Reddy SP and Kalvakolanu DV: Tumor-suppressive activity of the cell
death activator GRIM-19 on a constitutively active signal
transducer and activator of transcription 3. Cancer Res.
67:6212–6220. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gortat A, San-Roman MJ, Vannier C and
Schmidt AA: Single point mutation in Bin/Amphiphysin/Rvs (BAR)
sequence of endophilin impairs dimerization, membrane shaping, and
Src homology 3 domain-mediated partnership. J Biol Chem.
287:4232–4247. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kalakonda S, Nallar SC, Gong P, Lindner
DJ, Goldblum SE, Reddy SP and Kalvakolanu DV: Tumor suppressive
protein gene associated with retinoid-interferon-induced mortality
(GRIM)-19 inhibits src-induced oncogenic transformation at multiple
levels. Am J Pathol. 171:1352–1368. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sinkovics JG and Horvath JC: Natural and
genetically engineered viral agents for oncolysis and gene therapy
of human cancers. Arch Immunol Ther Exp (Warsz). 56 Suppl 1:3S–59S.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang MM, Yan LN, Li DH, Gou XH, Liu JW,
Su Z, Han L and Zhao LY: Inhibition of adenovirus-mediated gene
transfer of antisense matrix metalloproteinase-2 on hepatocellular
carcinoma growth in vivo. Zhonghua Gan Zang Bing Za Zhi.
13:671–674. 2005.(In Chinese). PubMed/NCBI
|
|
39
|
Alchanati I, Nallar SC, Sun P, Gao L, Hu
J, Stein A, Yakirevich E, Konforty D, Alroy I, Zhao X, et al: A
proteomic analysis reveals the loss of expression of the cell death
regulatory gene GRIM-19 in human renal cell carcinomas. Oncogene.
25:7138–7147. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chidambaram NV, Angell JE, Ling W, Hofmann
ER and Kalvakolanu DV: Chromosomal localization of human GRIM-19, a
novel IFN-beta and retinoic acid-activated regulator of cell death.
J Interferon Cytokine Res. 20:661–665. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang XY, Li M, Sun K, Chen XJ, Meng J, Wu
L, Zhang P, Tong X and Jiang WW: Decreased expression of GRIM-19 by
DNA hypermethylation promotes aerobic glycolysis and cell
proliferation in head and neck squamous cell carcinoma. Oncotarget.
6:101–115. 2015.PubMed/NCBI
|
|
42
|
Chao L, Wang X, Yang Y, Cui W, Xu J, Chen
H, Hao A and Deng X: Downregulation of gene expression and activity
of GRIM-19 affects mouse oocyte viability, maturation, embryo
development and implantation. J Assist Reprod Genet. 32:461–470.
2015. View Article : Google Scholar : PubMed/NCBI
|