Introduction
Gastric cancer, one of the most common malignant
tumors of the digestive tract, represents a serious health threat.
In China, gastric cancer is the second most common type of cancer,
and the third highest cause of death of malignant tumors (1). Despite the improvements in cancer
treatment in recent years, the prognosis remains unsatisfactory,
especially since there is no biomarker suitable for early diagnosis
(2). Therefore, the identification
of an effective biomarker, elucidation of the underlying mechanisms
of development, and the improvement of the treatment strategies are
necessary.
Recently, microRNAs (miRNAs/miRs; ~22 nucleotides
long), which negatively regulate target gene expression, attracted
a lot of research attention. Aberrant expression of miRNAs has been
identified in numerous cancers, and these molecules can act as
tumor-promoting or suppressor genes. Increasing body of evidence
demonstrates that the abnormal expression of miRNAs may be involved
in the development and progression of cancers in humans (3–5).
Moreover, various miRNAs were shown to be involved in the
development of gastric cancer, including miR-584-5p (6), miR-27a (7), and miR-545 (8). Recently, a novel miRNA, miR-4284, was
identified, and shown to promote the development of diffuse large
B-cell lymphoma (9), in addition
to its anti-tumor effects in glioblastoma (10). However, the expression of miR-4284
and its relationship with clinically observed digestive tract
alterations remain unclear, especially in gastric cancer.
Ten-eleven translocation 1 (TET1), a member of TET
family, was shown to be downregulated in different cancer types,
and to decrease cell proliferation and metastasis in different
cancer types, including breast (11), renal (12), and colon cancers (13). Furthermore, it was shown to
represent a direct target of other miRNAs, such as miR-29a and
miR-520b (14,15).
Therefore, in this study, we aimed to investigate
the expression, functions, and the underlying mechanisms of
miR-4284 in gastric cancer. We analyzed the expression of this
molecule in 40 paired gastric cancer tissue samples, and the
potential correlations with clinical features. Afterward, miR-4284
functions in gastric cancer cells in vitro were further
explored. Finally, we found that TET1 was a direct target of
miR-4284, which elucidated the potential mechanisms underlying the
observed effects.
Materials and methods
Clinical specimens
Forty pairs of frozen gastric cancer and the
corresponding normal tissue samples, preserved at −80°C in our
laboratory, were collected from January 2011 to August 2011 and
underwent pathological examination. All patients were followed-up
for at least 5 years following the tissue collection and received
no anti-tumor treatment before operation. All patients provided
informed consent and the study was approved by ethics committee of
Peking University People's Hospital (Beijing, China).
Cell lines and cultures
Human gastric cancer cell lines, AGS and NCI-N87 and
gastric mucosal normal cell line GES-1 were obtained from ATCC,
SGC-7901, HGC-27, were obtained from Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China), and FU97 was
purchased from JCRB. FU97 cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10 mg/l insulin, while
others were grown in RPMI-1640 medium (Thermo Fisher Scientific,
Inc.). All media were supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.), and the cells were
incubated at 37°C in the atmosphere with 5% CO2.
Cell transfection
The miR-4284 mimics, inhibitors, and negative
control (mimic NC or inhibitor NC) were purchased from Suzhou
GenePharma Co., Ltd. (Suzhou, China). After incubating
8×104 gastric cells in 12-well plates for 16 h, the
cells were transfected with miRNAs using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. The final concentrations of mimics and
inhibitors were 50 nM.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) and reversely transcribed using
transcription kit (Takara Biotechnology Co., Ltd., Dalian, China;
Tiangen, Biotech, Co., Ltd., Beijing, China) according to the
manufacturer's instruction. mRNA was performed with the SYBR-Green
PCR kit (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using the
CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.).
Primers for miR-4284 and U6 were synthesized by Tiangen
(Tiangen, Biotech, Co., Ltd.), while those for TET1 and
GAPDH were synthesized by Sangon Biotech Co., Ltd.
(Shanghai, China). The primers used were: TET1 forward:
5′-CTGGCTCAAACGAGGTCCAT-3′, reverse: 5′-TGCCATCACGTTAGCACACT-3′.
Expression levels were normalized to those of U6 or
GAPDH.
Colony formation
For colony formation assays, 1×103
gastric cancer single-cell suspensions were added to the 6-well
plates and cultured for 2 weeks. Colonies with at least 50 cells
were counted. The experiments were performed three times.
CCK-8 assays
To assess the proliferation, following the treatment
of cells with miRNA mimics or inhibitors for 24 h, 1,500 single
cells in 100 µl of medium were seeded into 96-well plates,
incubated for 1.5 h, after which CCK8 reagents (cat. no. C0038;
Dojindo Molecular Technologies, Kumamoto, Japan) were added.
Proliferation rates at 0, 24, 48, 72, and 96 h were determined by
measuring the absorbance at 450 nm using a microplate reader
(Bio-Rad Laboratories, Inc.). Each group was assayed five
times.
Migration and invasion assays
For migration assays, 4×104 cells in 200
µl of medium with 1% FBS were plated into the upper chamber,
separated by a membrane from the lower chamber (24-well insert;
8-µm pore size; Corning Costar, Corning, NY, USA), which contained
600 µl medium with 10% FBS. After 24 h, the membranes were stained
with 0.1% crystal violet and photographed.
For invasion assays, 8×104 cells were
seeded into the upper chambers. All other conditions remained as
described, except the addition of 50 µl Matrigel on the membranes
and incubation time of 48 h.
Western blot analysis
After washing the samples three times with
phosphate-buffered saline (PBS: Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China), cells were lysed with
radioimmunoprecipitation assay (RIPA) buffer at 4°C and collected.
Sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAGE) and blotting were performed as previously described
(16). Anti-TET1 antibody was
purchased from Santa Cruz Biotechnology, Inc. (1:1,000, cat. no.
sc-293186; Dallas, TX, USA).
miRNA target gene prediction and
Luciferase reporter assays
miR-4284 binding site at TET1 molecule was predicted
by Microrna (http://www.microma.org/). The mRNA
3′-UTR of TET1, which carrying the predicted binding site or mutant
binding site of miR-4284, was amplified by PCR and was inserted
between the XhoI and SacI restriction sites of the
pMIR-GLO™ Luciferase vector (Promega Corporation,
Madison, WI, USA). The cloning procedure was performed by
GenePharma. For Dual-Luciferase assay, cells were seeded into
96-well plates and co-transfected with Pmir-GLO-TET1-3′UTR
Luciferase vector or mutated sequences with 50 nM miR-4284 mimics
or NC. After 24 h incubation, luciferase activity was detected
using dual-luciferase reporter assay system (Promega., Ltd.,
Shanghai, China). Relative luciferase activities were normalized to
Renilla Luciferase activity levels.
Statistical analysis
All data were expressed as means ± standard
deviation (SD) and were analyzed using SPSS 18.0. Differences
between clinicopathological variables were assessed using
χ2 test analysis; the biological variables between the
groups were compared using Student's t-test; Multiple comparisons
using One-way ANOVA (post hoc is LSD). The overall survival was
analyzed by Kaplan-Meier method and the log-rank test and the
median was used to define the thresholds for miR-4284 expression.
P<0.05 was considered to indicate a statistically significant
difference.
Results
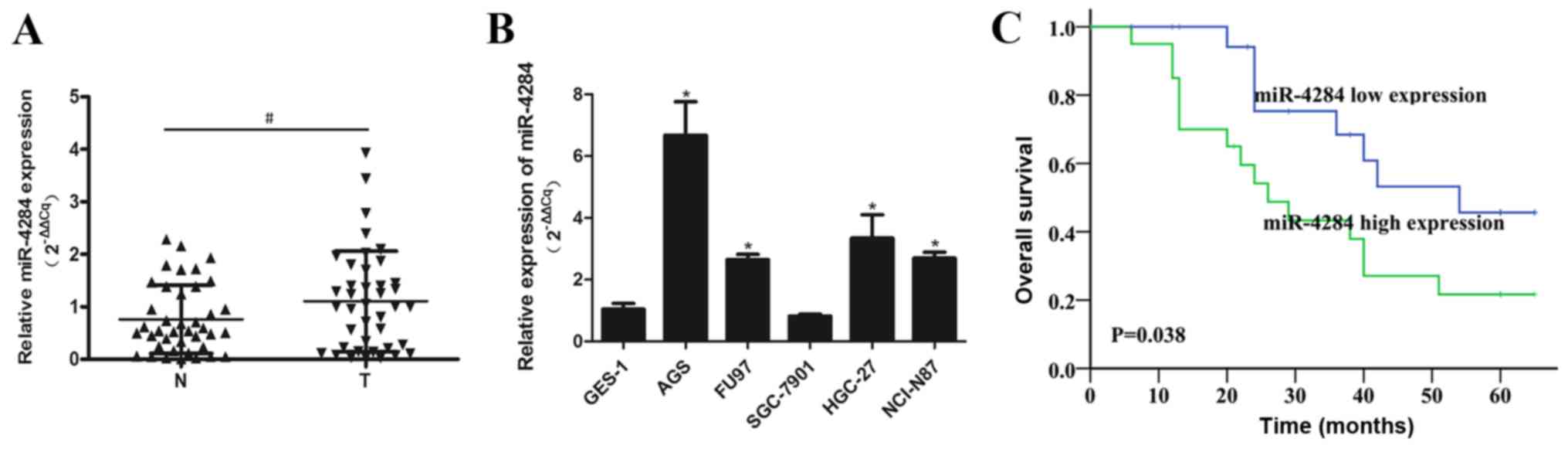
miR-4284 expression is upregulated in
gastric cancer tissues
miR-4284 expression was shown to be significantly
upregulated in gastric cancer tissues, compared with that in the
corresponding normal tissues (Fig.
1A). The levels of this miRNA were increased in gastric cancer
cells (AGS, FU97, HGC-27 and NCI-N87) in vitro, than those
in the normal GES-1 cells, with the exception of SGC-7901 cells
(Fig. 1B). Furthermore,
Kaplan-Meier analysis of the relationship between miR-4284
expression and gastric cancer patient prognosis showed that high
miR-4284 expression correlates with a significant decrease in
patient survival rate (P=0.038; Fig.
1C). Increased miR-4284 expression was shown to be
significantly associated with TNM stage (P=0.035) and distant
metastasis rate (P=0.022), but was not associated with age, gender,
tumor size, differentiation, or lymph node metastasis in gastric
cancer patients (Table I).
 | Table I.Patient characteristics and miR-4284
expression in gastric cancer tissues. |
Table I.
Patient characteristics and miR-4284
expression in gastric cancer tissues.
| Factors | No. of patients | miR-4284 expression
(mean ± SD) | P-value |
|---|
| Age (year) |
|
| 0.459 |
| ≤60 | 13 | 0.94±0.78 |
|
|
>60 | 27 | 1.18±1.03 |
|
| Sex |
|
| 0.837 |
| Male | 26 | 1.13±1.01 |
|
|
Female | 14 | 1.06±0.87 |
|
| Tumor size (cm) |
|
| 0.338 |
| ≤4 | 20 | 1.25±1.10 |
|
|
>4 | 20 | 0.96±0.79 |
|
| Tumor
differentiation |
|
| 0.456 |
|
Well/Moderate | 13 | 1.27±0.95 |
|
| Poor | 27 | 1.02±0.96 |
|
| TNM stage |
|
|
0.035a |
|
I+II | 18 | 0.77±0.60 |
|
|
III+IV | 22 | 1.38±1.11 |
|
| Lymph node
metastasis |
|
| 0.648 |
|
Positive | 17 | 1.02±0.92 |
|
|
Negative | 23 | 1.16±1.00 |
|
| Distant
metastasis |
|
|
0.022a |
|
Positive | 5 | 2.01±0.51 |
|
|
Negative | 35 | 0.98±0.94 |
|
miR-4284 promotes gastric cancer cell
proliferation, invasion, and migration
To assess miR-4284 effects in gastric cancer cells,
this molecule was overexpressed in SGC-7901 cells using miR-4284
mimics, and the efficiency of overexpression was confirmed by
RT-qPCR analysis (Fig. 2A;
P<0.05). The proliferation of SGC-7901 cells was shown to be
significantly increased following the treatment with miR-4284,
compared with that in cells treated with the NC (Fig. 2B and C; P<0.05). Additionally,
increased miR-4284 expression considerably enhanced the ability of
migration and invasion of SGC-7901 cells compared with the NC
(Fig. 2D; P<0.05).
Decreased miR-4284 expression inhibits
gastric cancer cell proliferation, invasion, and migration
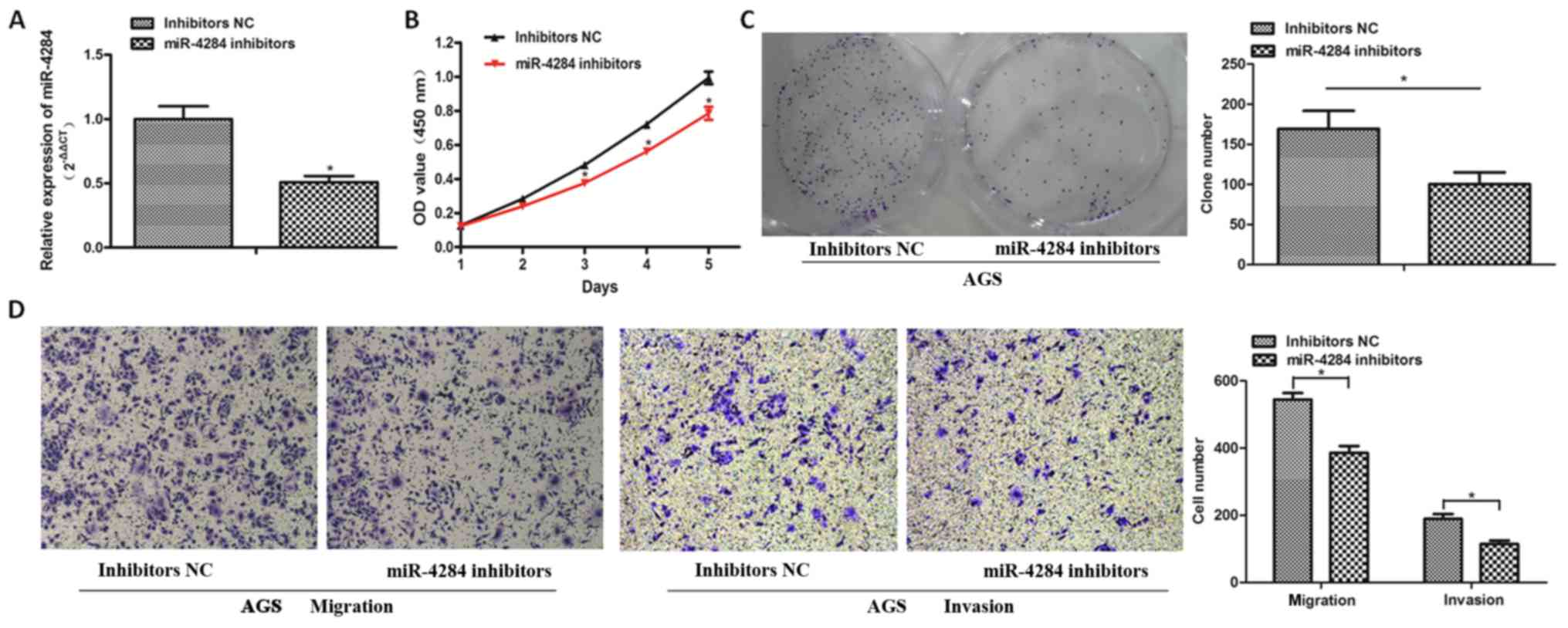
To explore the role of miR-4284 in gastric cancer
further, we inhibited the expression of miR-4284 in AGS cells using
inhibitors, which was confirmed by RT-qPCR analysis (Fig. 3A; P<0.05). CCK-8 and colony
formation assays showed that the proliferation of AGS cells
decreases following the suppression of miR-4284 expression,
compared with that in the NC group (Fig. 3B and C; P<0.05). The migration
and invasion of AGS cells treated with miR-4284 inhibitors were
significantly inhibited (Fig. 3D;
P<0.05).
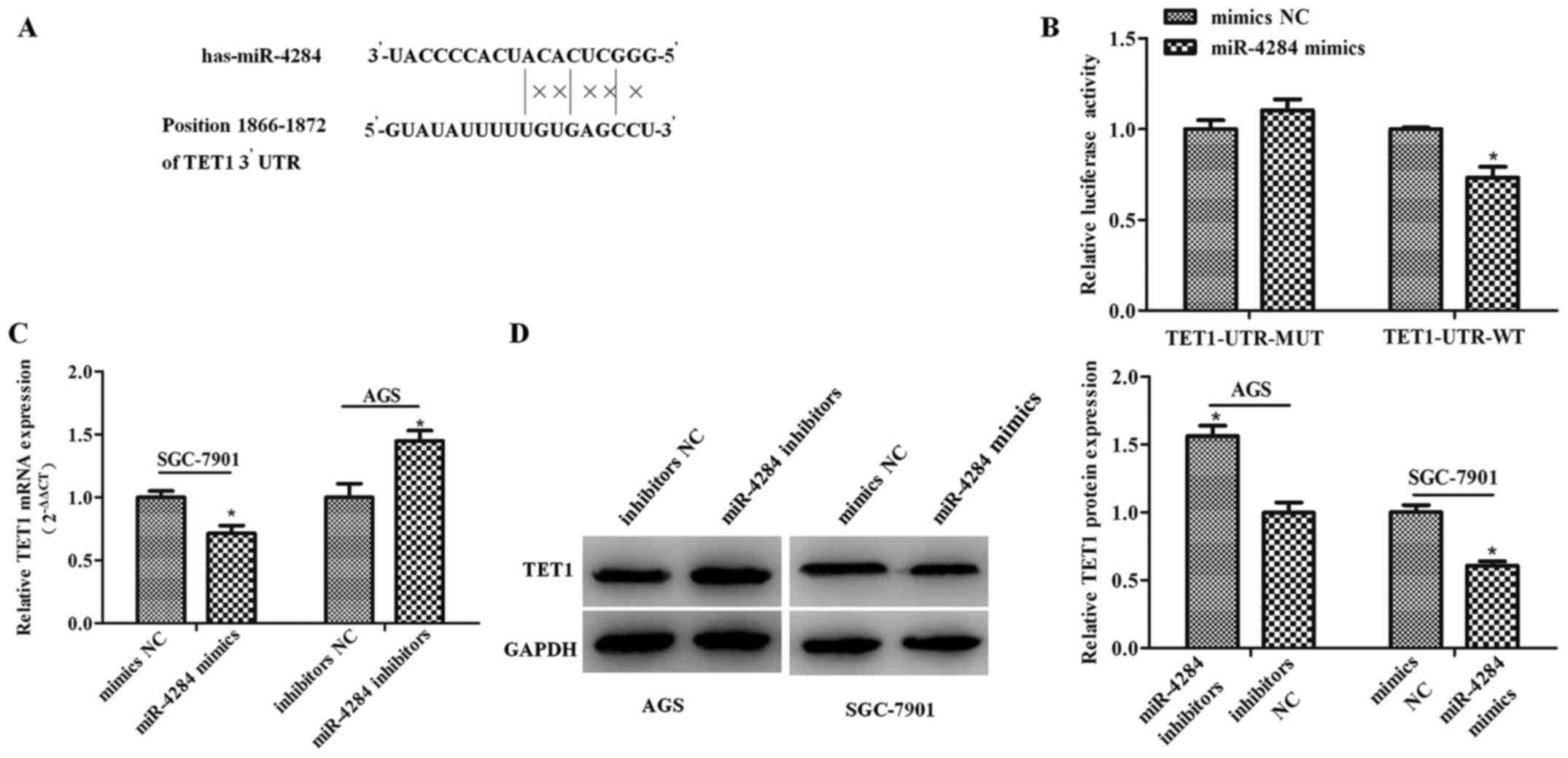
TET1 is a direct miR-4284 target
To elucidate the molecular mechanisms underlying
miR-4284 effects on the proliferation and migration of gastric
cancer cells, we employed microRNA.org to
identify miR-4284 target genes, which led to the identification of
TET1 as a direct target gene. Therefore, we cloned the
wild-type and mutant TET1 3′UTR sequences into a
dual-luciferase reporter (Fig.
4A), which showed that miR-4284 induces a significant decrease
in the relative luciferase activity of wild-type TET1 3′-UTR
(TET1-UTR-WT) (Fig. 4B;
P<0.05), compared with the control, whereas this activity in the
mutant group was not affected (Fig.
4B; P>0.05). Furthermore, TET1 mRNA and protein expression
following the treatment with miR-4284 mimics or inhibitors was
determined, showing that miR-4284 inhibits TET1 mRNA (Fig. 4C; P<0.05) and protein expression
(Fig. 4D; P<0.05).
The correlation between miR-4284 and
TET1 expression
We showed that the expression of TET1 in gastric
cancer tissue samples is significantly decreased compared with that
in the adjacent normal tissue samples (Fig. 5A; P<0.05). Pearson correlation
analysis showed a negative correlation between miR-4284 and TET1
expression levels in gastric cancer tissues (Fig. 5B; r=−0.319, P<0.05).
Discussion
In most gastric cancer cases, the cancer is already
in an advanced stage when diagnosed, with unsatisfactory prognosis
(17). The underlying molecular
mechanisms remain unclear, and no current prognostic biomarker is
effective.
The dysregulation of miRNA expression was shown to
play important roles in gastric cancer development, by affecting
cell proliferation, invasion, and migration (18–20).
Furthermore, miR-4284 was shown to be involved in physiological and
pathological process, including diffuse large B-cell lymphoma and
glioblastoma development (9,10).
However, the role of miR-4284 in gastrointestinal tumors,
especially in gastric cancer, has not been fully elucidated, which
is why we focused on determining the levels and biological
functions of this molecule in gastric cancer. We showed that
miR-4284 expression is significantly upregulated in gastric cancer
tissues in comparison with that in the matched normal tissues,
suggesting that miR-4284 may be a tumor-driving factor in gastric
cancer. Furthermore, our results show that high miR-4284 expression
in gastric cancer correlates with TNM stage, distant metastases,
and poor prognosis, indicating that miR-4284 may be a prognostic
and potentially an early diagnostic biomarker in gastric
cancer.
Furthermore, we analyzed miR-4284 expression levels
in five gastric cancer cell lines and in one normal gastric mucosa
epithelial cell line (GES-1), which showed that these levels are
significantly higher in all gastric cancer cells, except SGC-7901
cells, compared with those in GES-1 cells. We further overexpressed
miR-4284 in SGC-7901 cells, and inhibited its expression in AGS
cells, which were shown to have highest levels of miR-4284 in all
five gastric cancer cell lines. Increase in miR-4284 expression
significantly induced gastric cancer cell proliferation, while the
decrease in miR-4284 expression significantly inhibited it. Since
enhanced cell migration leads to tumor metastasis, this represents
a major factor affecting cancer prognosis (21). Here, cell invasion and migration
assays showed that the treatment of SGC-7901 cells with miR-4284
mimics and AGS cells with miR-4284 inhibitors led to an increase
and decrease, respectively, in invasiveness and migratory rate of
these cells. To the best of our knowledge, this is the first study
showing the roles of miR-4284 in the development of gastric
cancer.
The effects of miRNAs are exerted primarily through
the binding to the tumor-related genes, inhibiting their expression
(22). Therefore, we aimed to
identify potential miR-4284 target genes, and among a number of
potential targets, we focused on TET1, which was previously shown
to be a tumor suppressor. Here, TET1 expression in gastric cancer
tissues was shown to be significantly lower than that in the
corresponding normal tissues, consistent with previous studies
(23–25). To ascertain whether TET1 was a
direct target of miR-4284, luciferase reporter assay was performed,
and we showed that miR-4284 overexpression significantly decreased
the luciferase activity in the wild-type TET1 group, which
was not observed in the mutant group. Additionally, we
overexpressed miR-4284 in SGC-7901 cells and inhibited miR-4284
expression in AGS cells, which led to a significant decrease and
increase, respectively, in TET1 levels. TET1 expression was shown
to correlate negatively with miR-4284 levels in gastric cancer
tissues. These findings suggest that miR-4284 may contribute to
gastric cancer progression by targeting TET1, which is the first
time this potential mechanism has been described.
In conclusion, the results obtained here show
miR-4284 expression is significantly upregulated in gastric cancer
tissues and cells, and that this molecule may represent a novel
predictive and prognostic biomarker for gastric cancer. Moreover,
we elucidated miR-4284 roles in cell proliferation and migration.
However, further research, confirming miR-4284 as a potential
therapeutic target, is required.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Science Foundation of China (grant no. s 81372290,
81572379 and 81672375).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SW and ZLS conceived and designed the study. YSL
performed the experiments. HPJ, ZYL and ZW collected and analyzed
the clinical data. KWJ and YJY analyzed and interpreted the data.
YSL wrote the manuscript.
Ethics approval and consent to
participate
All patients provided their informed consent and the
study was approved by Ethics Committee of Peking University
People's Hospital.
Consent for publication
All patients provided their informed consent for
publication of the data.
Competing interest
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kim TH and Shivdasani RA: Stomach
development, stem cells and disease. Development. 143:554–565.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Han X, Wang X, Zhao B, Chen G, Sheng Y,
Wang W and Teng M: MicroRNA-187 inhibits tumor growth and
metastasis via targeting of IGF-1R in hepatocellular carcinoma. Mol
Med Rep. 16:2241–2246. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hou R, Wang D and Lu J: MicroRNA-10b
inhibits proliferation, migration and invasion in cervical cancer
cells via direct targeting of insulin-like growth factor-1
receptor. Oncol Lett. 13:5009–5015. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lu S, Wang MS, Chen PJ, Ren Q and Bai P:
miRNA-186 inhibits prostate cancer cell proliferation and tumor
growth by targeting YY1 and CDK6. Exp Ther Med. 13:3309–3314. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Q, Li Z, Wei S, Wang W, Chen Z, Zhang
L, Chen L, Li B, Sun G, Xu J, et al: Overexpression of miR-584-5p
inhibits proliferation and induces apoptosis by targeting WW
domain-containing E3 ubiquitin protein ligase 1 in gastric cancer.
J Exp Clin Cancer Res. 36:592017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu F, Li J, Guo N, Wang XH and Liao YQ:
miRNA-27a promotes the proliferation and invasion of human gastric
cancer MGC803 cells by targeting SFRP1 via Wnt/β-catenin signaling
pathway. Am J Cancer Res. 7:405–416. 2017.PubMed/NCBI
|
|
8
|
Huang X and Lu S: MicroR-545 mediates
colorectal cancer cells proliferation through up-regulating
epidermal growth factor receptor expression in HOTAIR long
non-coding RNA dependent. Mol Cell Biochem. 431:45–54. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tamaddon G, Geramizadeh B, Karimi MH,
Mowla SJ and Abroun S: miR-4284 and miR-4484 as putative biomarkers
for diffuse large B-cell lymphoma. Iran J Med Sci. 41:334–339.
2016.PubMed/NCBI
|
|
10
|
Yang F, Nam S, Brown CE, Zhao R, Starr R,
Ma Y, Xie J, Horne DA, Malkas LH, Jove R and Hickey RJ: A novel
berbamine derivative inhibits cell viability and induces apoptosis
in cancer stem-like cells of human glioblastoma, via up-regulation
of miRNA-4284 and JNK/AP-1 signaling. PLoS One. 9:e944432014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang L, Yu SJ, Hong Q, Yang Y and Zhao ZM:
Reduced expression of TET1, TET2, TET3 and TDG mRNAs are associated
with poor prognosis of patients with early breast cancer. PLoS One.
10:e01338962015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan M, He X and Xu X: Restored expression
levels of TET1 decrease the proliferation and migration of renal
carcinoma cells. Mol Med Rep. 12:4837–4842. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Neri F, Dettori D, Incarnato D, Krepelova
A, Rapelli S, Maldotti M, Parlato C, Paliogiannis P and Oliviero S:
TET1 is a tumour suppressor that inhibits colon cancer growth by
derepressing inhibitors of the WNT pathway. Oncogene. 34:4168–4176.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang W, Lu Z, Gao Y, Ye L, Song T and
Zhang X: miR-520b suppresses proliferation of hepatoma cells
through targeting ten-eleven translocation 1 (TET1) mRNA. Biochem
Biophys Res Commun. 460:793–798. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pei YF, Lei Y and Liu XQ: miR-29a promotes
cell proliferation and EMT in breast cancer by targeting ten eleven
translocation 1. Biochim Biophys Acta. 1862:2177–2185. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang B, Shen ZL, Jiang KW, Zhao G, Wang
CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, et al: MicroRNA-217
functions as a prognosis predictor and inhibits colorectal cancer
cell proliferation and invasion via an AEG-1 dependent mechanism.
BMC Cancer. 15:4372015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ang TL, Khor CJ and Gotoda T: Diagnosis
and endoscopic resection of early gastric cancer. Singapore Med J.
51:93–100. 2010.PubMed/NCBI
|
|
18
|
Gu H, Yang T, Fu S, Chen X, Guo L and Ni
Y: MicroRNA-490-3p inhibits proliferation of A549 lung cancer cells
by targeting CCND1. Biochem Biophys Res Commun. 444:104–108. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li N, Miao Y, Shan Y, Liu B, Li Y, Zhao L
and Jia L: miR-106b and miR-93 regulate cell progression by
suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell
Death Dis. 8:e27962017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang B, Wu H, Chai C, Lewis J, Pichiorri
F, Eisenstat DD, Pomeroy SL and Leng RP: MicroRNA-1301 suppresses
tumor cell migration and invasion by targeting the p53/UBE4B
pathway in multiple human cancer cells. Cancer Lett. 401:20–32.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eccles SA and Welch DR: Metastasis: Recent
discoveries and novel treatment strategies. Lancet. 369:1742–1757.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peter ME: Targeting of mRNAs by multiple
miRNAs: The next step. Oncogene. 29:2161–2164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fu HL, Ma Y, Lu LG, Hou P, Li BJ, Jin WL
and Cui DX: TET1 exerts its tumor suppressor function by
interacting with p53-EZH2 pathway in gastric cancer. J Biomed
Nanotechnol. 10:1217–1230. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Frycz BA, Murawa D, Borejsza-Wysocki M,
Marciniak R, Murawa P, Drews M, Kołodziejczak A, Tomela K and
Jagodziński PP: Decreased expression of ten-eleven translocation 1
protein is associated with some clinicopathological features in
gastric cancer. Biomed Pharmacother. 68:209–212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pei YF, Tao R, Li JF, Su LP, Yu BQ, Wu XY,
Yan M, Gu QL, Zhu ZG and Liu BY: TET1 inhibits gastric cancer
growth and metastasis by PTEN demethylation and re-expression.
Oncotarget. 7:31322–31335. 2016. View Article : Google Scholar : PubMed/NCBI
|