Introduction
Lung cancer, with a high incidence rate and poor
prognosis, is reported to be one of the most frequently-occurring
aggressive tumors, and is a principal cause of human mortality
around the world (1). Due to the
high invasiveness and metastatic ability, the 5-year survival rate
of lung cancer is <15%, which is lower than most common cancers
(2). In particular, non-small cell
lung cancer (NSCLC) accounts for >80% of all lung cancer cases
(3). Although it is frequently
considered to be ineffective or excessively toxic, the primary
therapeutic treatment for NSCLC is chemotherapy (4). Intensive research is required to
identify novel, effective and low-toxicity therapeutic methods.
Given the development of targeted agents which are able to target
specific molecular pathways in tumor cells, molecular targeted
therapy has become a promising therapeutic option for various types
of cancer. Thus, research into specific targets for NSCLC has
become a focus in recent decades.
MicroRNAs (miRNAs/miRs) are single-stranded small
(~24 nucleotides) non-coding endogenous RNAs that regulate the
expression of genes in a variety of cellular processes, by binding
to the 3′ untranslated region (UTR) of their target mRNAs for
degradation and translation suppression (5). A previous study indicated the
important roles of miRNAs in carcinogenesis (6). The miR-29 family is aberrantly
expressed in NSCLC cells. According to a previous study, the miR-29
family serves a notable role in lung tumor cellular processes
associated with poor prognosis (7). miR-29c is a miR-29 family member
(7). It was reported that miR-29c
was observably underexpressed in NSCLC tumor tissues, illustrating
that miR-29c may be able to suppress NSCLC tumorigenesis (8,9).
The growth of tumors is accompanied by stimulation
of angiogenesis. One of the primary factors in tumor angiogenesis
stimulation is vascular endothelial growth factor A (VEGFA)
(10). According to previous
research, high expression of VEGF is associated with the
proliferation and metastasis of several types of cancer (11). Through bioinformatics methods, it
was identified that VEGFA may be a gene target for miR-29c.
However, studies focusing on whether miR-29c is able to regulate
cell proliferation and cellular apoptosis in NSCLC tumors by
targeting VEGFA have not yet been reported.
In the present study, aberrant underexpression of
miR-29c and overexpression of VEGFA in NSCLC tumor tissues was
observed. Clinical sample investigation and fundamental research
were combined to examine the correlation between the expression of
miR-29c and VEGFA, and the regulatory mechanism of action of
miR-29c on NSCLC tumor progression.
Materials and methods
Specimens
A total of 30 cases of NSCLC tumor tissue samples
and the corresponding para-carcinoma tissue samples were obtained
from patients with NSCLC who were treated in Linyi People's
Hospital from January 2014 to January 2016; there were a total of
17 male and 13 female patients, and the mean age was
51±14-years-old. All the collected cases were pathologically
diagnosed as NSCLC without any preoperative radiotherapy and/or
chemotherapy. The reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) method was applied for the measurement of
the expression of miR-29c and VEGFA in the collected NSCLC tumor
tissue samples and the corresponding para-carcinoma tissue samples.
The correlation between miR-29c and VEGFA expression was
statistically analyzed. The present study was approved by the
ethics committee of Linyi People's Hospital (Linyi, China). Written
informed consent was obtained from all patients.
Cell culture
Human NSCLC cell lines including A549, NCI-H1299 and
H1650 were cultivated in RPMI-1640 medium (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine
serum (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in an
incubator (5% CO2, humidified). The RT-qPCR method was
applied for the measurement of the expression of miR-29c in the
above cell lines. The cell line with the lowest miR-29c expression
was chosen for the following experiments.
Cell transfection
Logarithmic growth phase NCI-H1299 cells were
trypsinized, washed and seeded in 96-well plates at a density of
8×103 cells/well, prior to being transfected with 10 nM
miR-29c mimics or miR-negative control (NC) mimics using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. Cells
were separated into three groups: i) Control group; ii) miR-29c
mimic group (transfected with miR-29c mimics); and iii) NC group
(transfected with miR-NC). Oligonucleotide sequences of miR-29c
mimic and miR-NC were as listed: miR-29c mimics forward,
5′-UAGCACCAUUUGAAAUCGGUUA-3′, reverse 5′-UAACCGAUUUCAAAUGGUGCUA-3′;
miR-NC forward, 5′-UCACAACCUCCUAGAAAGAGUAGA-3′ and reverse,
5′-UCUACUCUUUCUAGGAGGUUGUGA-3′. The oligonucleotides were
chemosynthesized at Extended Nature Biotechnology Co., Ltd.
(Shanghai, China). Total RNA was collected following 72 h. The
RT-qPCR method was applied to detect the transfection efficiency in
the three groups.
Cell proliferation analysis
A total of 72 h following cell transfection, an MTT
assay (Invitrogen; Thermo Fisher Scientific, Inc.) was used to
monitor the cell proliferation rate of each group. NCI-H1299 cells
were first washed with buffer (PBS, pH 7.4), trypsinized, washed,
counted and reseeded into a 96-well plate at a density of
1×104 cells/100 µl/well. A total of 10 µl MTT reagent
was added and the plate was cultured at 37°C in an incubator (5%
CO2, humidified) until a purple precipitate appeared.
Thereafter, 100 µl detergent solution (dimethyl sulfoxide) was then
used to dissolve the formazan crystals and the plate was incubated
at 37°C for 2 h away from light. Absorbance was detected at 570 nm
using a microplate reader.
Cellular apoptosis analysis
A total of 72 h following cell transfection, flow
cytometry with the Annexin V-fluorescein isothiocyanate
(FITC)/propidium iodide (PI) staining method was employed to
monitor the cellular apoptosis in each group. NCI-H1299 cells were
washed, trypsinized and resuspended in the staining solution
provided with the Annexin V-FITC Apoptosis Detection kit
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Following incubation for 1 h at 37°C,
cellular apoptosis was measured with a flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA). Cells with a positive
Annexin V-FITC signal and a negative PI signal were regarded as
apoptotic cells. The cell number at each phase was analyzed using
FloJo software version 7.6.3 (FloJi LLC, Ashland, OR, USA).
RT-qPCR analysis
Total RNA was extracted from cells using a miRNeasy
Mini kit (Qiagen GmbH, Hilden, Germany), according to the
manufacturer's protocol. The reverse transcription of VEGFA RNA and
miR-29c-3p was conducted using a TaqMan MicroRNA RT kit
(Invitrogen; Thermo Fisher Scientific, Inc.) at 42°C for 1 h,
according to the manufacturer's protocol. miScript SYBR®
Green PCR kit (Qiagen, Inc., Valencia, CA, USA) was employed for
RT-qPCR analysis, the amplification conditions were: Total volume
20 µl, initial denaturation 95°C for 10 min, then 45 cycles 95°C
for 15 sec, 60°C for 1 min, which was performed using a 7900HT Fast
Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). The The sequences of the primers used were: miR-29c-3p
forward, 5′-ACACTCCAGCTGGGTAGCACCATTTGA-3′, reverse,
5′-TGGTGTCGTGGAGTCG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; VEGFA forward,
5′-TTTCTGCTGTCTTGGGTGCATTGG-3′, reverse,
5′-ACCACTTCGTGATGATTCTGCCCT-3′ and GAPDH forward,
5′-ACACCCACTCCTCCACCTTT-3′ and reverse 5′-TTACTCCTTGGAGGCCATGT-3′.
U6 small RNA served as the internal control material for
miR-29c-3p, and the relative expression level of VEGFA RNA was
normalized to GAPDH using the 2−ΔΔCq quantification
cycle method (12).
Western blot analysis
Total proteins were extracted from cells using T-PER
Protein Extraction Reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Protein
concentration was determined using a Bicinchoninic Acid kit
(Beyotime Institute of Biotechnology, Shanghai, China). Proteins
(15 µg/lane) were resolved by 10% SDS-PAGE and transferred to a
polyvinylidene fluoride (PVDF) membrane. Subsequently, the PVDF
membrane was blocked in 5% non-fat milk in Tris Buffer Saline
containing 0.1% Tween-20, at room temperature for 1 h and blotted
with antibodies against GAPDH (1:1,000; cat. no. 5174; Cell
Signaling Technology, Inc. Danvers, MA, USA), VEGFA (1:1,000; cat.
no. 8065; Cell Signaling Technology, Inc.), phosphorylated (p)-PI3K
(1:1,000; ab151549; Abcam, Cambridge, MA, USA) and p-Akt (1:1,000;
cat. no. 4060; Cell Signaling Technology, Inc.) at 4°C overnight.
GAPDH served as the internal control material. Chemiluminescence
via an Echo-chemiluminescence Detection system (GE Healthcare,
Chicago, IL, USA) was measured following incubation with
anti-rabbit immunoglobulin G secondary antibody conjugated to
horseradish peroxidase (1:1,000; cat. no. 7074; Cell Signaling
Technology, Inc.) in room temperature for 2 h. The relative
expression of VEGFA, p-PI3K and p-Akt proteins was evaluated
statistically by Quantity One 4.6.2 (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Dual-luciferase reporter analysis
Through bioinformatics methods using TargetScan 7.1
(http://www.targetscan.org), it was
identified that VEGFA may be a target gene for miR-29c. Two types
of 3′UTR fragments (wild type and mutant type) of VEGFA were
inserted into the XhoI-PmeI restriction sites of the 3′UTR of the
Renilla luciferase gene of the dual-luciferase reporters psiCHECK2
(Promega Corporation, Madison, WI, USA) to obtain the wild type
reporter, VEGFA 3′UTR-WT, and the mutant reporter, VEGFA 3′UTR-Mut.
Cells (1×105 cells/well) were transfected with 50 ng
vector, 10 nM miR-29c-3p, and 1 µl Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.) in 100 µl Opti-Minimum Essential
medium (Gibco; Thermo Fisher Scientific, Inc.). At 72 h
post-transfection, luciferase activity of Firefly and Renilla was
monitored using a Dual-Luciferase Reporter Assay System (Promega
Corporation), according to the manufacturer's protocol. The final
results were normalized to the Renilla luciferase and analyzed
statistically.
Statistical analysis
All the above experiments were verified with
triplicate repetition. The final data were analyzed statistically
using SPSS 19.0 software (IBM Corp., Armonk, NY, USA). All data are
presented in the form of the mean ± standard deviation. Spearman's
correlation analysis was used to analyze the correlation between
VEGFA and miR-29c-3p. Comparisons between two groups were performed
using a Student's t-test, and differences among three groups were
analyzed via one-way analysis of variance followed by the
Student-Newman-Keuls test. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-29c and VEGFA expression in NSCLC
tumor tissues are negatively correlated
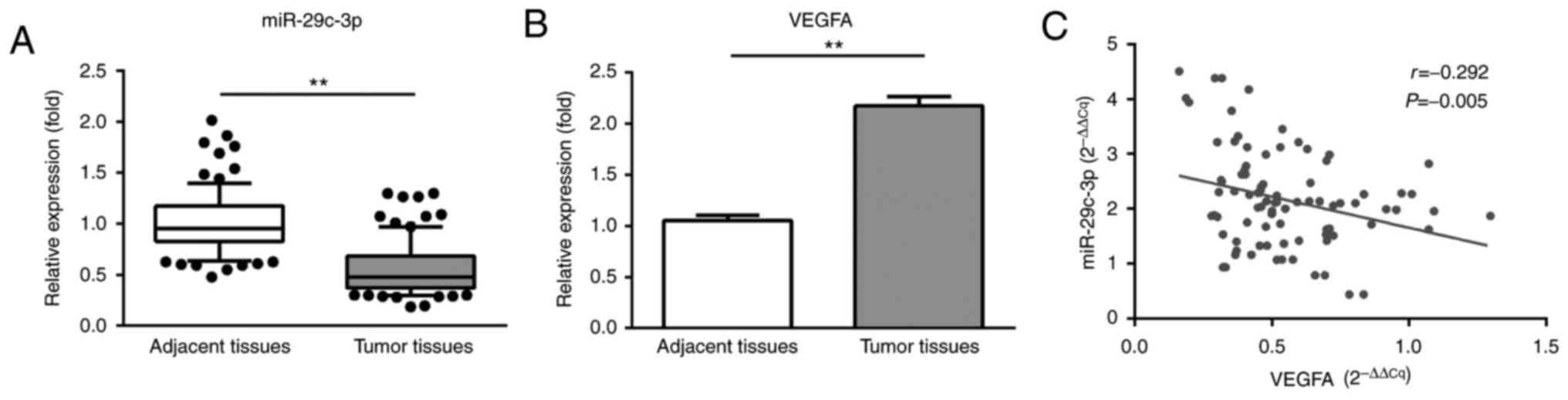
In order to examine the expression patterns of
miR-29c and VEGFA in NSCLC tumor tissues, 30 specimens from NSCLC
cases with their corresponding para-carcinoma normal tissues were
analyzed using the RT-qPCR method. As presented in Fig. 1, the analytical results indicated a
significant downregulation of miR-29c and a significant
upregulation of VEGFA in NSCLC tumor tissues, compared with the
corresponding normal tissues. A significant negative correlation
between the expression levels of miR-29c and VEGFA was observed in
NSCLC tumor tissues (P=0.005).
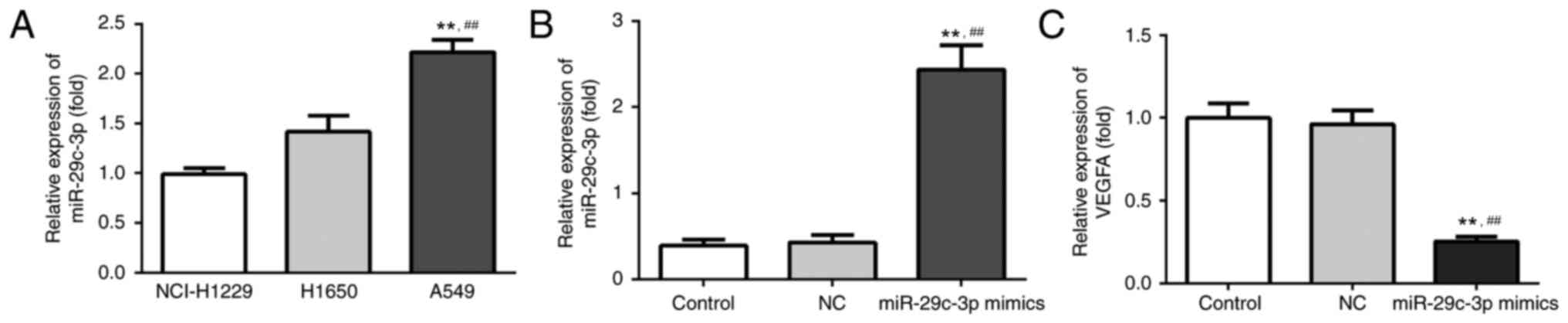
miR-29c expression in lung cancer cell
lines and VEGFA expression following transfection
In order to select the NSCLC cell line with the
lowest expression level of miR-29c-3p for further experiments, the
RT-qPCR method was employed to compare miR-29c expression levels in
the human NSCLC cell lines A549, NCI-H1299 and H1650. As presented
in Fig. 2A, the NCI-H1299 cell
line had the lowest relative expression level of miR-29c, and was
chosen for subsequent experiments. The relative expression of VEGFA
in three groups of NCI-H1299 cells was measured following
transfection. The results demonstrated no significant difference
between the VEGFA expression levels of the control and NC groups
(Fig. 2B and C). However, the
results demonstrated an upregulation of the expression of
miR-29c-3p (Fig. 2B) and a
downregulation of the expression of VEGFA (Fig. 2C) in the miR-29c mimic group, which
further confirmed the negative correlation between the expression
levels of miR-29c and VEGFA.
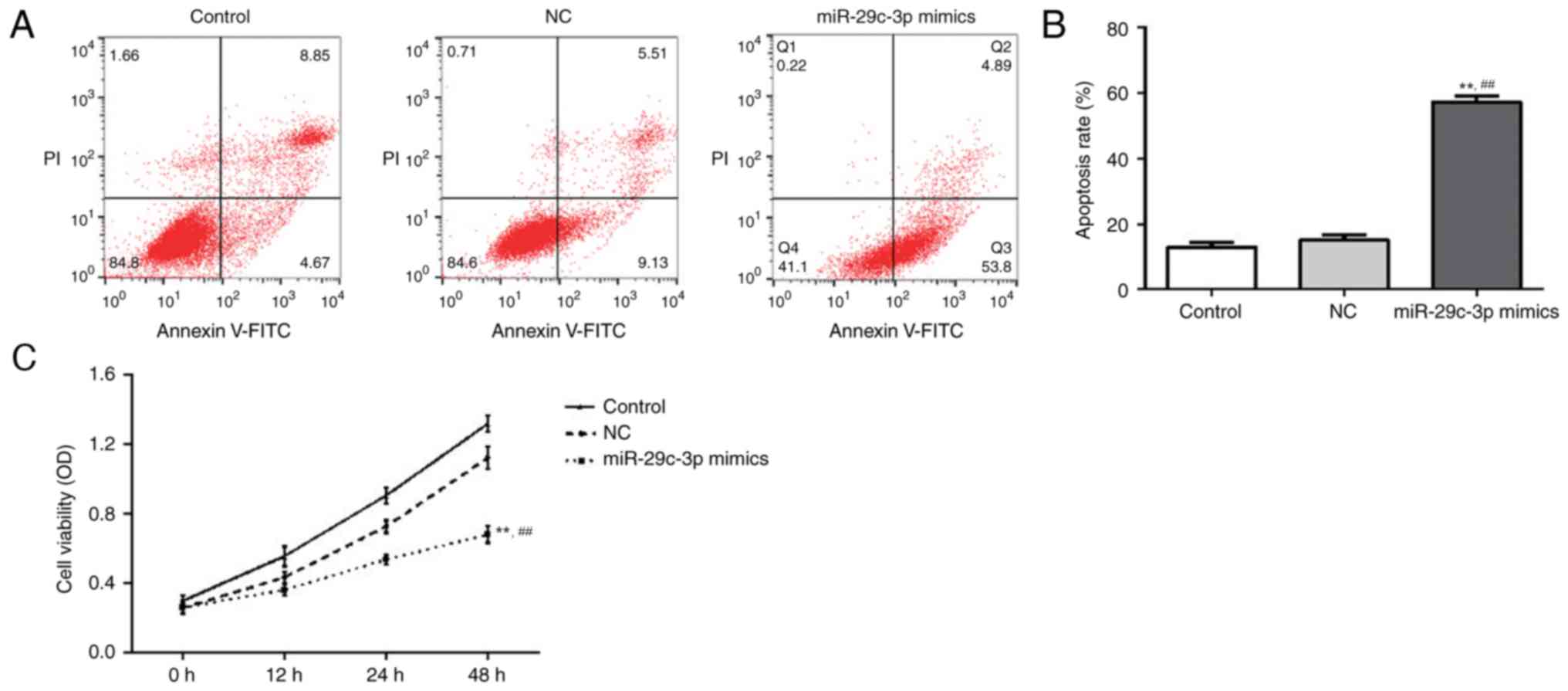
miR-29c expression affects lung cancer
cell proliferation and apoptosis
The influence of miR-29c expression on NSCLC cell
proliferation and apoptosis was further investigated using an MTT
assay and Annexin V-FITC apoptosis assay. From the apoptosis assay
results (Fig. 3A), the cellular
apoptosis rates of the control group, NC group and the miR-29c
mimic group were demonstrated to be 4.67, 9.13 and 53.8%,
respectively. The cellular apoptosis rate of miR-29c mimic group
was significantly higher compared with the other two groups
(Fig. 3B). This significant
difference indicated that the overexpression of miR-29c may
facilitate NSCLC cellular apoptosis. The growth curves from the MTT
assay illustrated the influence of miR-29c on NSCLC cell
proliferation (Fig. 3C). At 48 h,
there was a clear suppressive effect on cell viability in the
miR-29c mimic group compared with the other two groups, meaning
that the overexpression of miR-29c was able to inhibit NSCLC cell
proliferation.
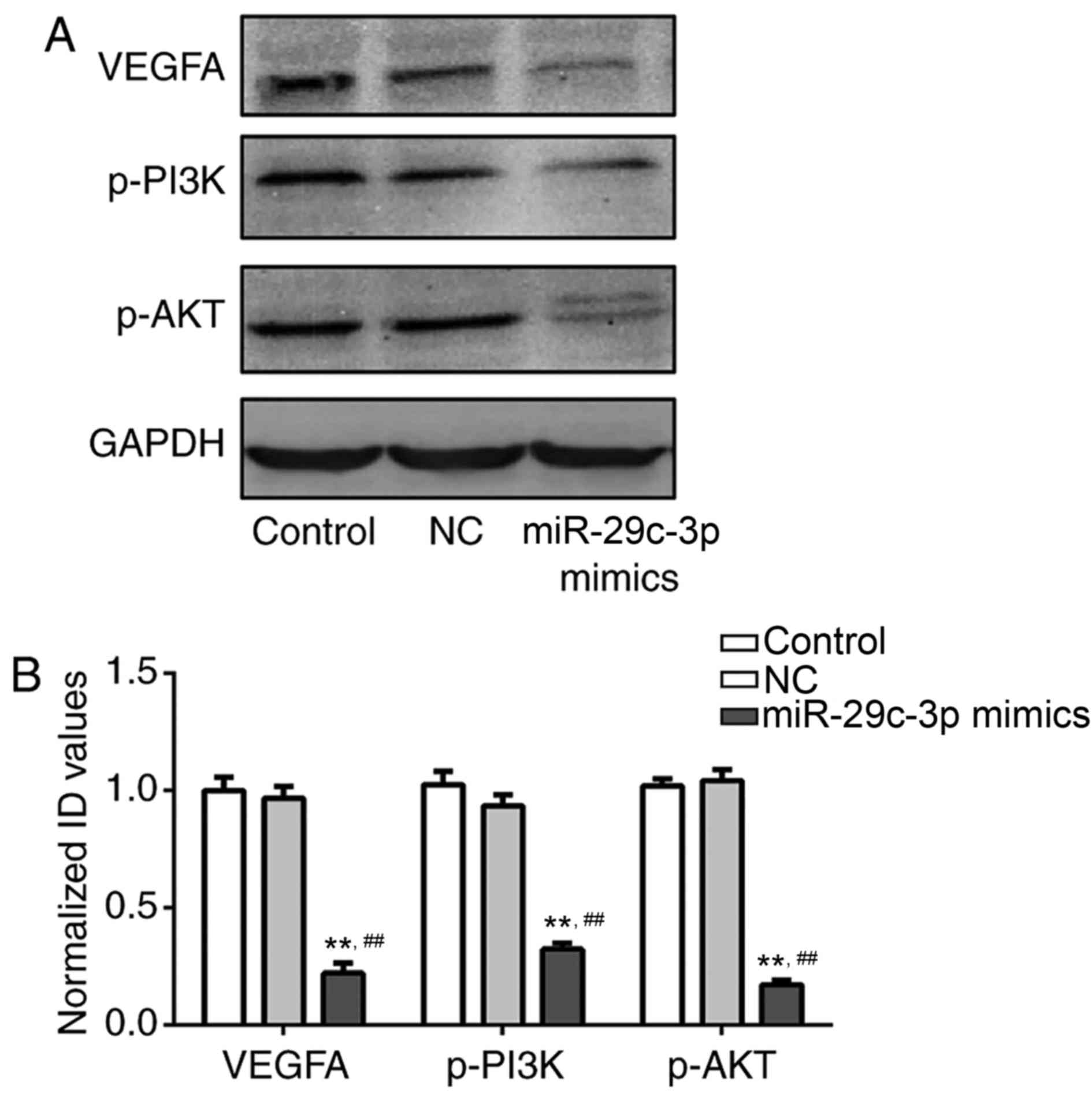
miR-29c regulates VEGFA expression and
the PI3K/Akt signaling pathway in lung cancer cells
To further examine the association between the
expression levels of miR-29c and VEGFA, and the possible regulatory
mechanism of miR-29c through the PI3K/Akt signaling pathway,
western blotting was used to measure the expression levels of VEGFA
protein and other signaling pathway-associated proteins in the
three groups. As presented in Fig.
4, no apparent variation was observed in the expression levels
of VEGFA, p-PI3K and p-Akt proteins in the control and NC groups.
However, in the miR-29c mimics group, the expression levels of
VEGFA, p-PI3K and p-Akt proteins were significantly downregulated
compared with the other two groups (P<0.01). This phenomenon
indicated that miR-29c was able to regulate VEGFA expression and
affect NSCLC cell activities via the PI3K/Akt signaling
pathway.
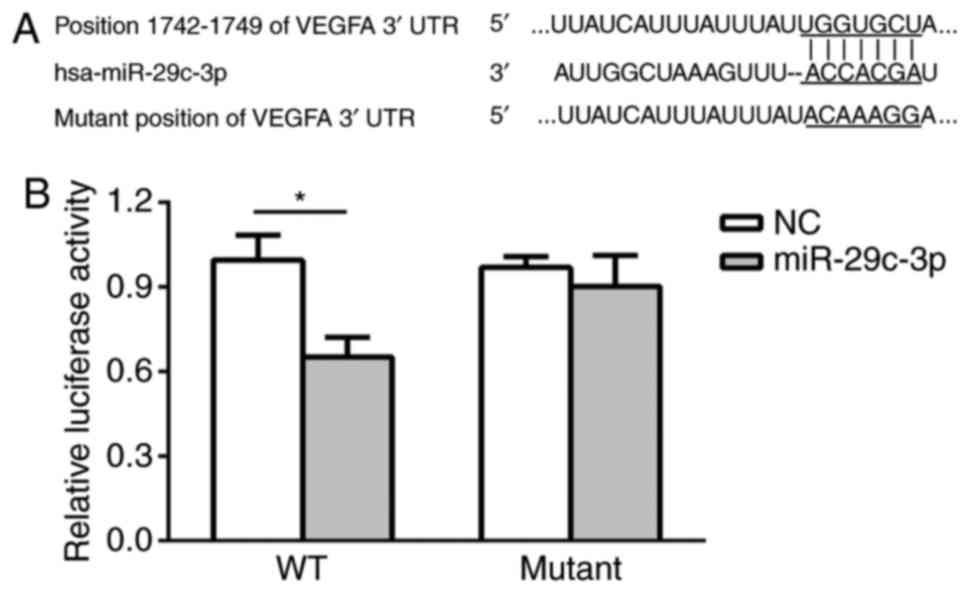
VEGFA is a direct gene target of
miR-29c
To verify the molecular basis for the regulatory
role of miR-29c in NSCLC cell activities observed above, it was
predicted that VEGFA may be a gene target for miR-29c using
bioinformatics analysis (Fig. 5A).
Thereafter, dual-luciferase reporter analysis was used to confirm
that VEGFA is a direct target for miR-29c. As presented in Fig. 5B, the relative luciferase activity
in the WT VEGFA 3′UTR group was significantly decreased in miR-29c
mimics-transfected cells compared with NC cells, while the relative
luciferase activity in the mutant VEGFA 3′UTR group exhibited no
notable decrease in cells transfected with miR-29c mimics. This
result suggested that VEGFA may be a direct gene target of
miR-29c.
Discussion
NSCLC has a median 5-year survival rate ranging
between 51% (stage IA) and 26% (stage IIIA), which is low compared
with other common tumors, including breast cancer and prostate
cancer (13). For early stage
NSCLC, complete surgical resection along with cisplatin-based
chemotherapy is the most frequently used therapeutic method,
although the efficacy of chemotherapy remains uncertain (14). Notably, molecular targeted therapy
for lung cancer has proven effective in dealing with specific
resistant tumors and certain targeted agents that may inhibit EGFR
have been approved for the treatment of NSCLC (15). Attention has been paid to the role
of miRs as biomarker-driven targeted agents in various types of
cancer (16). The miR-29 family,
namely miR-29a, miR-29b and miR-29c, have been reported to be of
importance in disease, including cardiac fibrosis, lung cancer,
nasopharyngeal carcinoma and Alzheimer's disease (8,17–19).
As mentioned previously, miR-29c has been proven to be a potential
biomarker for the detection of early stage NSCLC and a promising
targeted agent for the treatment of NSCLC (20,21).
However, the possible regulatory mechanism and corresponding
signaling pathway of miR-29c in NSCLC tumor development remain to
be elucidated. As an important factor in tumor growth, VEGFA is a
possible target for miR-29c. A series of studies have reported the
overexpression of VEGFA, which may contribute to poor prognosis in
a range of types of cancer (22–24).
Furthermore, it was reported that VEGFA was able to modulate the
metastasis and invasion of lung cancer via the PI3K/Akt pathway
(25). Based on this information,
the present study investigated the correlation between miR-29c and
VEGFA expression, and PI3K/Akt pathway-associated protein
expression in NSCLC tumors, to elucidate the potential regulatory
mechanism.
In the present study, the evident low expression of
miR-29c was found in NSCLC tumor tissues in comparison to adjacent
normal tissues. Correspondingly, the VEGFA expression level in
NSCLC tumor tissues was upregulated compared with normal tissues.
Statistical analysis demonstrated an evident negative correlation
in the expression levels of miR-29c and VEGFA in NSCLC tumor
tissues. To verify this observed negative correlation between
miR-29c and VEGFA expression, further study was performed via
transfection of miR-29c mimics into NSCLC tumor cells. It was
confirmed that the overexpression of miR-29c was able to suppress
VEGFA expression in the miR-29c mimics group compared with the
other two control groups. Subsequently, analysis of the influence
of miR-29c on NSCLC cell activities demonstrated that
overexpression of miR-29c was able to inhibit the proliferation and
promote the apoptosis of NSCLC cells. Since high expression of
VEGFA has been reported to be correlated with the proliferation and
metastasis of various types of tumor (11), it was hypothesized that miR-29c may
regulate NSCLC cell activities by targeting VEGFA. Through
bioinformatics analysis, it was predicted that VEGFA may be a
direct target for miR-29c. The dual-luciferase reporter analysis
demonstrated that miR-29c was able to bind the VEGFA 3′UTR directly
and thus suppress VEGFA expression. Furthermore, western blot
analysis results demonstrated that the expression level of VEGFA
protein was downregulated in the miR-29c mimics group compared with
the control groups. The expression levels of PI3K/Akt
pathway-associated proteins, including p-PI3K and p-Akt, were
downregulated in the miR-29c mimics group. These results revealed
for the first time, to the best of our knowledge, that miR-29c was
able to modulate NSCLC tumor activity, including proliferative and
apoptotic processes, by targeting VEGFA.
In conclusion, the functional role and possible
mechanism of miR-29c in NSCLC tumor progression were investigated
in the present study. Overall, it was demonstrated that miR-29c was
aberrantly underexpressed in NSCLC tumor tissues, and there was an
evident negative correlation between the expression levels of
miR-29c and VEGFA. In particular, the overexpression of miR-29c was
able to downregulate the expression level of VEGFA and decrease the
proliferation, as well as promoting the apoptosis, of NSCLC tumor
cells. Further experiments confirmed that VEGFA was a direct target
of miR-29c, and that miR-29c was able to affect NSCLC tumor cell
activities by modulating the PI3K/Akt signaling pathway. Although
more detailed analysis of the accurate regulatory mechanisms of
miR-29c in NSCLC tumor progression are required, the present study
identified an important miR and its functional role in NSCLC
tumors, and provided a basis for future clinical therapeutic
research into NSCLC tumors.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contribution
SZ designed and performed the experiments and
analyzed the data. CW performed the experiments and analyzed the
data. FY designed the experiments and composed the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Linyi People's Hospital (Linyi, China). Written
informed consent was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay JJ,
Ward E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu
Y, Chen Y, Xu L, Zen K, Zhang C and Shen H: Serum microRNA
signatures identified in a genome-wide serum microRNA expression
profiling predict survival of non-small-cell lung cancer. J Clin
Oncol. 28:1721–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peters S, Adjei AA, Gridelli C, Reck M,
Kerr K and Felip E: ESMO Guidelines Working Group: Metastatic
non-small-cell lung cancer: ESMO clinical practice guidelines for
diagnosis, treatment and follow-up. Ann Oncol. 23 Suppl
7:vii56–vii64. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Giovannetti E, Toffalorio F, De Pas T and
Peters GJ: Pharmacogenetics of conventional chemotherapy in
non-small-cell lung cancer: A changing landscape? Pharmacogenomics.
13:1073–1086. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Osada H and Takahashi T: MicroRNAs in
biological processes and carcinogenesis. Carcinogenesis. 28:2–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fabbri M, Garzon R, Cimmino A, Liu Z,
Zanesi N, Callegari E, Liu S, Alder H, Costinean S,
Fernandez-Cymering C, et al: MicroRNA-29 family reverts aberrant
methylation in lung cancer by targeting DNA methyltransferases 3A
and 3B. Proc Natl Acad Sci USA. 104:pp. 15805–15810. 2007;
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang HW, Wang EW, Li LX, Yi SH, Li LC, Xu
FL, Wang DL, Wu YZ and Nian WQ: A regulatory loop involving miR-29c
and Sp1 elevates the TGF-β1 mediated epithelial-to-mesenchymal
transition in lung cancer. Oncotarget. 7:85905–85916.
2016.PubMed/NCBI
|
|
9
|
Gu A, Lu J, Wang W, Shi C, Han B and Yao
M: Role of miR-497 in VEGF-A-mediated cancer cell growth and
invasion in non-small cell lung cancer. Int J Biochem Cell Biol.
70:118–125. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Claesson-Welsh L and Welsh M: VEGFA and
tumour angiogenesis. J Intern Med. 273:114–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Liu X, Zhang J, Li L and Liu C:
The expression and clinical significance of PI3K, pAkt and VEGF in
colon cancer. Oncol Lett. 4:763–766. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chuang JC, Neal JW, Niu XM and Wakelee HA:
Adjuvant therapy for EGFR mutant and ALK positive NSCLC: Current
data and future prospects. Lung Cancer. 90:1–7. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
NSCLC Meta-analyses Collaborative Group1,
. Arriagada R, Auperin A, Burdett S, Higgins JP, Johnson DH, Le
Chevalier T, Le Pechoux C, Parmar MK, Pignon JP, et al: Adjuvant
chemotherapy, with or without postoperative radiotherapy, in
operable non-small-cell lung cancer: Two meta-analyses of
individual patient data. Lancet. 375:1267–1277. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thomas A, Liu SV, Subramaniam DS and
Giaccone G: Refining the treatment of NSCLC according to
histological and molecular subtypes. Nat Rev Clin Oncol.
12:511–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tibaldi C, D'Incecco A and Lagana A:
MicroRNAs and targeted therapies in non-small cell lung cancer:
Minireview. Anticancer Agents Med Chem. 15:694–700. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Van Rooij E, Sutherland LB, Thatcher JE,
DiMaio JM, Naseem RH, Marshall WS, Hill JA and Olson EN:
Dysregulation of microRNAs after myocardial infarction reveals a
role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci USA. 105:pp.
13027–13032. 2008; View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Qiu F, Sun R, Deng N, Guo T, Cao Y, Yu Y,
Wang X, Zou B, Zhang S, Jing T, et al: miR-29a/b enhances cell
migration and invasion in nasopharyngeal carcinoma progression by
regulating SPARC and COL3A1 gene expression. PLoS One.
10:e01209692015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lei X, Lei L, Zhang Z, Zhang Z and Cheng
Y: Downregulated miR-29c correlates with increased BACE1 expression
in sporadic Alzheimer's disease. Int J Clin Exp Pathol.
8:1565–1574. 2015.PubMed/NCBI
|
|
20
|
Zhu W, He J, Chen D, Zhang B, Xu L, Ma H,
Liu X, Zhang Y and Le H: Expression of miR-29c, miR-93, and miR-429
as potential biomarkers for detection of early stage non-small lung
cancer. PLoS One. 9:e877802014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang H, Zhu Y, Zhao M, Wu C, Zhang P, Tang
L, Zhang H, Chen X, Yang Y and Liu G: miRNA-29c suppresses lung
cancer cell adhesion to extracellular matrix and metastasis by
targeting integrin β1 and matrix metalloproteinase2 (MMP2). PLoS
One. 8:e701922013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Goos JA, de Cuba EM, Coupé VM, Diosdado B,
Delis-Van Diemen PM, Karga C, Beliën JA, Menke-Van der Houven van
Oordt CW, Geldof AA, Meijer GA, et al: Glucose transporter 1
(SLC2A1) and vascular endothelial growth factor a (VEGFA) predict
survival after resection of colorectal cancer liver metastasis. Ann
Surg. 263:138–145. 2016.PubMed/NCBI
|
|
23
|
Mao D, Zhang Y, Lu H and Zhang H:
Molecular basis underlying inhibition of metastasis of gastric
cancer by anti-VEGFa treatment. Tumor Biol. 35:8217–8223. 2014.
View Article : Google Scholar
|
|
24
|
Geng L, Chaudhuri A, Talmon G, Wisecarver
JL and Wang J: TGF-beta suppresses VEGFA-mediated angiogenesis in
colon cancer metastasis. PLoS One. 8:e599182013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen CH, Lai JM, Chou TY, Chen CY, Su LJ,
Lee YC, Cheng TS, Hong YR, Chou CK, Whang-Peng J, et al: VEGFA
upregulates FLJ10540 and modulates migration and invasion of lung
cancer via PI3K/AKT pathway. PLoS One. 4:e50522009. View Article : Google Scholar : PubMed/NCBI
|