Introduction
Osteosarcoma (OS) is characterized by the production
of immature or osteoid bone, which is derived from mesenchymal
cells and has been become the most common type of malignant bone
tumor (1–3). OS frequently occurs in children and
young adults (4). Previous studies
have suggested that OS possesses high metastatic potential; ~80% of
the metastases arise in bone, lung and lymph nodes (5–7). At
present, OS treatment has significantly advanced, with options
including chemotherapy, radiotherapy and surgery; the survival rate
of patients with non-metastatic OS has improved by 60–70% (8), although the 5-year survival rate of
patients with OS exhibiting lung metastasis remains low (9–14).
Therefore, it is essential to identify the molecular mechanisms of
OS.
Transcription factor SOX (SOX) family proteins
contain a highly conserved high-mobility group (HMG) domain, which
regulates their capacity for DNA binding (15–17).
Based on the sequences of the HMG domains, SOX proteins are
classified into groups A-H (15,16).
In addition, SOX proteins are expressed in several cell lineages,
and serve essential roles in the differentiation of developing
tissues and cell fate determination. Depending on the target
sequences and interacting partners, SOX proteins may act as
transcriptional repressors or activators to regulate the expression
of various genes (17). SOX6
belongs to the D group and recognizes the (A/T)(A/T) CAA(A/T)
highly conserved sequence. Previous studies have demonstrated that
SOX6 serves tumor-suppressive functions in various human
malignancies, including hepatocellular carcinoma (18,19),
esophageal squamous cell carcinoma (20) and chronic myeloid leukemia
(21). Additionally, SOX6 serves
different roles in different types of cancer; for example, SOX6
transcriptionally activates suppressor of cytokine signaling 3
(SOCS3) in primary erythroid cells (21). SOX6 transcriptionally suppresses
cyclin D1 in pancreatic β-cells (22); however, the underlying mechanisms
of the role of SOX6 in OS requires further investigation.
In the present study, the expression levels of SOX6
were downregulated in OS, which was associated with tumor size,
metastasis and poor prognosis of OS; SOX6 additionally suppressed
cell proliferation. In OS, SOX6 acts as tumor suppressor and
inhibits cell migration, invasion and epithelial-mesenchymal
transition (EMT) via transcriptional regulation of TWIST1.
Materials and methods
Cell lines and cell culture
The human immortalized osteoblast cell line hFOB1.19
and human OS cell lines MG-63 and U2OS were purchased from the
American Type Culture Collection (Manassas, VA, USA). MG-63 and
U2OS cells were cultured in RPMI 1640 (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum
(FBS; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) at 37°C with
5% CO2. hFOB1.19 cells were cultured in Dulbecco's
modified Eagle's medium (DMEM)/Ham's F-12 (HyClone; GE Healthcare
Life Sciences) supplemented with 10% FBS and geneticin (400 mg/ml)
at 34°C with 5% CO2.
Cell transfection
For transfection with the pcDNA3.1-SOX6 plasmid,
cells were cultured to 60–70% confluence, and transfected with 2.5
µg vector (pcDNA3.1) or pcDNA3.1-SOX6 (YouBio, Hunan, China) using
the polyethylenimine reagent (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to the manufacturer's protocol. For
transfection with SOX6 small interfering (si)RNA, cells were
cultured to 30–40% confluence, and transfected with 5 µl scramble
siRNA or SOX6 siRNA (Sigma-Aldrich; Merck KGaA) using the
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. The
sequence of SOX6 siRNA as follows: SOX6 siRNA#1:
5′-GGAAGAGACUUACGAAUUAGA-3′, SOX6 siRNA#2:
5′-GGAUCUCGCUGGAAAUCAAUG-3′. The more efficient sequence was
subsequently selected for further experiments. A total of 48 h
following transfection, cells were collected and used for
subsequent experiments.
Tissue specimens
OS tissues and adjacent normal tissues were obtained
from patients who were diagnosed with OS between May 2013 and
August 2016. All patients received no treatment with radiotherapy
or chemotherapy prior to surgical excision; tissue samples were
immediately frozen in liquid nitrogen prior to use. The present
study was approved by the institutional ethics committee of Changyi
People's Hospital (Shandong, China) and written informed consent
was obtained from patients.
Cell counting kit-8 (CCK-8) assay
A CCK-8 assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) was used to detect cell proliferation. Briefly, a
total of 3×103 treated cells were suspended in 200 µl
DMEM and placed into a 96-well plate; 20 µl CCK-8 solution was
added to each well and cells were cultured at 37°C for 60 min. The
optical density was measured at 450 nm using a spectrophotometer
(SpectraMax 190; Molecular Devices, Sunnyvale, CA, USA). All assays
were performed in triplicate.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
RNA was isolated from cells and tumor specimens
using an EasyPure RNA kit (Transgen Biotech Co., Ltd. Beijing,
China), according to the manufacturer's protocol. RT was performed
using EasyScript First-Strand cDNA Synthesis SuperMix (Transgen
Biotech Co., Ltd) at 25°C for 10 min, 42°C for 15 min and 85°C for
3 min, followed by cooling at 4°C. RT-qPCR was performed with the
SYBR Premix Ex Taq™ (Takara Bio, Inc., Otsu, Japan) using the
Thermal Cycler Dice Detection System (Takara Bio, Inc.). GAPDH was
used as an internal control. The relative expression level of genes
was measured using the 2−ΔΔCq method (23). The following primer sequences were
used: SOX6 forward, 5′-CCTCTACCTCACCACATAAGC-3′; reverse,
5′-TCCACCACATCGGCAAGA-3′. E-cadhein forward,
5′-CCTCGAACTATATTCTTCTGTGAG-3′; reverse,
5′-TAGATTCTTGGGTTGGGTCG-3′. α-catenin forward,
5′-TGTTACACAGGTTACAACCCT-3′; reverse, 5′-GCAGCCTTCATCAAATCACC-3′.
N-cadherin forward, 5′-GTCATCACAGTGACAGATGTC-3′; reverse,
5′-TTCAAAGTCGATTGGTTTGACC-3′. Fibronectin forward,
5′-CCGAACAGAAATTGACAAACCA-3′; reverse, 5′-CTTTAGGGCGATCAATGTTGG-3′.
GAPDH forward, 5′-TCTCTGATTTGGTCGTATTGG-3′; reverse,
5′-CATGTAAACCATGTAGTTGAGGTC-3′.
Western blotting
Cells were harvested and lysed using
radioimmunoprecipitation assay buffer supplemented with a protein
inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany) for
30 min on ice, and centrifuged at 18,000 × g for 10 min at 4°C.
Protein concentration was measured using a Bicinchoninic Acid kit
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. A total of 45 µg protein lysate was separated via 10%
SDS-PAGE and transferred onto a nitrocellulose filter membrane.
Following blocking with 5% skimmed milk at room temperature for 1
h, the membranes were incubated with the indicated primary antibody
at 4°C overnight and with horseradish-peroxidase-conjugated
secondary antibodies at room temperature for 1 h (1:5,000; Abcam,
Cambridge, MA, USA; cat. nos. 6728 and 6721). The blots were
visualized with an enhanced chemiluminescence kit (Beyotime
Institute of Biotechnology, Haimen, China). All assays were
performed in triplicate. The primary antibodies were as follows:
SOX6 (1:3,000; cat. no. 64946; Abcam); β-actin (1:5,000; cat. no.
8226; Abcam), EMT kit (1:1,000; cat. no. 9782; Cell Signaling
Technology, Danvers, MA, USA).
Wound healing assay
Treated cells (~1×106) were placed into
6-well plates, and the cell monolayer was gently scratched with a
20 µl pipette tip when cells were cultured to 90–100% confluence.
The cells were washed with cold PBS three times to remove the
detached cells. Wound healing capacity was evaluated via the ratio
of wounded area at the time points of 48 and 0 h post-scratching.
All assays were performed in triplicate.
Transwell assays
A Transwell assay was performed to estimate the
effect of SOX6 on cell invasion. A total of 100 µl Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) was coated on the top
chambers (6.5 mm diameter, 8 µm pore size; Corning Incorporated,
Corning, NY, USA) and incubated at 37°C for 30 min. Subsequently, a
total of 3×104 cells were suspended in 600 µl serum-free
RPMI-1640 and placed to the upper chamber. A total of 600 µl
RPMI-1640 containing 10% FBS was added to the lower chamber.
Following 18 h of incubation at 37°C with 5% CO2, the
invaded cells were fixed with 4% paraformaldehyde at room
temperature for 10 min and stained with 0.5% crystal violet at room
temperature for 10 min. The cells on the top membranes were removed
with a cotton swab and the number of invaded cells was counted with
a light microscope (five fields randomly selected). All assays were
performed in triplicate.
Chromatin immunoprecipitation (ChIP)
and quantitative ChIP (qChIP) assays
The U2OS cells were fixed with 1% formaldehyde,
followed by numerous intervals of sonication (20 kHz, amplitude
setting, 40%; 20 cycles, 1 sec on and 1 sec off). For ChIP, the
immunoprecipitated DNA fragments were quantified by PCR. The
following thermocycling conditions were used for PCR: Initial
denaturation for 2 min at 98°C; 30 cycles of 98°C for 30 sec, 58°C
for 30 sec and 72°C for 1 min. The following primer sequences were
used: Twist1 forward, 5′-CGAGATGAGACATCACCCAC-3′; reverse,
5′-TAACAATTCGTCCTCCCAAA-3′; Snail forward,
5′-GTTCTGCCCTTCAGGTTGGT-3′; reverse, 5′-AGGCTGTAACACGGCTCCAT-3′;
Slug forward, 5′-ACTACCAGCAAATAAATACC-3′; reverse,
5′-AACTGGAACCTGGAGTAAAA-3′. For qChIP, the immunoprecipitated DNA
fragments were quantified by qPCR. The following thermocycling
conditions were used: Initial denaturation for 5 min at 95°C; 30
cycles of 95°C for 30 sec, 58°C for 30 sec and 72°C for 1 min. The
following primer sequences were used: Twist1 forward,
5′-AAGGGATGGACCTGAAACGG-3′; reverse, 5′-GGCAAACTGGAAGCAGCAAA-3;
Snail forward, 5′-GAAGGAACGGGTGCTCTTGG-3′; reverse,
5′-TGGCATTGACGAGGGAAACG-3. Slug forward,
5′-CTGGATTATGCCTCTGTGAT-3′; reverse,
5′-TGGTATTTATTTGCTGGTAG-3′.
Dual-luciferase activity assay
The promoter region (−2000 to +200) of TWIST1 was
amplified and sub-cloned into the pGL-3 basic vector (Promega
Corporation, Madison, WI, USA). Subsequently, plasmids were
transfected into U2OS cells along with vector (2 µg) or SOX6 (0.5,
1 or 2 µg) using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), following the manufacturer's protocol. In
addition, the Renilla gene was co-transfected as an internal
control. Following transfection for 24 h, the cells were lysed and
the dual-luciferase reporter assay was conducted using the
dual-luciferase assay system (Promega Corporation), according to
the manufacturer's protocol. All assays were performed in
triplicate.
Statistical analysis
All statistical analyses were performed with SPSS
18.0 software (SPSS, Inc, Chicago, IL, USA). The results of
experiments are presented as the mean ± standard deviation.
Kaplan-Meier analysis followed by log-rank test was used to analyze
the correlation between SOX6 expression and survival. The
differences between two groups were determined with a Student's
t-test. One-way analysis of variance followed by Tukey's test was
used to analyze the differences between multiple groups. P<0.05
was considered to indicate a statistically significant
difference.
Results
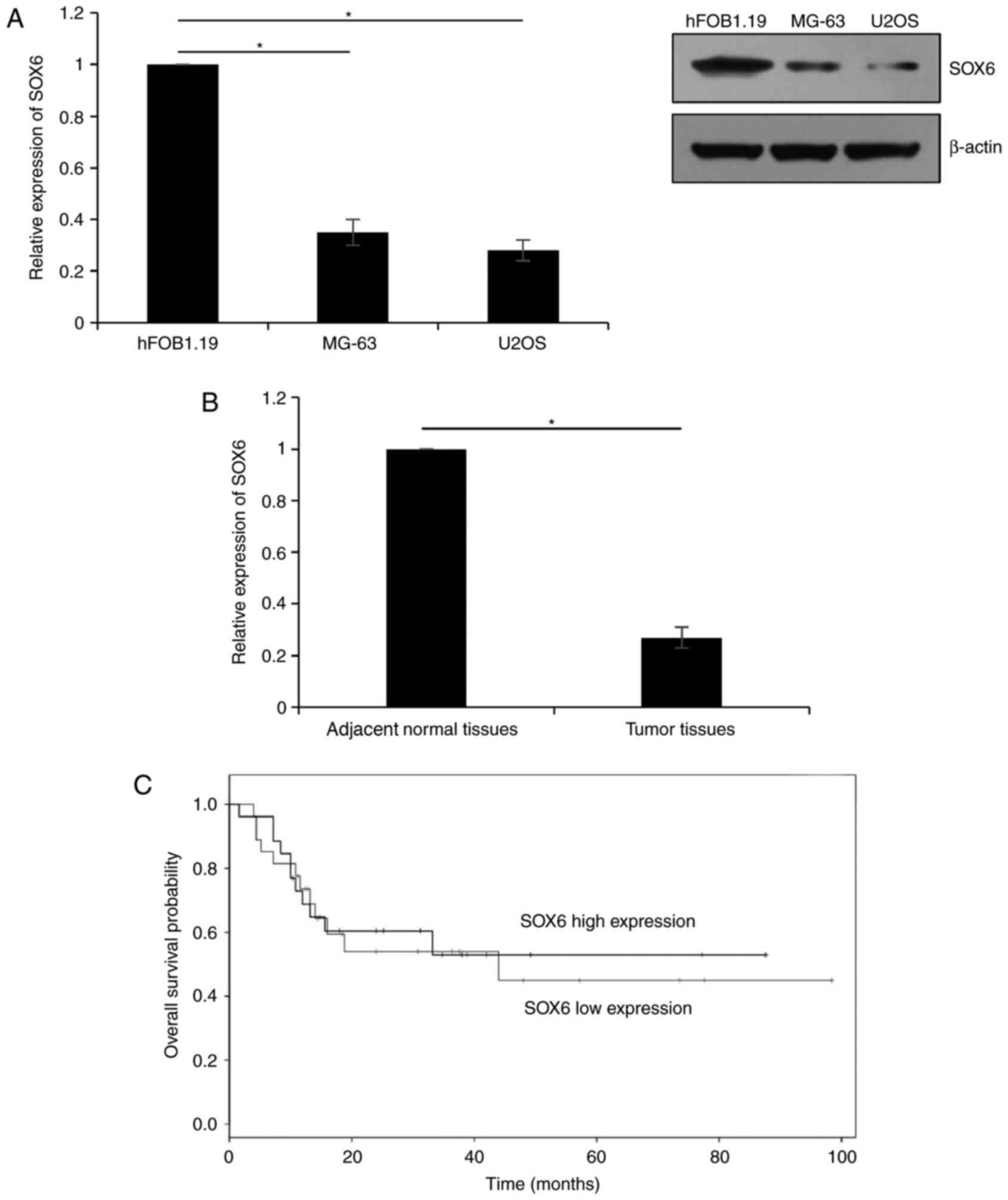
SOX6 is frequently downregulated in OS
tissues and cell lines
To investigate the function of SOX6 in OD, the
expression of SOX6 in OS cell lines, MG-63 and U2OS, was analyzed
via RT-qPCR and western blotting; the osteoblast cell line hFOB1.19
was used as a control group. The results of the present study
demonstrated that the expression levels of SOX6 in MG-63 and U2OS
cells were markedly lower compared with in hFOB1.19 cells
(P<0.05; Fig. 1A).
Subsequently, the downregulation of SOX6 expression levels within
OS tissues was determined by RT-qPCR using a total of 42 pairs of
OS tissues and adjacent normal tissues. As presented in Fig. 1B, similar to the expression in
cells, SOX6 was downregulated in OS tissues compared with normal
tissues (P<0.05). Analysis of SOX6 expression levels and patient
clinical information revealed that low expression levels of SOX6
were associated with the development of tumor metastasis. In
addition, the expression of SOX6 was associated with tumor size and
tumor stage (Table I).
Subsequently, the correlation between SOX6 expression and survival
was analyzed using a survival curve, which revealed that patients
with low expression levels of SOX6 had a poor prognosis (P<0.05;
Fig. 1C).
 | Table I.Clinical pathological variables in 42
patients with osteosarcoma. |
Table I.
Clinical pathological variables in 42
patients with osteosarcoma.
|
|
| SOX6 protein
expression |
|
|---|
|
|
|
|
|
|---|
| Variables | Total (n=42) | Low (n=28) | High (n=14) | P-value |
|---|
| Gender |
|
|
|
|
| Male | 22 | 15 | 7 | 0.827 |
|
Female | 20 | 13 | 7 |
|
| Age, years |
|
|
|
|
|
<20 | 25 | 16 | 9 | 0.657 |
| ≥20 | 17 | 12 | 5 |
|
| Tumor size |
|
|
|
|
| ≥5
cm | 28 | 22 | 6 | 0.021 |
| <5
cm | 14 | 6 | 8 |
|
| Differentiation |
|
|
|
|
|
Well/moderate | 20 | 14 | 6 | 0.662 |
|
Poor | 22 | 14 | 8 |
|
| TNM stage |
|
|
|
|
|
I–II | 20 | 9 | 11 | 0.005 |
|
III–IV | 22 | 19 | 3 |
|
| Metastasis |
|
|
|
|
|
Yes | 22 | 18 | 4 | 0.029 |
| No | 20 | 10 | 10 |
|
Downregulation of SOX6 facilitates
migration and invasion in U2OS
As presented in Table
I, SOX6 may suppress the migratory and invasive abilities of OS
cells. This was verified via the overexpression and silencing of
SOX6 within U2OS cells expressing the plasmid pcDNA3.1-SOX6 or SOX6
siRNA, respectively. Western blotting and RT-qPCR were used to
demonstrate SOX6 expression (P<0.05, P<0.001; Fig. 2A). The siSOX6#2 was more efficiency
than siSOX6#1, so siSOX6#2 was used for subsequent experiments.
Wound healing analysis was performed to detect the effects of SOX6
on cell migration, which revealed that the ectopic expression of
SOX6 markedly reduced the migratory ability of U2OS cells; however,
the inhibition of SOX6 significantly improved the migratory ability
of U2OS cells (P<0.05; Fig.
2B). In addition, the Transwell assay demonstrated that
overexpression of SOX6 markedly decreased the number of invaded
cells, and the inhibition of SOX6 markedly increased the number of
invasive cells (P<0.05, P<0.01; Fig. 2C). Therefore, the findings of the
present study suggested that the downregulation of SOX6 facilitated
migration and invasion of U2OS cells.
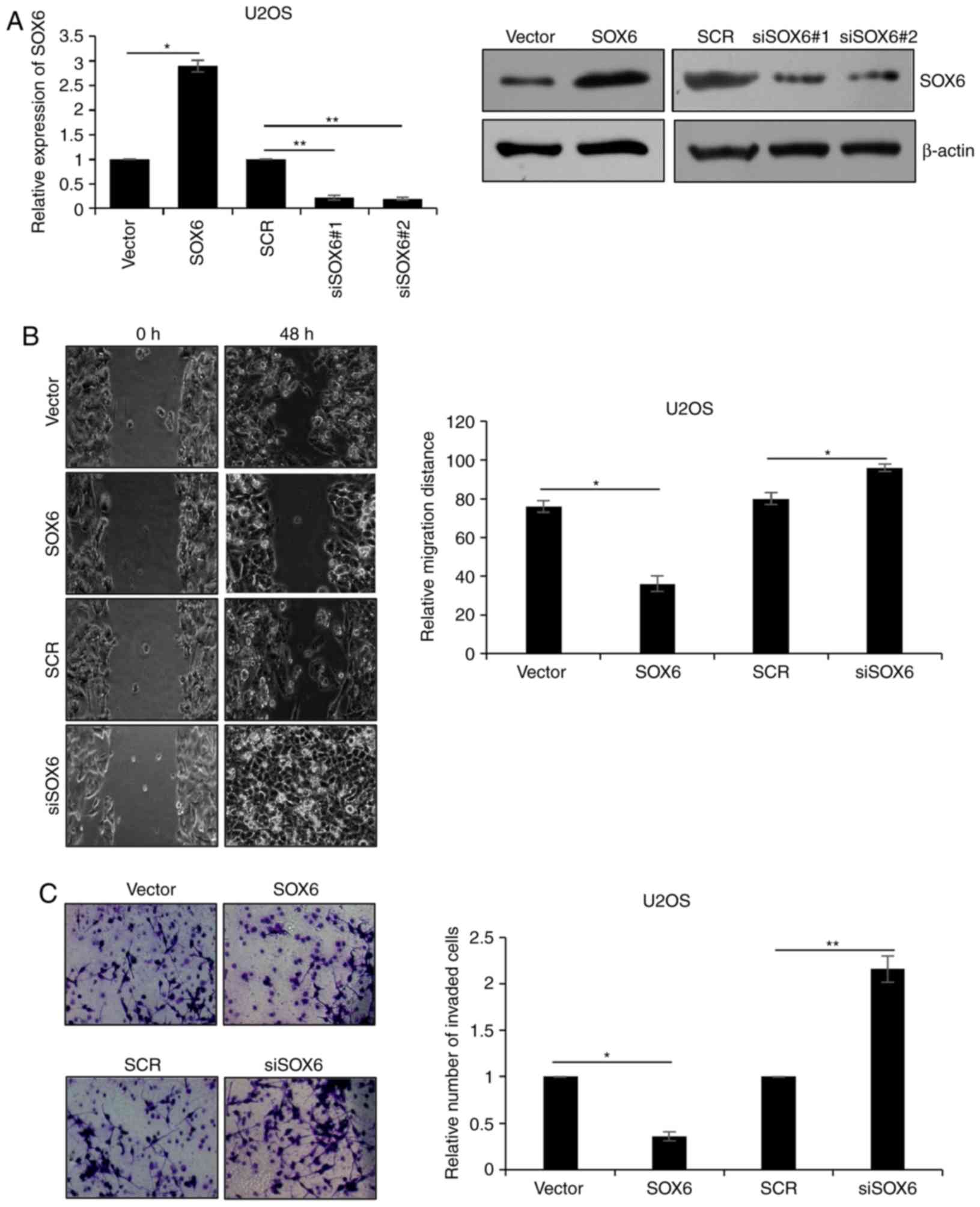 | Figure 2.Downregulation of SOX6 facilitates
migration and invasion in U2OS cells. (A) U2OS cells were
transfected with vector or pcDNA3.1-SOX6, SCR or SOX6 siRNA #1 and
#2, respectively. Following transfection for 48 h, the mRNA and
protein levels of SOX6 were detected using reverse
transcription-quantitative polymerase chain reaction and western
blotting. Vector and SCR were used as controls. (B) U2OS cells were
transfected with vector or SOX6, SCR or #2 SOX6 siRNA. Following
transfection for 48 h, a wound healing assay was used to determine
the effect of SOX6 on cell migration. (C) U2OS cells were
transfected with vector or SOX6, SCR or siSOX6. Following
transfection for 48 h, Transwell assays were used to determine the
effect of SOX6 on cell invasion. *P<0.05; **P<0.01. SCR,
scrambled small interfering RNA; siRNA, small interfering RNA;
SOX6, transcription factor SOX6; siSOX6, siRNA#1 and 2
(magnification, ×40). |
Modulation of SOX6 expression within
OS cells affects cell proliferation
As presented in Table
I, the expression of SOX6 was associated with tumor size. In
the present study, the effects of SOX6 on cell proliferation were
investigated via colony formation and CCK-8 assays. The results of
the colony formation assay revealed that the ectopic expression of
SOX6 notably decreased the number of U2OS colonies compared with
the control. Conversely, the inhibition of SOX6 expression markedly
increased the number of colonies compared with the control
(P<0.05; Fig. 3A). Similar
results were observed within MG-63 cells (P<0.05; Fig. 3B). Additionally, CCK-8 analysis
illustrated that the overexpression of SOX6 reduced cell
proliferation and that the inhibition of SOX6 increased the
proliferative abilities of U2OS and MG-63 cells (P<0.05,
P<0.01; Fig. 3C and D). These
results indicated that the modulation of SOX6 expression within OS
cells affected proliferation.
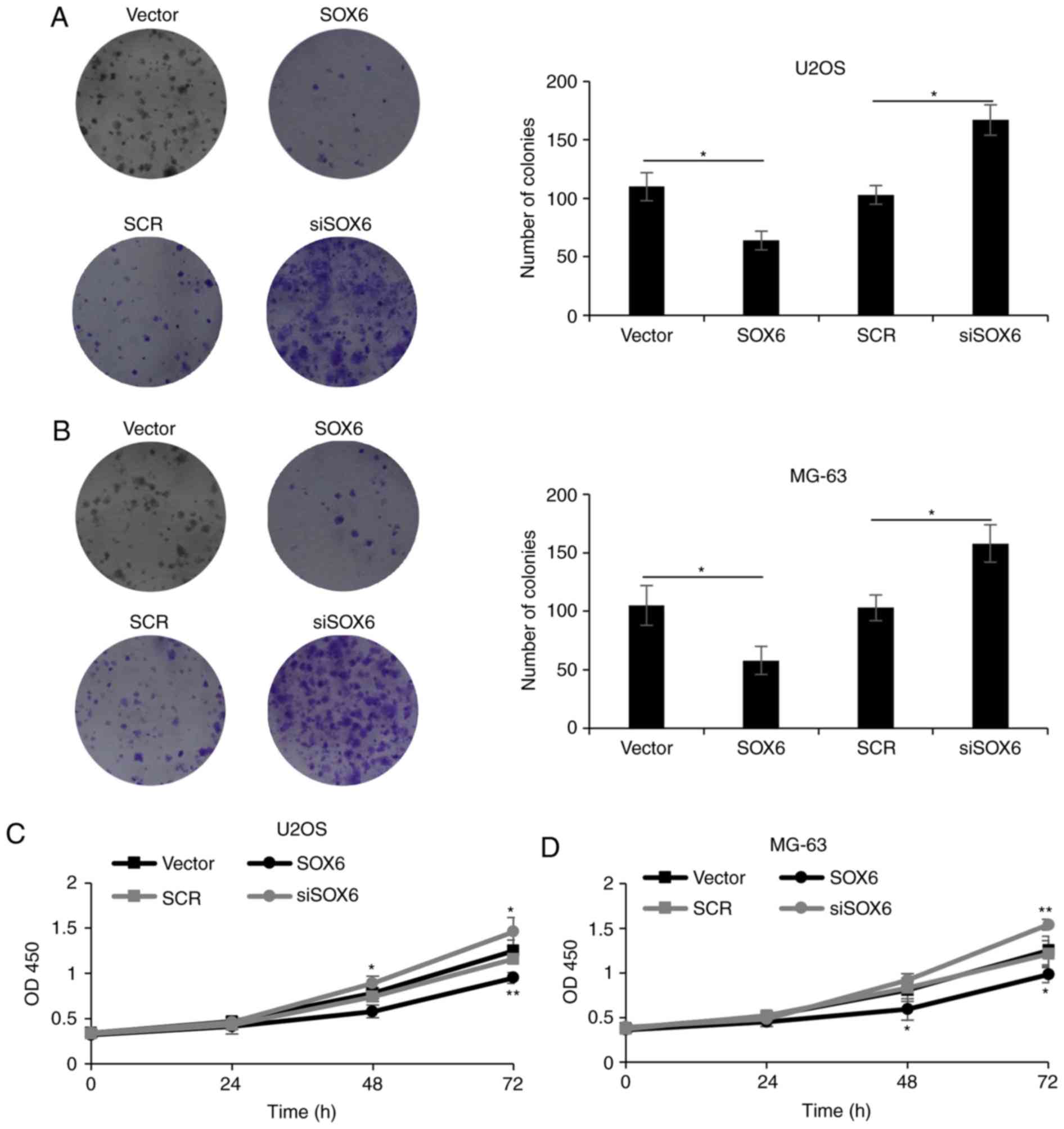 | Figure 3.Modulation of SOX6 expression in
osteosarcoma cells impacts upon their proliferation. (A) U2OS cells
and (B) MG-63 cells were transfected with vector or SOX6, SCR or
siSOX6. Following transfection for 48 h, a colony formation assay
was used to determine the effect of SOX6 on cell proliferation.
*P<0.05. (C) U2OS cells were transfected with vector or SOX6,
SCR or siSOX6. Following transfection for 48 h, cell viability was
measured on the indicated days using the CCK-8 assay. (D) MG-63
cells were transfected with vector or SOX6, SCR or siSOX6.
Following transfection for 48 h, cell viability was measured on the
indicated days using the CCK-8 assay. *P<0.05; **P<0.01 vs.
vector group. CCK-8, cell counting kit-8; SCR, scrambled short
interfering RNA; SOX6, transcription factor SOX6; siSOX6, siRNA#1
and 2. |
SOX6 suppresses EMT of OS cells via
the regulation of TWIST1
Previous reports have demonstrated that EMT promotes
cancer development via migration and invasion (24); SOX5, in addition to SOX9, has been
reported to regulate EMT (25,26).
To determine whether SOX6 suppressed the migration and invasion of
U2OS cells via EMT regulation, markers of EMT were detected
following the overexpression or downregulation of SOX6 in U2OS
cells.
As presented in Fig.
4A, the mRNA and protein expression levels of mesenchymal
markers, including N-cadherin and fibronectin, were significantly
decreased when SOX6 was overexpressed, although the expression of
epithelial markers, including E-cadherin and α-catenin, was
significantly increased (P<0.05). However, inhibition of SOX6
significantly downregulated the expression of mesenchymal markers
and upregulated epithelial marker expression levels (P<0.05;
Fig. 4B). To further investigate
the underlying mechanism of EMT modulation via SOX6, the expression
levels of EMT-associated transcription factors, including TWIST1,
zinc finger protein SNAI2 (Slug) and zinc finger protein SNAI1
(Snail1) were analyzed. The results of the present study revealed
the SOX6 exerted little effect on the expression levels of Slug and
Snail1; however, the expression of TWIST1 was regulated by SOX6
(P<0.05; Fig. 4C and D).
Additionally, the expression of TWIST1 was detected in MG-63 and
U2OS cells using RT-qPCR analysis; the osteoblast cell line
hFOB1.19 served as the control. The results of the present study
revealed that the expression levels of TWIST1 were upregulated
within MG-63 and U2OS cells (P<0.05; Fig. 4E); therefore, SOX6 may regulate EMT
via TWIST1 modulation.
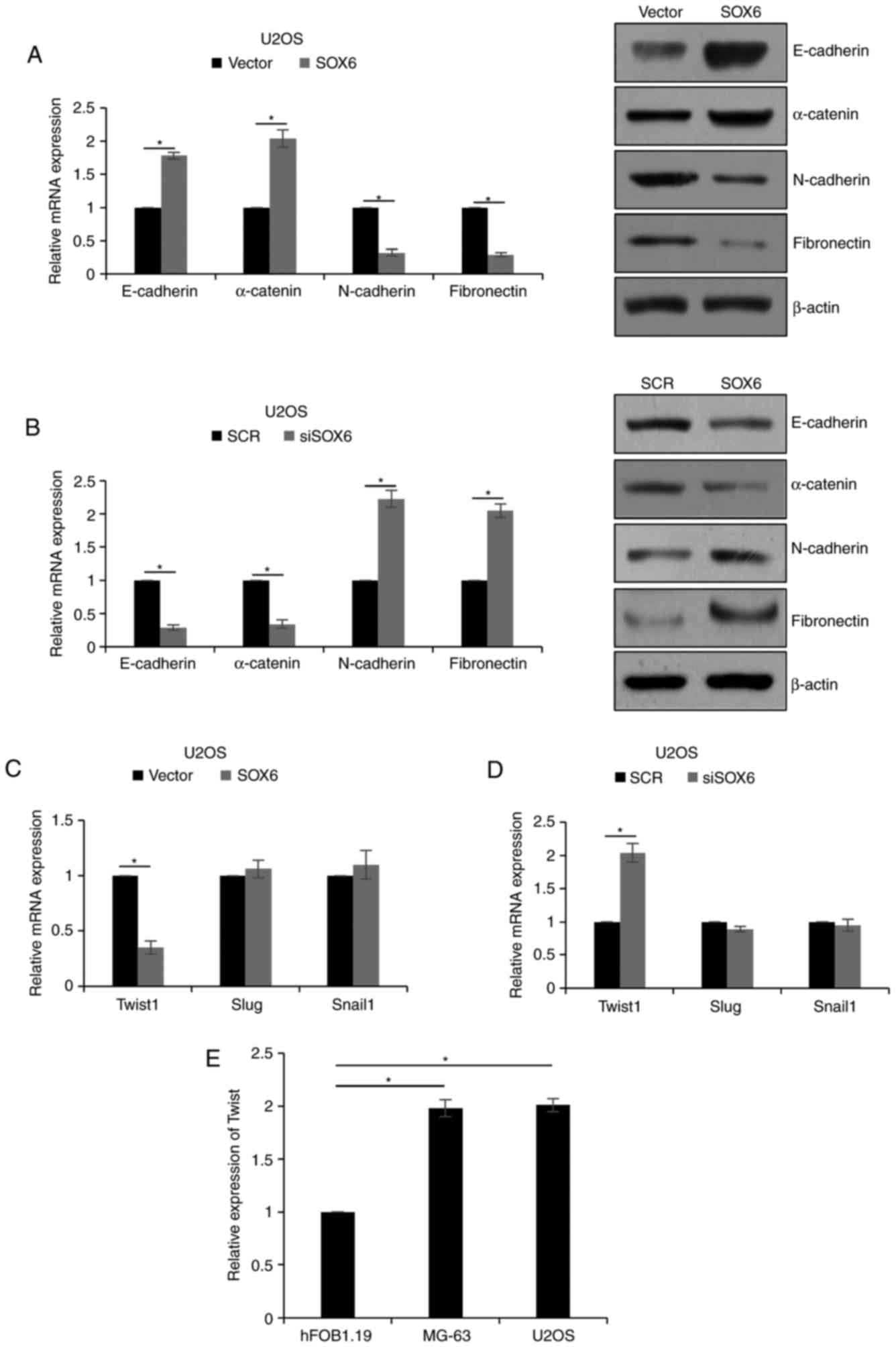 | Figure 4.SOX6 suppresses EMT in osteosarcoma
cells via the regulation of TWIST1. (A) SOX6 was overexpressed in
U2OS cells. Following transfection for 48 h, the expression of
epithelial markers and mesenchymal markers was detected by RT-qPCR
and western blotting. (B) SOX6 was knocked down in U2OS cells.
Following transfection for 48 h, the expression of epithelial
markers and mesenchymal markers was detected by RT-qPCR and western
blotting. (C) SOX6 was overexpressed in U2OS cells. Following
transfection for 48 h, the expression of EMT-associated
transcription factors was detected by RT-qPCR. (D) SOX6 was knocked
down in U2OS cells. Following transfection for 48 h, the expression
of EMT-associated transcription factors was detected RT-qPCR. (E)
The expression of TWIST1 in human osteosarcoma cell lines MG-63 and
U2OS, and the normal osteoblastic cell line hFOB 1.19, was detected
using RT-qPCR. *P<0.05. EMT, epithelial-mesenchymal transition;
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; SCR, scrambled short interfering RNA; SOX6, transcription
factor SOX6; siSOX6, short interfering RNA#1 and 2; Slug, zinc
finger protein SNAI2; Snail1, zinc finger protein SNAI1; TWIST1,
twist-related protein 1. |
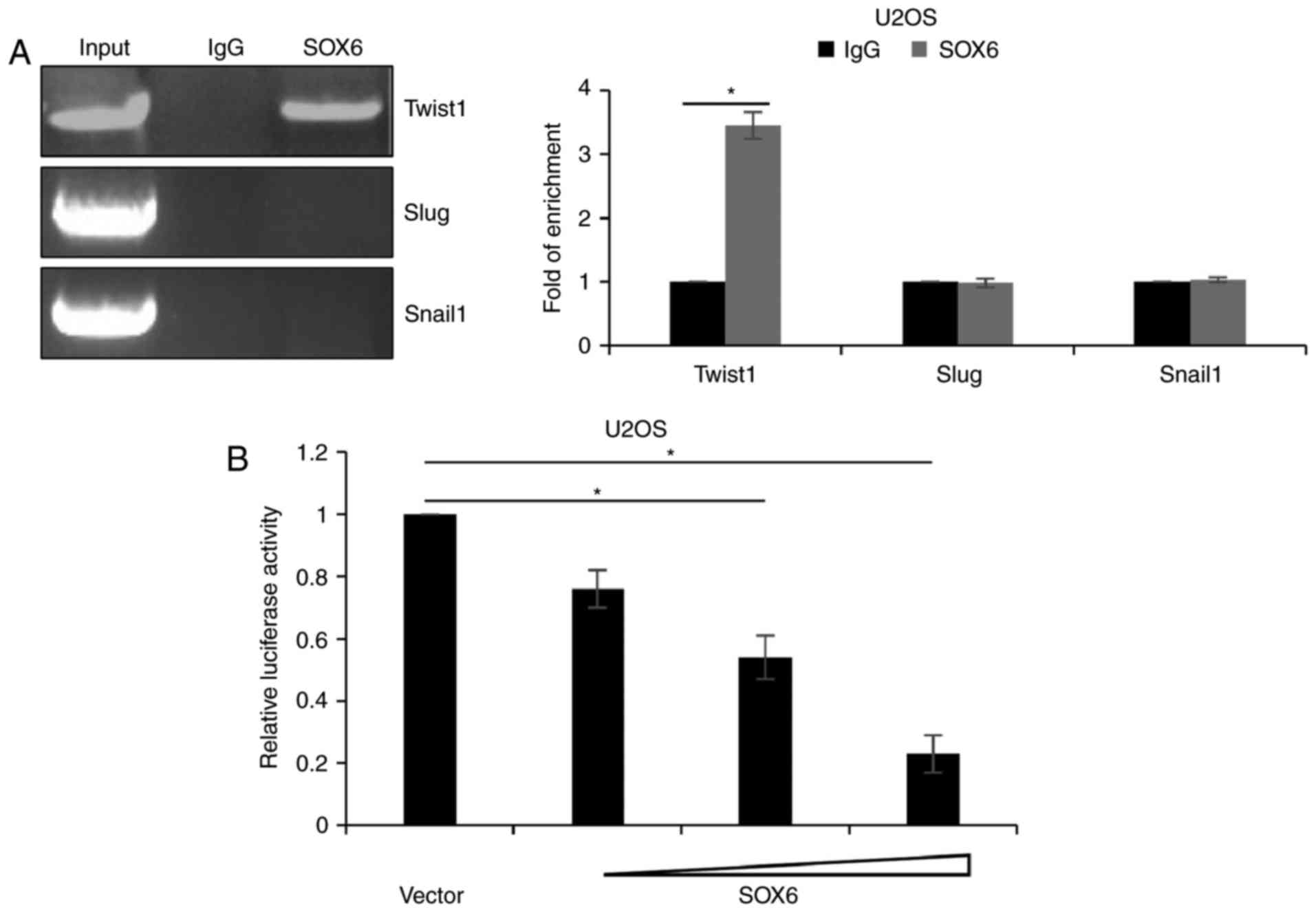
SOX6 transcriptionally regulates
TWIST1 in OS cells
As SOX6 serves as a transcription factor, whether
SOX6 is transcriptionally regulated was investigated. Chromatin
immunoprecipitation (ChIP) and quantitative ChIP analysis was used
to determine whether SOX6 bound to the promoter region of TWIST1.
The results of the present study revealed that SOX6 directly bound
to the promoter region of TWIST1 (P<0.05; Fig. 5A). Subsequently, a dual luciferase
activity assay was performed, which revealed that transfection with
SOX6 markedly decreased the luciferase activity of the pGL3-TWIST1
group (P<0.05; Fig. 5B);
therefore, SOX6 may have transcriptionally inhibited TWIST1.
Discussion
Numerous reports have reported that SOX6 is
downregulated in a number of types of cancer and acts as a tumor
suppressor (18–20). However, the underlying mechanism of
SOX6 in OS requires further investigation. The present study
reported that the SOX6 expression levels were notably downregulated
in OS, which was negatively-associated with tumor size, metastasis
and tumor, node, metastasis stages. In addition, patients with low
expression levels of SOX6 exhibited poor prognosis.
In the present study, numerous functional
experiments, including a wound healing assay and a Transwell assay
were performed to detect the effects of SOX6 on cell migration and
invasion. The results suggested that SOX6 suppressed the migration
and invasion of OS cells. EMT is a complex cellular process that
has been associated with cancer migration and invasion (24). To improve understanding of the
underlying mechanism of SOX6 in OS cell migration and invasion,
SOX6-mediated regulation of EMT was investigated. The results of
the present study revealed that the ectopic expression of SOX6
reduced the expression of mesenchymal markers, N-cadherin and
fibronectin; however, the expression of epithelial markers,
E-cadherin and α-catenin, were increased. In addition, SOX6
inhibition increased the expression of mesenchymal markers,
N-cadherin and fibronectin, whereas the expression of epithelial
markers, E-cadherin and α-catenin, was decreased. EMT is regulated
by a variety of transcription factors, including Snail1, TWIST1,
Slug and zinc finger E-box-binding homeobox 1 (27–30).
ChIP and qChIP analysis, and the dual-luciferase activity assay,
demonstrated that SOX6 transcriptionally suppressed TWIST1. Further
investigation is required to elucidate the detailed binding site of
SOX6 on the TWIST1 promoter, which may require interactions with
unknown proteins. Therefore, the results of the present study
demonstrated that, SOX6 suppressed EMT via the regulation of TWIST1
in OS.
Previous studies have reported that SOX6 suppresses
cell proliferation by transcriptionally suppressing cyclin D1 in
pancreatic β-cells (23) or
transcriptionally activating SOCS3 in primary erythroid cells
(22). Additionally, numerous
microRNAs promote cell proliferation by targeting SOX6 (31,32);
however, further investigation is required to understand whether
SOX6 may suppress cell proliferation in OS. The present study
reported that the expression of SOX6 was negatively correlated with
tumor size. A colony formation assay and a CCK-8 assay revealed
that SOX6 additionally inhibited cell proliferation in OS; however,
the underlying mechanism is yet to be investigated.
In the present study, SOX6 expression levels were
downregulated in OS, indicating the tumor-suppressing roles of
SOX6; low expression levels were associated with poor prognosis in
OS. Additionally, SOX6 inhibited cell proliferation in OS cells,
which suggested the potential role of SOX6 as a biomarker for
OS.
References
|
1
|
Bielack SS: Osteosarcoma: Time to move on?
Eur J Cancer. 46:1942–1945. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheng DD, Yu T, Hu T, Yao M, Fan CY and
Yang QC: MiR-542-5p is a negative prognostic factor and promotes
osteosarcoma tumorigenesis by targeting HUWE1. Oncotarget.
6:42761–42772. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wagle S, Park SH, Kim KM, Moon YJ, Bae JS,
Kwon KS, Park HS, Lee H, Moon WS, Kim JR and Jang KY: DBC1/CCAR2 is
involved in the stabilization of androgen receptor and the
progression of osteosarcoma. Sci Rep. 5:131442015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Damron TA, Ward WG and Stewart A:
Osteosarcoma, chondrosarcoma and Ewing's sarcoma: National Cancer
Data Base Report. Clin Orthop Relat Res. 459:40–47. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Byun BH, Kong CB, Lim I, Kim BI, Choi CW,
Song WS, Cho WH, Jeon DG, Koh JS, Lee SY and Lim SM: Comparison of
(18)F-FDG PET/CT and (99 m)Tc-MDP bone scintigraphy for detection
of bone metastasis in osteosarcoma. Skeletal Radiol. 42:1673–1681.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dirik Y, Çınar A, Yumrukçal F and Eralp L:
Popliteal lymph node metastasis of tibial osteoblastic
osteosarcoma. Int J Surg Case Rep. 5:840–844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yanagawa T, Saito K and Takagishi K: Risk
factors for lymphatic metastasis of malignant bone and soft-tissue
tumors: A retrospective cohort study of 242 patients. Medicine
(Baltimore). 93:e2252014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu H, Tang J, Tang M and Cai H:
Upregulation of SOX9 in osteosarcoma and its association with tumor
progression and patients' prognosis. Diagn Pathol. 8:1832013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bi Y, Jing Y and Cao Y: Overexpression of
miR-100 inhibits growth of osteosarcoma through FGFR3. Tumour Biol.
36:8405–8411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang W, Zhou X and Wei M: MicroRNA-144
suppresses osteosarcoma growth and metastasis by targeting ROCK1
and ROCK2. Oncotarget. 6:10297–10308. 2015.PubMed/NCBI
|
|
11
|
Rainusso N, Wang LL and Yustein JT: The
adolescent and young adult with cancer: State of the art-bone
tumors. Curr Oncol Rep. 15:296–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gorlick R, Anderson P, Andrulis I, Arndt
C, Beardsley GP, Bernstein M, Bridge J, Cheung NK, Dome JS, Ebb D,
et al: Biology of childhood osteogenic sarcoma and potential
targets for therapeutic development: Meeting summary. Clin Cancer
Res. 9:5442–5453. 2003.PubMed/NCBI
|
|
13
|
Kager L, Zoubek A, Pötschger U, Kastner U,
Flege S, Kempf-Bielack B, Branscheid D, Kotz R, Salzer-Kuntschik M,
Winkelmann W, et al: Primary metastatic osteosarcoma: Presentation
and outcome of patients treated on neoadjuvant cooperative
osteosarcoma study group protocols. J Clin Oncol. 21:2011–2018.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wan J and Zhang X, Liu T and Zhang X:
Strategies and developments of immunotherapies in osteosarcoma.
Oncol Lett. 11:511–520. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kamachi Y and Kondoh H: Sox proteins:
Regulators of cell fate specification and differentiation.
Development. 140:4129–4144. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wegner M: From head to toes: The multiple
facets of Sox proteins. Nucleic Acids Res. 27:1409–1420. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bowles J, Schepers G and Koopman P:
Phylogeny of the SOX family of developmental transcription factors
based on sequence and structural indicators. Dev Biol. 227:239–255.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie Q, Chen X, Lu F, Zhang T, Hao M, Wang
Y, Zhao J, McCrae MA and Zhuang H: Aberrant expression of microRNA
155 may accelerate cell proliferation by targeting sex-determining
region Y box 6 in hepatocellular carcinoma. Cancer. 118:2431–2442.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Guo X, Yang M, Gu H, Zhao J and Zou L:
Decreased expression of SOX6 confers a poor prognosis in
hepatocellular carcinoma. Cancer Epidemiol. 37:732–736. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qin YR, Tang H, Xie F, Liu H, Zhu Y, Ai J,
Chen L, Li Y, Kwong DL, Fu L and Guan XY: Characterization of
tumor-suppressive function of SOX6 in human esophageal squamous
cell carcinoma. Clin Cancer Res. 17:46–55. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cantù C, Ierardi R, Alborelli I, Fugazza
C, Cassinelli L, Piconese S, Bosè F, Ottolenghi S, Ferrari G and
Ronchi A: Sox6 enhances erythroid differentiation in human
erythroid progenitors. Blood. 117:3669–3679. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iguchi H, Urashima Y, Inagaki Y, Ikeda Y,
Okamura M, Tanaka T, Uchida A, Yamamoto TT, Kodama T and Sakai J:
SOX6 suppresses cyclin D1 promoter activity by interacting with
beta-catenin and histone deacetylase 1, and its down-regulation
induces pancreatic beta-cell proliferation. J Biol Chem.
282:19052–19061. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sannino G, Armbruster N, Bodenhöfer M,
Haerle U, Behrens D, Buchholz M, Rothbauer U, Sipos B and Schmees
C: Role of BCL9 L in transforming growth factor-β (TGF-β)-induced
epithelial-to-mesenchymal-transition (EMT) and metastasis of
pancreatic cancer. Oncotarget. 7:73725–73738. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang D, Han S, Wang X, Peng R and Li X:
SOX5 promotes epithelial-mesenchymal transition and cell invasion
via regulation of Twist1 in hepatocellular carcinoma. Med Oncol.
32:4612015.PubMed/NCBI
|
|
26
|
Huang J and Guo L: Knockdown of SOX9
Inhibits the Proliferation, Invasion, and EMT in Thyroid Cancer
Cells. Oncol Res. 25:167–176. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Medici D, Hay ED and Olsen BR: Snail and
Slug promote epithelial-mesenchymal transition through
beta-catenin-T-cell factor-4-dependent expression of transforming
growth factor-beta3. Mol Biol Cell. 19:4875–4887. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alves CC, Carneiro F, Hoefler H and Becker
KF: Role of the epithelial-mesenchymal transition regulator Slug in
primary human cancers. Front Biosci (Landmark Ed). 14:3035–3050.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kojc N, Zidar N, Gale N, Poljak M, Fujs
Komlos K, Cardesa A, Höfler H and Becker KF: Transcription factors
Snail, Slug, Twist, and SIP1 in spindle cell carcinoma of the head
and neck. Virchows Arch. 454:549–555. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Preca BT, Bajdak K, Mock K, Lehmann W,
Sundararajan V, Bronsert P, Matzge-Ogi A, Orian-Rousseau V,
Brabletz S, Brabletz T, et al: A novel ZEB1/HAS2 positive feedback
loop promotes EMT in breast cancer. Oncotarget. 8:11530–11543.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Z and Wang Y: MiR-96 targets SOX6 and
promotes proliferation, migration and invasion of hepatocellular
carcinoma. Biochem Cell Biol. July 17–2017.doi:
10.1139/bcb-2017-0183. (Epub ahead of print). View Article : Google Scholar
|
|
32
|
Li YC, Li CF, Chen LB, Li DD, Yang L, Jin
JP and Zhang B: MicroRNA-766 targeting regulation of SOX6
expression promoted cell proliferation of human colorectal cancer.
Onco Targets Ther. 8:2981–2988. 2015. View Article : Google Scholar : PubMed/NCBI
|