Introduction
Cancer is one of the most common causes of death in
developed countries (1), and
recent researches showed that gut bacteria seriously influenced the
treatment and outcomes for patients with advanced malignancies
(2). Researchers provided strong
evidence of the role of stool microbiota in response and resistance
to immunotherapy (3–5), and Bacteroidales and
Bifidobacterium greatly increased the number and activity of
immune cells, which then promoted the efficacy of anti-CTLA-4 and
anti-PD-L1 therapy (6).
Do the microbiota inside tumours also play an
important role in cancer development and therapy? It has been
recognized for more than 60 years that the tumour microenvironment
creates an ideal condition for the growth of anaerobic and some
facultative anaerobic bacteria (7), and several approaches to developing
anaerobic bacteria for tumour therapies have been described
(8–10). Until now, the anaerobic species of
Bifidobacterium longum, lactic acid bacteria, Clostridium
novyi and Salmonella typhimurium have been used as an
anticancer protein-delivery agent to repress tumour growth for
their selective growth in the hypoxic environment of large solid
tumours (11–14). However, two clinical trials
indicated that Salmonella failed to fully colonize human
tumours, conflicting with the observation made for murine models
for some unknown reasons (15,16).
As we know, the mouse model is a powerful tool in
biological evaluation, while the species differences between humans
and mice often result in the failure of some drug evaluations. Some
researchers have indicated that functional differences might be the
reason for the failure of two clinical Salmonella trials
(15,16), therefore it is crucial to find the
bacteria that are common to both human and mice tumours, which may
serve as sound anticancer protein-delivery agents.
So far, though medical microbiologists have relied
on culture techniques for decades, these culture methods can only
be used for identifying the ‘culturable’ bacteria that grow
relatively quickly and easily in laboratory media (17,18)
because of their various nutritional, pH, temperature and oxygen
requirements, and the interspecies competition and different
concentrations of bacteria within a plate also increase
difficulties with screening (19,20).
To determine the full panoply of organisms within tumours,
high-throughput sequencing methods may be useful since they can
detect almost all the DNA signatures of micro-organisms within a
specific environment, even some of them with low numbers or a
dormant metabolic state (21–23).
In the present study, the high-throughput sequencing
method was applied and the bacterial diversity in human and mice
tumours was compared. In addition, the common bacteria in human and
mice tissues were identified and these might be used as the
potential strain to solve the host's problems of incompatibility in
relation to bacteriotherapy of cancer.
Materials and methods
Ethical statement
Patient samples were obtained with written informed
consent in accordance with the Ethics Committee requirements at the
participating institutes and the Declaration of Helsinki.
Permission to carry out the study was obtained from the
Institutional Review Board (IRB) of the Second Affiliated Hospital
of Nanchang University.
This study was carried out in strict accordance with
the recommendations in the Guide for the Care and Use of Laboratory
Animals of the National Institutes of Health. The protocol was
approved by the Committee on the Ethics of Animal Experiments of
the Second Affiliated Hospital of Nanchang University.
Artificial tumour samples in a mice
model (M-Artificial tumour, M.AT group)
H22 cells (BALB/c background, ATCC, USA) were
cultured in a humidified incubator at 37°C under 5% CO2
in RPMI 1640 supplemented with 10% heat-inactivated foetal bovine
serum, 2 mmol/l glutamine, 100 U/ml penicillin and 100 U/ml
streptomycin (14).
Mice (6 SPF-class KM male mice weighing from 18 to
22 g, 8 weeks) were kept under standardized conditions (22–24°C
temperature; 55±15% humidity) in a 12 h light/12 h dark cycle in
groups of three in an enriched environment (social enrichment,
structured bedding, mouse house with nested material). Food and tap
water were provided ad libitum. To establish a solid tumour
model, each mouse was inoculated with 0.2 ml H22 tumour cell
suspension (1×106/ml PBS) by s.c. injection to the right
flank. At the end of 2 weeks, the mice were sacrificed and soaked
in 75% ethanol for 5 min in a biosafety cabinet, and the tumour
xenografts (one tumor per mouse, ranging from 2,500 to 3,000
mm3, no signs of distress and no tumor ulceration was
observed) were sterilely removed and stored at −20°C for future use
(24).
Spontaneous tumour samples in mice
[M-tumour group (M.T group)]
Mice (6 SPF-class KM mice, aged from 8 to 50 weeks)
with spontaneous tumors (one tumor per mouse, ranging from 2,000 to
3,100 mm3, no signs of distress and no tumor ulceration
was observed) were provided by the Experimental Animal Center of
Nanchang University, and were kept under standardized conditions
(22–24°C temperature; 55±15% humidity) in a 12 h light/12 h dark
cycle in groups of three in an enriched environment (social
enrichment, structured bedding, mouse house with nested material).
Food and tap water were provided ad libitum. The tumour
samples were sterilized separately from the mice and stored at
−20°C for future use (n=6).
Tumour samples from patients [H-tumour
group (H.T group)]
From a total of six patients with diagnosed liver
cancer (without bacterial infection, viral and alcoholic
hepatitis), tumour samples were collected by tumour excision
surgery. Then the samples were stored in disposable sterile tubes
and immediately transferred into the refrigerator at −20°C for
bacterial DNA extraction.
Total genomic DNA extraction and
high-throughput sequencing
Sample DNA was extracted from the samples using a
TIANamp Genomic DNA kit (Tiangen Biotech Co., Ltd., Beijing, China)
according to the manufacturer's instructions. To avoid the loss of
microbial diversity caused by the small amount of bacteria in the
tumours, all the collected tumours in the same group were mixed and
cut into small pieces and homogenized in 500 µl GA with 0.2 g micro
glass beads and centrifuged at 1,000 g for 2 min. Then the mixtures
were transferred into new columns of 1.5 ml. A total of 20 µl of
proteinase K solution (20 mg/ml) was added, vortexed to mix, and
then incubated at room temperature for approximately 5 min. The
Buffer GB provided in the kit (200 µl) was added and vortexed for 5
min. The samples were then placed in a temperature of 70°C for 10
to 15 min. Then 200 µl of ethyl alcohol were added and agitated for
15 sec. The mixtures were transferred into Spin Columns CB3 with a
2 ml collection tube and centrifuged at 10,000 g for 30 sec. The
flow-through liquid was discarded. Then 500 µl of buffer GD was
added and centrifuged at 10,000 g for 30 sec. After discarding the
flow-through liquid, 600 µl of wash buffer PW was added and
centrifuged at 10,000 g for 30 sec. Placed spin columns CB3 for 30
min and added 100 µl of elution buffer at room temperature for 2
min. The column was then centrifuged for 2 min at 10,000 g and
stored at −20°C for future analysis.
Total genomic DNA was amplified using a 515f/806r
primer set that amplifies the V4 region of the 16S rDNA gene
(25,26). The forward primer contains a 6 bp
error-correcting barcode unique to each sample. DNA was amplified
and 16S rRNA tag-encoded high-throughput sequencing was carried out
using the Illumina MiSeq platform at Beijing Novogene
Bioinformatics Technology Co., Ltd. (Beijing, China).
Bioinformatics and multivariate
statistics
Paired-end reads from the original DNA fragments
were merged using FLASH to merge paired-end reads when at least
some of the reads overlapped the read generated from the opposite
end of the same DNA fragment, and paired-end reads were assigned to
each sample according to the unique barcodes.
Then, sequence analysis was performed utilizing the
UPARSE software package using the UPARSE-operational taxonomic unit
(OTU) and UPARSE-OTUref algorithms. In-house Perl scripts were used
to analyse α (within samples) and β (among samples) diversity.
Sequences with ≥97% similarity were assigned to the same OTUs. A
sequence was picked as a representative for each OTU, and the RDP
classifier was used to annotate taxonomic information for each
representative sequence. Cluster analysis was preceded by
unweighted UniFrac distance using the QIIME software package.
Results
Shared genera and bacterial
communities in human and mice tumours
To compare the microbes in human and mice tumours,
16S rRNA amplicon sequencing analysis was used to sequence the V4
hypervariable region. The sequencing data was filtered to get the
valid data, and all the effective tags of all samples were
clustered and those sequences with over 97% similarity were
considered as one OTU. In total, 380,296 usable raw sequences
(1,893 unique sequences) and 678 OTUs were obtained from all the
samples with an average of 226 OTUs per group (Table I). Moreover, the Chao1 index and
Shannon index were nearly saturated and the rarefaction curve of
every sample was able to enter the plateau phase, which indicated
that the sequencing data was reasonable and the species composition
was highly uniform in each group (data not shown).
 | Table I.Number of total tags, taxon tags,
unclassified tags, unique tags and operational taxonomic units from
tumours by high-throughput sequencing. |
Table I.
Number of total tags, taxon tags,
unclassified tags, unique tags and operational taxonomic units from
tumours by high-throughput sequencing.
| Group | Total tags | Taxon tags | Unclassified
tags | Unique tags | OTUs |
|---|
| M.AT | 133,150.00 | 132,546.00 | 0.00 |
604.00 | 244.00 |
| M.T |
55,113.00 |
54,667.00 | 8.00 |
438.00 | 226.00 |
| H.T | 192,033.00 | 191,182.00 | 0.00 |
851.00 | 208.00 |
| Mean | 126,765.30 | 126,131.70 | 2.67 |
631.00 | 226.00 |
| Total | 380,296.00 | 378,395.00 | 8.00 | 1,893.00 | 678.00 |
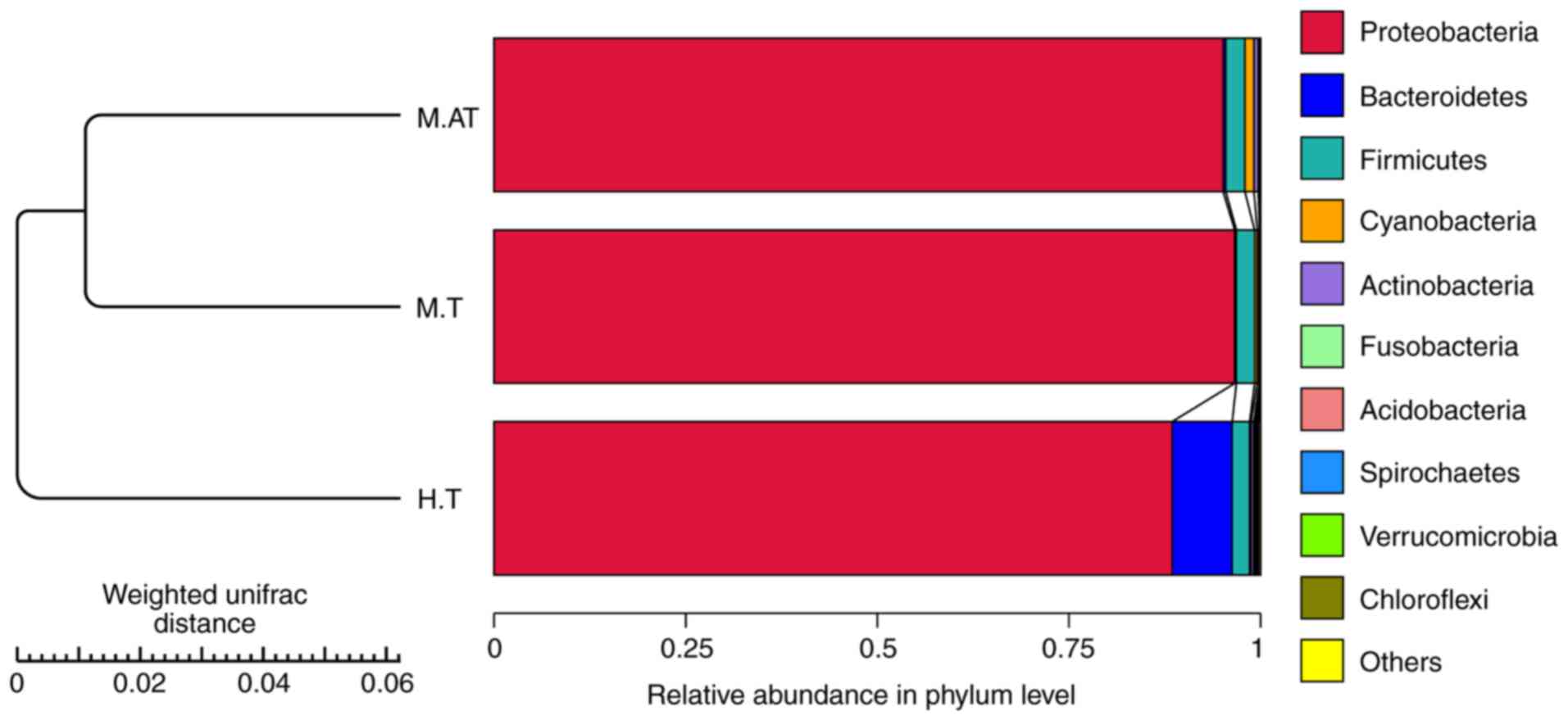
At the phylum level, the data of the top ten
micro-organism populations was analysed. As shown in Fig. 1, Proteobacteria constituted
the predominant phylum in the M.AT, M.T and H.T groups, which
accounted for 95.8, 97 and 89%, respectively, of the total
sequencing number in these three groups.
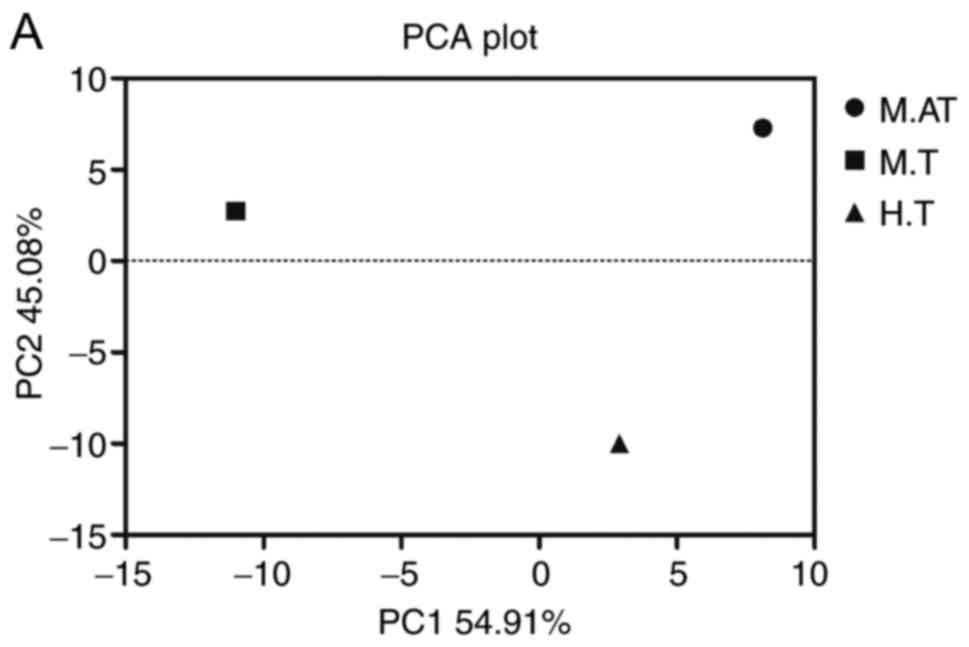
The β diversity of the microbial
community in human and mice tumours
Principal component analysis (PCA) of both human and
mice tumours was conducted, and a closer distance of samples
indicated a more similar bacterial composition. As shown in
Fig. 2A, though the M.AT, M.T and
H.T groups were scattered far from each other, the heatmap of β
diversity indicated that the microbial community among mice tumours
(M.AT vs. M.T, 0.102) was closer than that of M.AT vs. H.T (0.114)
and M.T vs. H.T (0.135) (Fig.
2B).
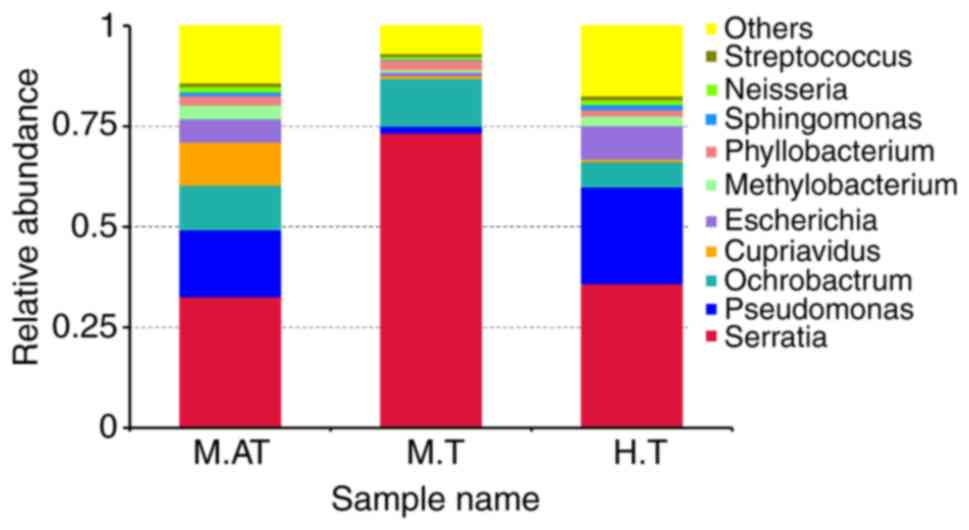
Compositions and relative abundance of
bacterial communities in human and mice tumours
To further investigate the relative microbial
abundance in the M.AT, M.T and H.T groups, the top 10 genera were
clustered. As shown in Fig. 3, the
dominant bacterial genera in the M.AT group were Serratia
(32.64%), Pseudomonas (16.62%), Ochrobactrum
(11.08%), Cupriavidus (10.72%), Methylobacterium
(3.45%) and Phyllobacterium (2.33%), while Serratia
(73.32%), Pseudomonas (1.72%), Ochrobactrum (11.90%)
and Phyllobacterium (2.36%) were predominant in the M.T
group. For the H.T group, Serratia (35.85%),
Pseudomonas (24.10%), Ochrobactrum (6.28%),
Escherichia (8.32%), Methylobacterium (2.38%),
Phyllobacterium (1.59%) and Sphingomonas (1.35%) were
identified as the dominant bacteria. In addition, the bacteria
classified as ‘others’ had accounted for 14.21, 6.90 and 17.38% of
the total sequencings.
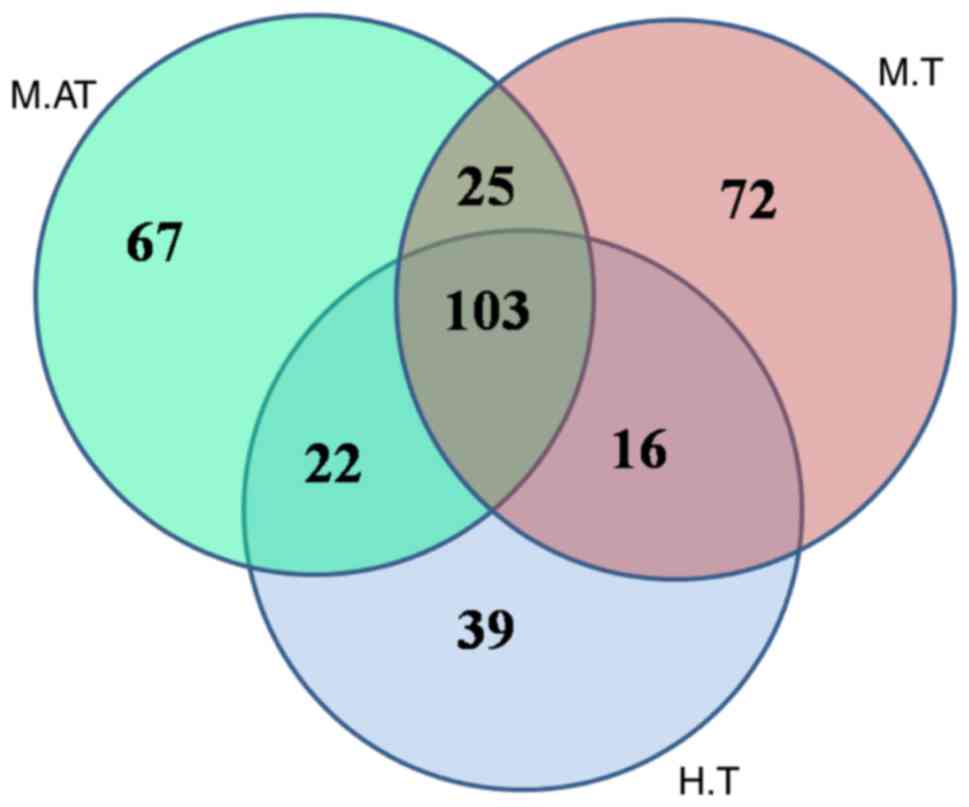
The specificity of bacterial
communities in human and mice tumours
The Venn figure.reflecting the difference between
human and mice tumours depicted, as shown in Fig. 4, that there were 217, 216 and 180
OTUs in the M.AT, M.T and H.T groups, respectively. A comparison
between all groups indicated that 103 common OTUs had been
identified, of which 2, 0, 1 and 0 OTUs belonged to
Lactobacillus, Salmonella, Escherichia and
Clostridium, respectively. Therefore, the no detection of
Salmonella and Clostridium in either human or mouse
tumours suggests that these two strains were not a good choice for
anticancer therapies.
Discussion
Tumours are composed of necrotic, hypoxic and
hypoxic areas offering a perfect niche for the growth of anaerobic
bacteria (10). Since Coley's
original work treating cancer patients with Streptococcus
pyogenes over 100 years ago, a variety of anaerobic bacteria
have been considered for this purpose but failed in part because of
poor reproducibility and unacceptable toxicity (27–29).
More recent work involved attenuated strains of S.
typhimurium, while the phase 1 clinical trials of S.
typhimurium received limited efficacy, though the bacterium
could be safely administered and targeted at tumours in both dogs
and human patients (16,30). Moreover, recent studies indicated
that bacteria imbalance (dysbiosis) did affect oncogenesis, tumour
progression and cancer therapy (3,6,31).
Unfortunately, little work has been done to explore the microbial
diversity inside tumours and clarify their potential roles in
tumour development and therapy.
To the best of our knowledge, the distribution of
bacteria, e.g., Bifidobacterium longum, lactic acid bacteria,
Clostridium novyi and Salmonella typhimurium, was mainly
affected by the volume of solid tumours rather than tumour sites
and types (32,33), so the microbial diversity in solid
tumours in humans and mice was compared using high-throughput
sequencing. For convenience of sampling, liver cancer, the second
leading cause of cancer death, was collected both from humans and
mice. Moreover, as the low microbial amount and rich tumour stroma
and blood vessels would lead to a huge loss of DNAs, which would
finally reduce the biodiversity of the bacterial community in
tumours, all the samples collected from the same different tissues
were mixed and a mean number of 378,395 usable sequences and the
saturated Chao1 index and Shannon index (Table I) ensured their reliability for
future analysis. Even so, more advanced DNA extraction method will
be applied in our further work to expand the sample size and
sustain the individual difference, and the fluorescence in
situ hybridization (FISH) will be used to locate the bacterial
position in liver tumors.
As shown in Fig. 1,
Proteobacteria accounted for 95.8, 97 and 89%, respectively,
of the total sequencing number in the M.AT, M.T and H.T groups.
Proteobacteria are a major group of gram-negative bacteria
and include a wide variety of pathogens, such as Escherichia,
Salmonella, Vibrio, Helicobacter and Yersinia, and some
of this phylum is responsible for nitrogen fixation (34). However, this group has been
detected as the dominant population in the human intestine and oral
cavity (26,35), so much more work needs to be done
to explore its potential effects on human health or diseases
(including cancer).
In Fig. 2, the
widely scattered PCA symbols of M.AT, M.T and H.T showed an
obviously low similarity in the microbial community in these three
groups, though the β diversity index indicated that the microbial
community in mice was much closer than that in different species.
Therefore, the obviously different microbial community between
humans and mice (even the microbial communities of spontaneous
tumours and artificial tumours were obviously different) indicated
that the tumour size and microenvironment (e. g. oxygen and pH)
might create different survival environments for different
micro-organisms, and the low similarity among the M.AT, M.T and H.T
groups partly explained the failure of the phase 1 clinical trials
of S. typhimurium.
Then, data of the top ten micro-organism populations
was analysed at the genus level. Just as expected, the microbial
composition in human tumours and artificial mice tumours was
obviously different, however Serratia (35.85 vs. 32.64%),
Pseudomonas (24.10 vs. 16.62%), Ochrobactrum (6.28
vs. 11.08%) had been identified as the dominant bacteria in human
tumours and artificial tumours. Interestingly, the microbial
composition of the M.T group was obviously different to that of the
H.T and M.AT groups at the genus level, and Serratia and
Ochrobactrum accounted for 73.32 and 11.90% of the total
sequencings, respectively (Fig.
3). As we know, Serratia is a genus of facultative
anaerobic bacteria of the Enterobacteriaceae family, and can be
distinguished from other members of the Enterobacteriaceae family
by their unique production of three enzymes: DNase, lipase and
gelatinase (36).
Pseudomonas belongs to the Pseudomonadaceae family
containing 191 valid species, and they demonstrate a great deal of
metabolic diversity, and consequently are able to colonize a wide
range of niches (37).
Interestingly, Streptococcus, the first genus used to treat
cancer patients 100 years ago, accounted for 0.7–1.0% of the total
number in these three groups.
To find the common bacteria between human and mice
tumours, the bacterial compositions were compared. In Fig. 4, the Venn figure reflected 103
common OTUs in the M.AT, M.T and H.T groups, and only two and one
OTUs belonged to Lactobacillus and Escherichia, and
no OTUs belonging to Salmonella, Bifidobacteria and
Clostridium were found. As we know, Streptococcus,
Lactobacillus, Escherichia, Salmonella, Bifidobacteria and
Clostridium have been used as vectors for gene therapy of
cancer (10,38–40),
therefore the existence of Streptococcus, Lactobacillus and
Escherichia both in humans and mice showed their potential
value for animal and human verification. Especially for
Lactobacillus, whose most members had been used in the
preparation of fermented foods and these microorganisms were
generally considered safe (41).
In conclusion, our results indicated that various
bacteria exist both in human and mice tumours, and the
significantly different bacterial composition in artificial mice
tumours, spontaneous mice tumours and human tumours may partly
explain the sound effect of attenuated Salmonella in murine
models and the unsatisfactory observation in human tumours.
However, our work indicated that the common bacteria of
Streptococcus, Lactobacillus and Escherichia both in
human and mice tumours might be used as potential strains in the
immunotherapy and bacteriotherapy of cancers.
Acknowledgements
The authors would like to thank Dr Bin Xie (Jiangxi
University of Traditional Chinese Medicine) and Miss Lin Xie
(Nanchang University) for providing the mouse and human tumors, as
well as all of the members in Dr Chen's Laboratory (Institute of
Translational Medicine) for their generous help and valuable
discussion.
Funding
The present study was supported by grants from The
National Natural Science Foundation of China (grant nos. 81503364
and 31560264) and Excellent Youth Foundation of Jiangxi Scientific
Committee (grant no. 20171BCB23028), and Science and Technology
Plan of Jianxi Health Planning Committee (grant no. 20175526).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TC designed the experiments, analysed the data and
wrote the manuscript. FZ, MZ, YW and CL performed the experiments.
All authors discussed the results and commented on the final
manuscript.
Ethics approval and consent to
participate
The protocol was approved by the Committee on the
Ethics of Animal Experiments of the Second Affiliated Hospital of
Nanchang University (Jiangxi, China). Patient samples were obtained
with written informed consent in accordance with the Ethics
Committee's requirements.
Consent for publication
Written informed consent was obtained from all
participants.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Williams NS, Bailey H, Bulstrode CJ, Love
RM and O'Connell PR: Bailey & Love's Short Practice of Surgery.
CRC Press; Boca Raton, FL: 2008, View
Article : Google Scholar
|
|
2
|
Postow MA, Chesney J, Pavlick AC, Robert
C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK,
Agarwala SS, et al: Nivolumab and ipilimumab versus ipilimumab in
untreated melanoma. N Engl J Med. 372:2006–2017. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sivan A, Corrales L, Hubert N, Williams
JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B,
Alegre ML, et al: Commensal Bifidobacterium promotes
antitumor immunity and facilitates anti-PD-L1 efficacy. Science.
350:1084–1089. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hoos A: Development of immuno-oncology
drugs-from CTLA4 to PD1 to the next generations. Nat Rev Drug
Discov. 15:235–247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vétizou M, Pitt JM, Daillère R, Lepage P,
Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong
CP, et al: Anticancer immunotherapy by CTLA-4 blockade relies on
the gut microbiota. Science. 350:1079–1084. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maynard CL, Elson CO, Hatton RD and Weaver
CT: Reciprocal interactions of the intestinal microbiota and immune
system. Nature. 489:231–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei MQ, Mengesha A, Good D and Anné J:
Bacterial targeted tumour therapy-dawn of a new era. Cancer Lett.
259:16–27. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gur C, Ibrahim Y, Isaacson B, Yamin R,
Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N,
Coppenhagen-Glazer S, et al: Binding of the Fap2 protein of
Fusobacterium nucleatum to human inhibitory receptor TIGIT
protects tumors from immune cell attack. Immunity. 42:344–355.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Frahm M, Felgner S, Kocijancic D, Rohde M,
Hensel M, Curtiss R III, Erhardt M and Weiss S: Efficiency of
conditionally attenuated Salmonella enterica serovar
Typhimurium in bacterium-mediated tumor therapy. MBio.
6:pii: e00254. –15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roberts NJ, Zhang L, Janku F, Collins A,
Bai RY, Staedtke V, Rusk AW, Tung D, Miller M, Roix J, et al:
Intratumoral injection of Clostridium novyi-NT spores
induces antitumor responses. Sci Transl Med. 6:249ra1112014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zeuthen LH, Christensen HR and Frøkiær H:
Lactic acid bacteria inducing a weak interleukin-12 and tumor
necrosis factor alpha response in human dendritic cells inhibit
strongly stimulating lactic acid bacteria but act synergistically
with gram-negative bacteria. Clin Vaccine Immunol. 13:365–375.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fox M, Lemmon M, Mauchline M, Davis TO,
Giaccia AJ, Minton NP and Brown JM: Anaerobic bacteria as a
delivery system for cancer gene therapy: In vitro activation of
5-fluorocytosine by genetically engineered clostridia. Gene Ther.
3:173–178. 1996.PubMed/NCBI
|
|
13
|
Low KB, Ittensohn M, Le T, Platt J, Sodi
S, Amoss M, Ash O, Carmichael E, Chakraborty A, Fischer J, et al:
Lipid A mutant Salmonella with suppressed virulence and TNFv
induction retain tumor-targeting in vivo. Nat Biotechnol. 17:37–41.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao M, Yang M, Li XM, Jiang P, Baranov E,
Li S, Xu M, Penman S and Hoffman RM: Tumor-targeting bacterial
therapy with amino acid auxotrophs of GFP-expressing Salmonella
typhimurium. Proc Natl Acad Sci USA. 102:pp. 755–760. 2005;
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nemunaitis J, Cunningham C, Senzer N, Kuhn
J, Cramm J, Litz C, Cavagnolo R, Cahill A, Clairmont C and Sznol M:
Pilot trial of genetically modified, attenuated Salmonella
expressing the E. coli cytosine deaminase gene in refractory
cancer patients. Cancer Gene Ther. 10:737–744. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Toso JF, Gill VJ, Hwu P, Marincola FM,
Restifo NP, Schwartzentruber DJ, Sherry RM, Topalian SL, Yang JC,
Stock F, et al: Phase I study of the intravenous administration of
attenuated Salmonella typhimurium to patients with
metastatic melanoma. J Clin Oncol. 20:142–152. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
McGuckin M, Goldman R, Bolton L and
Salcido R: The clinical relevance of microbiology in acute and
chronic wounds. Adv Skin Wound Car. 16:12–25. 2003. View Article : Google Scholar
|
|
18
|
Thomson PD: Immunology, microbiology, and
the recalcitrant wound. Ostomy Wound Manage. 46 1A Suppl:77S–84S.
2000.PubMed/NCBI
|
|
19
|
Whitley R: The new age of molecular
diagnostics for microbial agents. N Engl J Med. 358:988–989. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Davies CE, Hill KE, Wilson MJ, Stephens P,
Hill CM, Harding KG and Thomas DW: Use of 16S ribosomal DNA PCR and
denaturing gradient gel electrophoresis for analysis of the
microfloras of healing and nonhealing chronic venous leg ulcers. J
Clin Microbiol. 42:3549–3557. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kobayashi N, Bauer TW, Tuohy MJ, Lieberman
IH, Krebs V, Togawa D, Fujishiro T and Procop GW: The comparison of
pyrosequencing molecular Gram stain, culture, and conventional Gram
stain for diagnosing orthopaedic infections. J Orthop Res.
24:1641–1649. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Zhang M, Pang X, Zhao Y, Wang L
and Zhao L: Structural resilience of the gut microbiota in adult
mice under high-fat dietary perturbations. ISME J. 6:1848–1857.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang
X, Jia W, Cai S and Zhao L: Structural segregation of gut
microbiota between colorectal cancer patients and healthy
volunteers. ISME J. 6:320–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao A, Nunez-Cruz S, Li C, Coukos G,
Siegel DL and Scholler N: Rapid isolation of high-affinity human
antibodies against the tumor vascular marker Endosialin/TEM1, using
a paired yeast-display/secretory scFv library platform. J Immunol
Methods. 363:221–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun M, Xiao T, Ning Z, Xiao E and Sun W:
Microbial community analysis in rice paddy soils irrigated by acid
mine drainage contaminated water. Appl Microbiol Biotechnol.
99:2911–2922. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu X, Wu X, Qiu L, Wang D, Gan M, Chen X,
Wei H and Xu F: Analysis of the intestinal microbial community
structure of healthy and long-living elderly residents in Gaotian
Village of Liuyang City. Appl Microbiol Biotechnol. 99:9085–9095.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Coley WB: The treatment of inoperable
sarcoma by bacterial toxins (the mixed toxins of the
Streptococcus erysipelas and the Bacillus
prodigiosus). Proc R Soc Med. 3:(Surg Sect). pp. 1–48. 1910;
PubMed/NCBI
|
|
28
|
Coley WB: The treatment of malignant
tumors by repeated inoculations of erysipelas. With a report of ten
original cases. 1893. Clin Orthop Relat Res. 262:3–11. 1991.
|
|
29
|
Hoffman RM and Zhao M: Methods for the
development of tumor-targeting bacteria. Expert Opin Drug Discov.
9:741–750. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thamm DH, Kurzman ID, King I, Li Z, Sznol
M, Dubielzig RR, Vail DM and MacEwen EG: Systemic administration of
an attenuated, tumor-targeting Salmonella typhimurium to
dogs with spontaneous neoplasia: Phase I evaluation. Clin Cancer
Res. 11:4827–4834. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Iida N, Dzutsev A, Stewart CA, Smith L,
Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S,
et al: Commensal bacteria control cancer response to therapy by
modulating the tumor microenvironment. Science. 342:967–970. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee CH: Engineering bacteria toward tumor
targeting for cancer treatment: Current state and perspectives.
Appl Microbiol Biotechnol. 93:517–523. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Singh R and Paterson Y: Listeria
monocytogenes as a vector for tumor-associated antigens for cancer
immunotherapy. Expert Rev Vaccine. 5:541–552. 2006. View Article : Google Scholar
|
|
34
|
Stackebrandt E, Murray R and Trüper H:
Proteobacteria classis nov., a name for the phylogenetic
taxon that includes the ‘purple bacteria and their relatives’. Int
J Syst Bacteriol. 38:321–325. 1988. View Article : Google Scholar
|
|
35
|
Diaz PI, Dupuy A, Abusleme L, Reese B,
Obergfell C, Choquette L, Dongari-Bagtzoglou A, Peterson DE, Terzi
E and Strausbaugh LD: Using high throughput sequencing to explore
the biodiversity in oral bacterial communities. Mol Oral Microbiol.
27:182–201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grimont PA and Grimont F: The genus
Serratia. Annu Rev Microbiol. 32:221–248. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Euzéby JP: List of bacterial names with
standing in nomenclature: A folder available on the internet. Int J
Syst Bacteriol. 47:590–592. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Baban CK, Cronin M, O'Hanlon D, O'Sullivan
GC and Tangney M: Bacteria as vectors for gene therapy of cancer.
Bioeng Bugs. 1:385–394. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zu C and Wang J: Tumor-colonizing
bacteria: A potential tumor targeting therapy. Crit Rev Microbiol.
40:225–235. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bolhassani A and Zahedifard F: Therapeutic
live vaccines as a potential anticancer strategy. Int J Cancer.
131:1733–1743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang W, Wang C, Huang C, Yu Q, Liu H,
Zhang C and Pei X: Construction and expression of food-grade
β-galactosidase gene in Lactococcus lactis. Curr Microbiol.
62:639–644. 2011. View Article : Google Scholar : PubMed/NCBI
|