Introduction
Malignant tumors of the vulva represent a very small
number of female genital malignancies, 80–90% of vulvar cancers are
vulvar squamous cell carcinomas (VSCCs). There will be about 6020
new patients in 2017, and 1150 people are expected to die. The
incidence is increasing by 0.6% annually in the last 10 years
(1). Women with distant metastases
(5%) and local diffusion (31%) have bad prognosis. The 5 year
survival rates were 17 and 57%, respectively (1). Thus it is of important clinical
essentiality to research the occurrence and development of VSCC. In
the present case, many scholars have begun to focus on the
microRNAs (miRNAs).
miRNAs prevent translation or promote mRNA
degradation (2). miRNA plays a key
role in the carcinogenesis and evolution by accommodating
epithelial-mesenchymal transition (EMT), oncogenic signaling
pathways and metastasis (3).
miRNAs repress target gene expression and usually perform important
functions in cancers.
Based on our previous findings (4), miR-3147 was markedly increased in
VSCC tissues, but the underlying mechanism is still unknown. Smad4
is responsible for regulating the TGF-β/Smad signaling pathway and
it was downregulated in VSCC samples (4). Based on the target prediction program
miRanda, Smad4 is a target gene for miR-3147, but the association
between miR-3147 and Smad4 in VSCC requires further investigation.
In the research, we performed functional studies to find the roles
of miR-3147 and Smad4 in VSCC. Smad4 was confirmed to be a target
gene of miR-3147.
Materials and methods
Experimental samples
Twenty VSCC and adjacent non-dysplastic tissues were
obtained from the patients admitted to the Department of
Gynaecology at our hospital between 2010 and 2017. Every sample of
VSCC was negative in HPV, and Linear Array HPV Genotyping (Roche
Applied Science, Pleasanton, CA, USA) was used for the approach HPV
testing. According to the standard of World Health Organization
(WHO), the diagnosis in histopathology was conducted. The grade of
tumor was determined according to the new version of the
International Federation of Gynecology and Obstetrics (FIGO) system
published in 2010; the stage of tumor was identified by the 7th TNM
categorization of the Union for International Cancer Control
(UICC). For the stages of tumors that diagnosed before 2009, they
were identified again based the latest version. The frozen
specimens were microscopically identified by two pathologists.
Clinicopathological data were retrospectively collected. Patients
without preoperative radiotherapy or chemotherapy were chosen. The
research was performed based on the Helsinki declaration. Prior to
surgery, all subjects provided written informed consent to
participate in the study. The Ethics Committee of the First
Affiliated Hospital of China Medical University (Liaoning, China)
approved the present study.
Cell culture and transfection
Acquirement of the A431 cell line was made from
ATCC, in addition to culturing in RPMI 1640 with 10% fetal bovine
serum at a temperature of 37°C and a humid atmosphere of 5%
CO2. Using the Dharmacon miRIDIAN miR-3147 mimics
(miR-3147), the transfection of the cells was carried out, together
with the negative control (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) at an eventual amounting to be 100 nmol/l. A small
interfering RNA targeting Smad4 (siR-Smad4; sc-29,484) was attained
from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA).
Transfections were conducted with the help of Lipofectamine 2000 in
accordance with the protocol of the manufacturer.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA extraction was conducted by making use of
TRIzol reagent (Takara Bio, Inc., Shiga, Japan) in accordance with
the guidelines of the manufacturer. Stem-loop RT-PCR was performed
for the purpose of testing the miR-3147 expression (5). Complementary DNA (cDNA) synthesis was
conducted according to a Gene Amp PCR System 9700 (Applied
Biosystems; Thermo Fisher Scientific, Inc.). We calculated the mRNA
expressions with the 2−ΔΔCq approach. Furthermore, the
primers for miRNAs and mRNAs are shown (Table I).
 | Table I.Primers used in reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primers used in reverse
transcription-quantitative polymerase chain reaction.
| Gene | Forward primers
(5′-3′) | Reverse primers
(5′-3′) |
|---|
| U6 |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
| miR-3147 |
GGUUGGGCAGUGAGGAGGGUGUGA |
TACGAGGTTGGGCAGTGAGGAGGGTGT |
| Smad4 |
CGCTTTTGTTTGGGTCAACT |
CCCAAACATCACCTTCACCTC |
| E-cadherin |
TGCTGTTTCTGGTTTCTGTTGG |
CTTCTCCGTATTCTCCTCCCT |
| N-cadherin |
TTTGGGGAGGGGTAAAAGTTC |
AAGAAACAGGCCACCCCTTT |
| Vimentin |
CGGTTGAGACCAGAGATGGA |
TGCTGGTACTGCACTGTTGG |
| P21 |
TGTCCGTCAGAACCCATG |
TGGGAAGGTAGAGCTTGG |
| PAI-1 |
GAGACAGGCAGCTCGGATTC |
GGCCTCCCAAAGTGCATTAC |
| Bax |
ATGGACGGGTCCGGGGAGCAG |
CATGATGGTTCTGATCAGT |
| Bim |
GCCTTCAACCACTATCTCA |
ATCCAGCTCGGTGTCTTCT |
| GAPDH |
AAGGTGAAGGTCGGAGTCAAC |
GGGTCATTGATGGCAACAATA |
Western blotting
Cells were harvested, together with the calculation
of protein concentration through the use of a protein assay kit
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). Three days
subsequent to the transfection with the miR-3147 mimics or
siRNA-Smad4, the total protein was obtained (6). In respect of immunoblotting, the
membrane was incubated with antibodies against E-cadherin (1:500,
ab15148; Abcam, Cambridge, UK), N-cadherin (1:500, ab18203; Abcam),
Vimentin (1:500, ab24525; Abcam), matrix metalloproteinase 2
(MMP-2; 1:300; BIOSS, Beijing, China), MMP-9 (1:300; BIOSS) or
GAPDH (1:10,000, ab181602; Abcam). Rinsing of the membrane was
performed, followed by incubation with anti-mouse or anti-rabbit
IgG (H+L)-HRP conjugate (1:10,000; Invitrogen; Thermo Fisher
Scientific, Inc.) antibody. We made use of the Image J software
(National Institute of Health, Bethesda, MD, USA) in order to
analyze the results.
MTT test
Cell proliferation assay was assessed by MTT test.
Ninety-six well plates were seeded with 5×104 cells per
well. At the appointed time after transfection (0, 48, and 96 h),
20 µl MTT (Sigma-Aldrich, St. Louis, MO, USA) was put into the
well, followed by the incubation of the cells at a temperature of
37°C for a period of 4 h. The liquid was abandoned and then we put
l50 µl dimethylsulfoxide (DMSO; Sigma-Aldrich) to lysis the
formazan. Measurement of the optical density was also taken at 490
nm by using a microplate spectrophotometer.
Cell migration and invasion
For the purpose of testing cell migration, a
Transwell test was conducted. 48 h of time following the
transfection, addition of 5×104 cells was made into the
above chambers of plates by an untreated membrane. Subsequent to 24
h of incubation, addition of 4% paraformaldehyde was made into the
chambers, followed by a fixation using the crystal violet. We
carried out the calculations of the cells, which passed all across
the membrane. As for the invasion experiments, Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) was used on the above
chamber. The other mechanisms possessed similarities with the
migration experimentation. All the tests were repeated in
triplicate.
Distribution of cell cycle
More than 1×104 cells were trypsinized,
collected, washed twice, as well as positioned in 70% ethanol for a
time period of 24 h. The cells were washed twice with PBS,
following the incubation using 400 µl RNase (0.25 mg/ml) at a
temperature of 37°C for 1 h, proceeding to treatment with propidium
iodide (40 µg/ml). Determination of the cell cycle distribution was
made through flow cytometry. The experiments were conducted for
three times.
Luciferase reporter assay
The fragment of the human Smad4 with (wild-type) or
without (mutant) the miR-3147 binding site at the 3′-untranslated
region (3′-UTR) was cloned and inserted into the pGL3-basic
luciferase report plasmid (Promega Corporation, Madison, WI, USA)
to generate luciferase reporter vectors, Smad4 3′-UTR-wt and Smad4
3′-UTR-mut. The 293T cells were treated in 96-well plates at 5,000
cells per well and permitted to stay for 24 h prior to
transfection. Then, transfection of the miR-3147 mimics or miR-NC
was performed into the 293T cells with the use of Lipofectamine
2000 with 100 ng of Smad4 3′-UTR-wt or Smad4 3′-UTR-mut, in
addition to 10 ng of pRL-TK Renilla plasmid (Promega Corporation).
Following the incubation for a time period of 48 h, determination
of the luciferase activities was performed with the help of a Dual
Luciferase Reporter system (Promega Corporation).
Statistical method
The presentation of the data has been made as the
means ± SD. Furthermore, we made use of the SPSS 19.0 software
(SPSS, Inc., Chicago, IL, USA) for the purpose of analyzing the
findings. A P-value of <0.05 was thought to be statistically
significant. Analysis of the significance of differences in the
mean values was carried out with the application of Student's
t-test. The use of a two-sided Fisher's exact test was made for the
purpose of determining the association between the expression level
of miR-3147 and clinicopathological data.
Results
RT-qPCR analysis
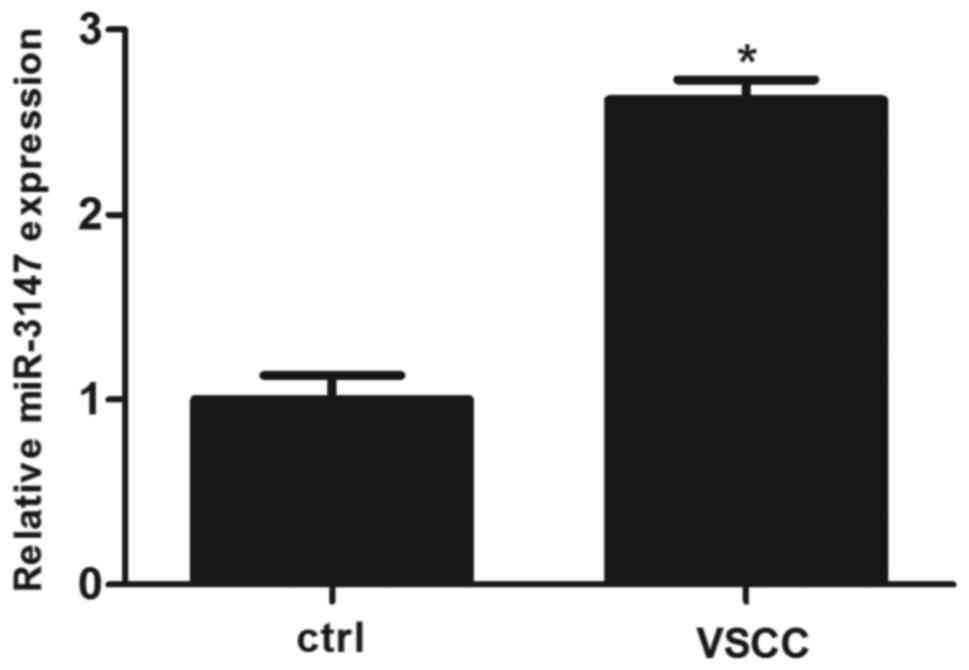
A RT-qPCR analysis of miR-3147 in VSCC as well as
surrounding non-dysplastic tissues was conducted. The expression of
miR-3147 was significantly increased in VSCC tissues
(P=8.67×10−5; Fig.
1).
Relationship between miR-3147 and
clinicopathological parameters
The 75th percentile of the 2−ΔΔCt values
was considered to be the cut-off value for tissues with low or high
expressions of miR-3147 (7). The
expression level of miR-3147 was not related to age, tumor
differentiation, vascular invasion, FIGO stage, tumor size and
lymph node metastasis, but was positively related to the depth of
invasion (P=0.023; Table II). The
results showed that the upregulation of miR-3147 contributes to the
invasion of VSCC.
 | Table II.Associations between the expression
of microRNA-3147 and clinical pathology. |
Table II.
Associations between the expression
of microRNA-3147 and clinical pathology.
| Variable | Number of cases
(n) | Low expression
(n) | High expression
(n) | P-value |
| Age (years) |
|
|
| 0.178 |
|
25–69 | 10 | 7 | 3 |
|
|
70–85 | 10 | 4 | 6 |
|
|
Differentiation |
|
|
| 0.961 |
|
Well | 10 | 4 | 6 |
|
|
Moderate | 7 | 3 | 4 |
|
|
Poor | 3 | 1 | 2 |
|
| Vascular
invasion |
|
|
| 0.531 |
|
Yes | 3 | 1 | 2 |
|
| No | 17 | 9 | 8 |
|
| Lymph node
metastasis |
|
|
| 0.436 |
| No | 15 | 6 | 9 |
|
|
Yes | 5 | 3 | 2 |
|
| FIGO staging |
|
|
| 0.286 |
| I | 10 | 2 | 8 |
|
| II | 7 | 4 | 3 |
|
|
III | 3 | 1 | 2 |
|
| Tumor diameter
(cm) |
|
|
| 0.989 |
|
0.3–2.5 | 10 | 3 | 7 |
|
|
2.6–4.0 | 7 | 2 | 5 |
|
|
4.1–20.0 | 3 | 1 | 2 |
|
| Depth of invasion
(mm) |
|
|
| 0.023a |
|
0.0–4.0 | 10 | 2 | 8 |
|
|
4.1–8.0 | 7 | 6 | 1 |
|
|
8.1–40.0 | 3 | 2 | 1 |
|
Effects of miR-3147 on vulvar
carcinoma in vitro
In comparison with the negative control, the
proliferation rate of the cells that had undergone transfection
with the miR-3147 mimics amounted to be significantly higher at 96
h (P=0.007; Fig. 2A). Selection of
MMP-2 as well as MMP-9 was made for the purpose of testing if
miR-3147 was capable of affecting the genes associated with tumor
invasion. MMP-2 and MMP-9 were increased after transfection
(Fig. 2B). For the fact that Smad4
is termed as a crucial mediator in TGF-β signal pathway, the
inhibition of Smad4 by miR-3147 is most likely to decrease the
expressions of downstream target genes in this pathway. Our results
displayed that miR-3147 transfection resulted into the inhibition
of the mRNA levels of p21 (P=0.007), PAI-1 (P=0.011), Bax (P=0.007)
and Bim (P=0.002; Fig. 2C),
indicating miR-3147 could regulate Smad4-mediated signaling
pathway. The miR-3147 mimics markedly increased the migration
(P=0.002; Fig. 2D) and invasion
(P=0.015; Fig. 2E) of A431 cells.
Cell cycle results revealed the fact that the cells in the G1 phase
were notably dropped from 62.34±1.62% to 57.87±1.78% (P=0.032;
Fig. 2F). In addition, the cells
in the S phase were remarkably added from 24.84±1.07% to
27.70±0.94% at 48 h (P=0.004; Fig.
2F).
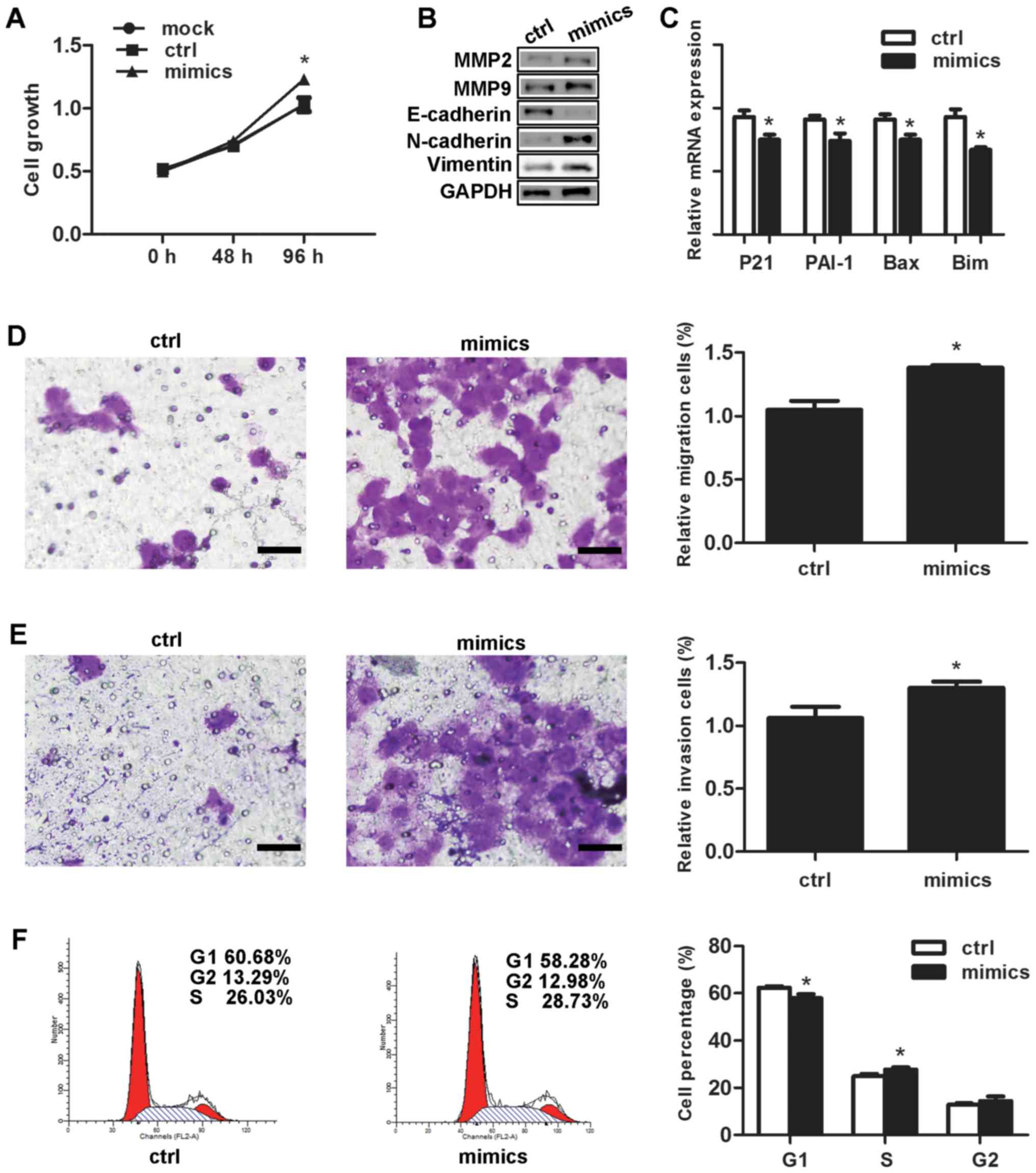 | Figure 2.Effects of miR-3147 on vulvar
carcinoma in vitro. Overexpression of miR-3147 (A) increased
the rate of growth, (B) promoted EMT and increased levels of MMP-2
and MMP-9, (C) decreased downstream target genes of Smad4, (D)
increased migration, (E) increased invasion (scale bars, 200 µm)
and (F) promoted G1/S progression. *P<0.05 vs. ctrl. miR,
microRNA; Ctrl, control; EMT, epithelial-mesenchymal transition;
MMP, matrix metalloproteinase; Bax, B-cell lymphoma 2-associated X
protein; Bim, B-cell lymphoma-2-like protein 11; PAI-1, plasminogen
activator inhibitor-1. |
Effects of Smad4 on vulvar carcinoma
in vitro
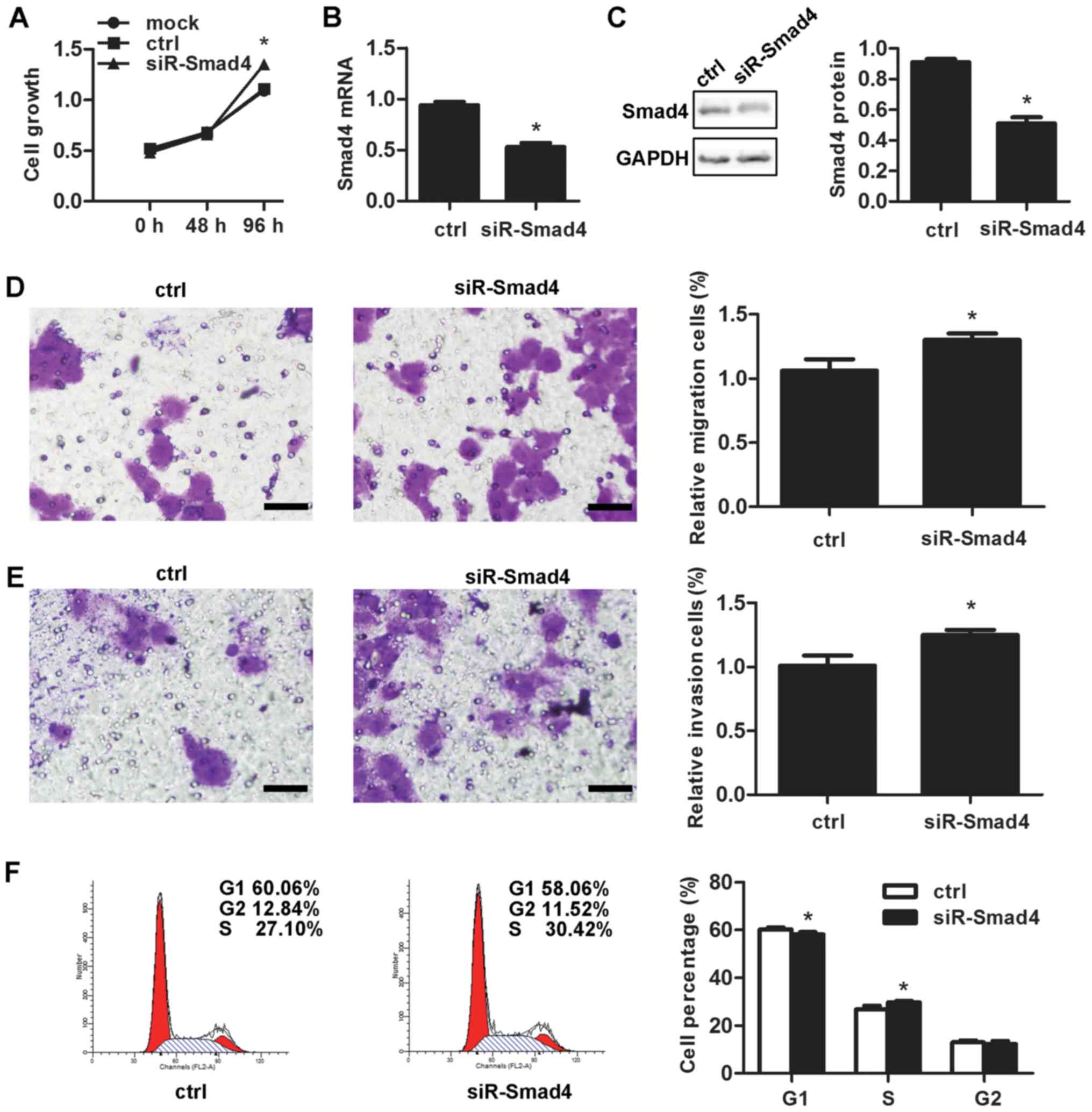
The siR-Smad4 was performed to discuss the effect of
Smad4 in vulvar cancer. Smad4 knockdown resulted into a surprising
promotion of the cell proliferation (P=0.001; Fig. 3A). siR-Smad4 transfection led to
the downregulation of the mRNA as well as protein expression levels
of Smad4 by 47.67±5.51% (P=1.18×10−4; Fig. 3B) and 40.00±1.73%
(P=7.07×10−5; Fig. 3C),
respectively. Smad4 knockdown also markedly increasd migration
(P=0.003; Fig. 3D) and invasive
ability (P=0.011; Fig. 3E), in
addition to decreasing cells in the G1 stage from 60.24±0.83% prior
to knockdown to 58.05±1.04% at 48 h subsequent to the knockdown
(P=0.003; Fig. 3F).
The effect of miR-3147 in EMT
The transfection of the miR-3147 mimics into A431
cells was performed. After 4 days, no obvious change in the shape
of cells was observed. miR-3147 transfection downregulated
E-cadherin protein expression, in addition to upregulating the
protein expressions of N-cadherin as well as Vimentin (Fig. 2B). The findings revealed the fact
that miR-3147 could take part in the mechanism of EMT in vulvar
cancer.
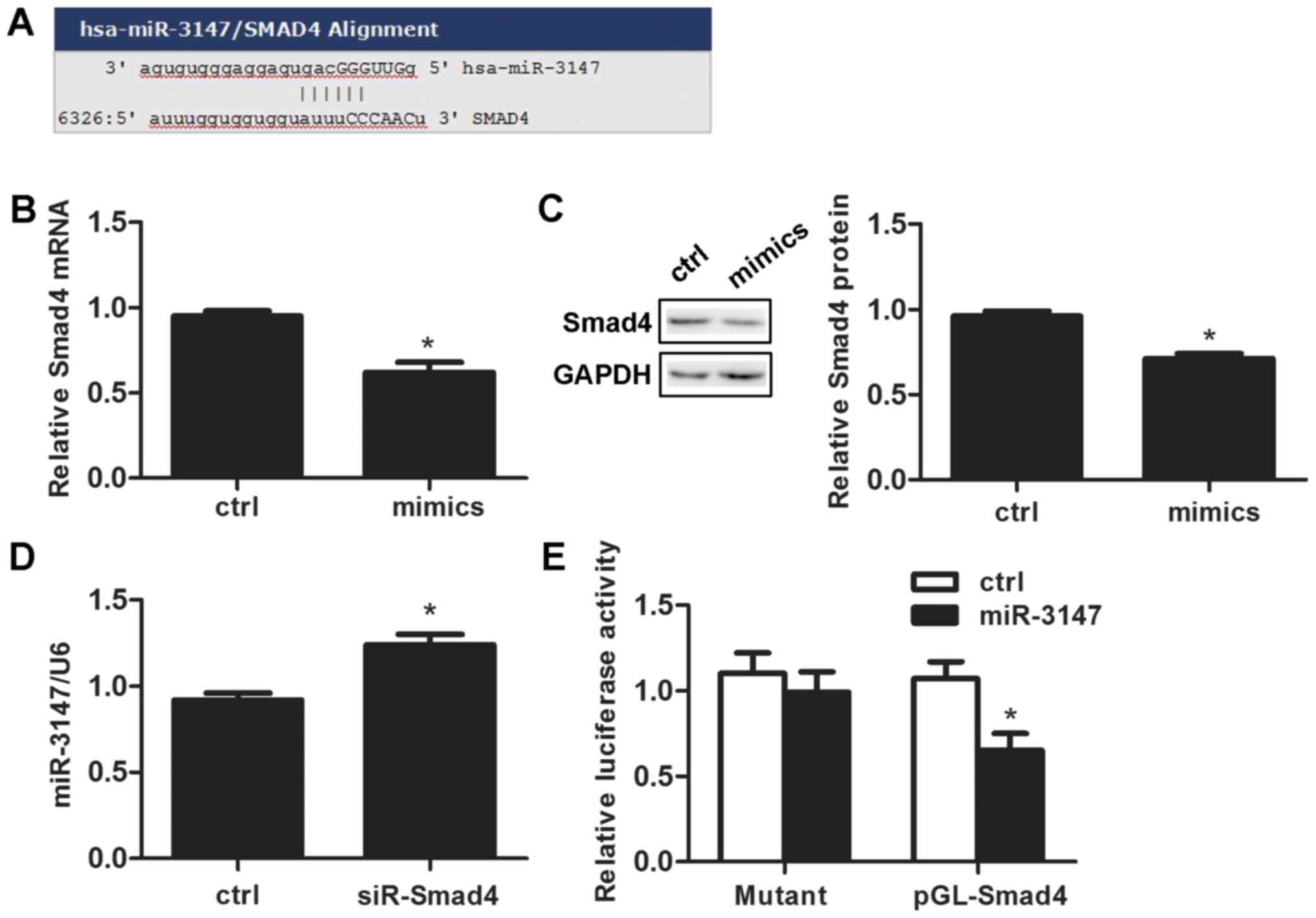
miR-3147 targets Smad4
We searched the target prediction website miR and a
(http://www.microrna.org/microrna/home.do) and
predicted that the miR-3147 promoter contains an Smad4 binding site
(Fig. 4A). miR-3147 transfection
reduced Smad4 mRNA expression by 33.0±7.0% after 48 h (P=0.001;
Fig. 4B). The change of Smad4
protein was consistent with that of mRNA (P=2.75×10−4;
Fig. 4C). Silencing Smad4 promoted
the expression level of miR-3147 (P=0.001; Fig. 4D). Dual-luciferase reporter assays
results indicated that the expression of luciferase reporter gene
linked with part sequence of the 3′UTR of Smad4 gene was
downregulated by the overexpression of miR-3147 in A431 cells
(P=0.004; Fig. 4E), and the
downregulation behavior was abolished when the miR-3147 binding
site in 3′UTR of Smad4 was mutated (P=0.571; Fig. 4E).
Discussion
VSCC is a relatively rare disease. Standard therapy
for VSCC has been described in the recently developed National
Comprehensive Cancer Network (NCCN) compendium for VSCC.
Nonetheless, advanced VSCC carries a poor prognosis and it is less
responsive to cytotoxic agents than other solid tumors. Patients
with advanced vulvar carcinoma experience significantly shorter
overall survival (OS) durations than those with other metastatic or
recurrent solid tumors treated with novel phase I therapeutics
(8). Because of this poor
prognosis in advanced disease, the development of satisfactory
improvements is warranted and necessary (9,10).
miRNA is widely studied in many fields of medicine
including cancers. The identification of 79 miRNAs was made by de
Melo Maia et al (11),
which revealed surprisingly varied expression levels in vulvar
cancer in comparison with control speciments. In addition, they
also explored some links between special miRNAs and the clinical
pathological data (11).
Furthermore, a miRNA sponge was also verified by them, which was
most likely to appear as an efficient method for diagnosing and
treating this cancer (12). It has
been reported that miR-223-5p works as an oncogene by targeting p63
in vulvar cancer (13). miR-590-5p
drives promotion of cellular malignant conduct with the help of the
target gene TGFβRII in VSCC (4).
In this study, we discussed the mechanism of miR-3147 in VSCC.
There were relatively fewer studies of miR-3147. It
has been reported to be upregulated in cervical squamous cancer
(14) and melanoma (15), while the specific mechanism of
action of miR-3147 in tumors has not been studied. In our study, 20
fresh VSCC tissues were collected to test the expression of
miR-3147 by RT-qPCR. The expression of miR-3147 was markedly
increased in VSCC tissues and miR-3147 expression was positively
related to the depth of invasion. We next increased miR-3147 by
transfection with the miR-3147 mimics in VSCC cell line A431.
miR-3147 increased vulvar cancer cell proliferation, migration,
invasion and G1/S progression (Fig.
2). These results reinforce that miR-3147 functions as an
oncogene in vulvar carcinoma.
When classify the VSCCs, we can summarize two
different kinds. The first one is HPV-associated, which accounts
for 20 to 50% in the whole cases. The second one is
HPV-independent. In epidemiology, clinics, pathology and molecular
mechanism, all of them have different features. While A431 cells
are HPV negative, we studied the expression of miR-3147 in HPV
negative samples and A431 cells to avoid viral interference in
miRNA pathways. As we know, no information about miR-3147 and the
infection of HPV has been reported in other tumors. 25 different
expressions of miRNAs have been found between HPV-positive vulvar
cancers and HPV-negative vulvar cancers (11). By changing the expressions of
miRNAs, the HPVs have oncogenic properties. This conclusion has
been proved through some HPV-related researches such as cervical
cancer. Researchers thought that there was a connection between
cancer progression and the expressions of miR-92a and miR-378 in
HPV-positive cervical cancer tissue samples (16). At the same time, there is a report
showing that the overexpression of miR-155 was linked to the
increasing danger of cervical cancer in HPV E6/E7 mRNA positive
tissues (17). There are lot of
factors that can distinguish HPV-positive head and neck squamous
cell carcinomas (HNSCC) patients from HPV-negative HNSCC patients.
For example, a panel includes miR-9, miR-134, miR-196b, miR-210 and
miR-455 (18).
Smad4 poses to be a vital regulatory factor in the
TGF-β/Smad signaling pathway (19), and the downregulation of Smad4 is
also indicative of advanced tumor stage and a worse prognosis
(20). Smad4 performs the function
of a tumor suppressor gene in a great number of cancers, for
instance pancreatic cancer (21),
colorectal cancer (22), lung
cancer (23), gastric
adenocarcinoma (24), cervical
cancer (25), HNSCC (26) and breast cancer (27). Smad4 was reported to be oncogenic
in other cancers such as hepatocellular carcinoma (28), suggesting that its role might be
cell-context-dependent. Smad4 has been validated as a direct target
of several oncogenic miRNAs, including miR-224 (29), miR-20a-5p (30) miR-1285 (31), miR-210 (25) and miR-130a/301a/454 (32). Consistent with these results, our
study found the down-expression of Smad4 in A431 cells can simulate
the oncogenic role of miR-3147 (Fig.
3), revealing that Smad4 acts as a tumor suppressor in VSCC.
TGF-β/Smad signaling pathway as well as miRNAs participate in
various cellular processes. However, the association between the
TGF-β/Smad signaling pathway and miR-3147 in VSCC is not fully
understood. We found the miR-3147 mimics could suppress the mRNA
expressions of downstream target genes of Smad4, indicating that
miR-3147 negatively regulates the TGF-β signaling pathway via the
suppression of Smad4.
Cell migration and invasion are complicated
biological processes that have key effects on the progression of
cancer. EMT has a key role in the progression and metastasis of
tumors. Cancer cells are capable of spreading and invading other
tissues when they experience EMT (33). We increased miR-3147 in A431 and
tested the expressions of key molecules in EMT. E-cadherin
decreased but N-cadherin and Vimentin increased, revealing that the
miR-3147 mimics could promote EMT (Fig. 2B). Current studies have confirmed
that certain MMPs promote the progression of EMT. We also identify
this view. We found that miR-3147 promoted MMP-2 and MMP-9
expression (Fig. 2B).
A prediction was validated by the application of the
dual luciferase reporter assay, which suggested that the miR-3147
promoter sequence possesses a Smad4 binding site. The
down-expression of Smad4 led to the promotion of miR-3147 mRNA
expression (Fig. 4D). Furthermore,
the miR-3147 mimics exhibited a decrease in the mRNA as well as
protein expressions of Smad4 (Fig. 4B
and C) that evidences the fact that miR-3147 targets Smad4
directly in VSCC.
In this article, we pointed out that miR-3147 can
play as an oncomiR by promoting vulvar cancer cells invasion and
metastasis. In this context, we can argue that miR-3147 has a great
potential on treatment. The restriction of this oncomiR in high
miR-3147/low Smad4 patients via synthetic miRNA silencing
mechanisms including antimiR nucleotides might be an equipment for
inhibiting offense from the tumor. In addition, it might become a
chance for the therapy of VSCC. Further studies should be followed
in this direction. The goals are not only to examine if our
findings can be applied in a wider range, but also to build in
vitro and in vivo inhibition models for miR-3147 in
VSCC.
In conclusion, we transfected the miR-3147 mimics
and siR-Smad4 into A431 cells whereby we discovered that
introducing an increase in the expression of miR-3147 and a
decrease in the expression of Smad4 could result into promoting
vulvar cancer cell proliferation, migration, invasion and G1/S
progression. miR-3147 performs the function of an oncogene for
playing a key biological function by targeting Smad4 in VSCC. As
for the etiology and progression of vulvar carcinoma,
miR-3147/Smad4 may be a new research direction. For example, making
detection of the expression levels of miR-3147 or Smad4 in the
blood or tissues of patients is likely to be used for early
detection and prognostic evaluation. Therefore, our study may
contribute to the diagnosis and treatment of VSCC in the
future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
XHY analyzed and interpreted the patients' data, and
was a major contributor in writing the manuscript. FG performed the
histological examination of the tissues. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of China Medical
University. All subjects provided written informed consent to
participate in the study.
Consent for publication
All subjects provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
VSCC
|
vulvar squamous cell carcinoma
|
|
miRNA
|
microRNA
|
|
EMT
|
epithelial-mesenchymal transition
|
|
WHO
|
World Health Organization
|
|
FIGO
|
International Federation of Gynecology
and Obstetrics
|
|
UICC
|
Union for International Cancer
Control
|
|
cDNA
|
complementary DNA
|
|
MMP-2
|
matrix metalloproteinase 2
|
|
DMSO
|
dimethylsulfoxide
|
|
NCCN
|
National Comprehensive Cancer
Network
|
|
OS
|
overall survival
|
|
HNSCC
|
head and neck squamous cell
carcinomas
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sha D, Lee AM, Shi Q, Alberts SR, Sargent
DJ, Sinicrope FA and Diasio RB: Association study of the let-7
miRNA-complementary site variant in the 3′ untranslated region of
the KRAS gene in stage III colon cancer (NCCTG N0147 Clinical
Trial). Clin Cancer Res. 20:3319–3327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hiramoto H, Muramatsu T, Ichikawa D,
Tanimoto K, Yasukawa S, Otsuji E and Inazawa J: miR-509-5p and
miR-1243 increase the sensitivity to gemcitabine by inhibiting
epithelial-mesenchymal transition in pancreatic cancer. Sci Rep.
7:40022017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang XH and Wu X: miRNA expression profile
of vulvar squamous cell carcinoma and identification of the
oncogenic role of miR-182-5p. Oncol Rep. 35:398–408. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acid Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J,
Lu W, Wan X, Ma D and Xie X: Progressive miRNA expression profiles
in cervical carcinogenesis and identification of HPV-related target
genes for miR-29. J Pathol. 224:484–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen ZL, Zhao XH, Wang JW, Li BZ, Wang Z,
Sun J, Tan FW, Ding DP, Xu XH, Zhou F, et al: microRNA-92a promotes
lymph node metastasis of human esophageal squamous cell carcinoma
via E-cadherin. J Biol Chem. 286:10725–10734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fu S, Shi N, Wheler J, Naing A, Janku F,
Piha-Paul S, Gong J, Hong D, Tsimberidou A, Zinner R, et al:
Characteristics and outcomes for patients with advanced vaginal or
vulvar cancer referred to a phase I clinical trials program: The MD
Anderson Cancer Center experience. Gynecol Oncol Res Pract.
2:102015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Economopoulou P, Agelaki S, Perisanidis C,
Giotakis EI and Psyrri A: The promise of immunotherapy in head and
neck squamous cell carcinoma. Ann Oncol. 27:1675–1685. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tewari KS, Sill MW, Long HJ III, Penson
RT, Huang H, Ramondetta LM, Landrum LM, Oaknin A, Reid TJ, Leitao
MM, et al: Improved survival with bevacizumab in advanced cervical
cancer. N Engl J Med. 370:734–743. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Melo Maia B, Lavorato-Rocha AM,
Rodrigues LS, Coutinho-Camillo CM, Baiocchi G, Stiepcich MM, Puga
R, de A Lima L, Soares FA and Rocha RM: microRNA portraits in human
vulvar carcinoma. Cancer Prev Res (Phila). 6:1231–1241. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Melo Maia B, Ling H, Monroig P, Ciccone
M, Soares FA, Calin GA and Rocha RM: Design of a miRNA sponge for
the miR-17 miRNA family as a therapeutic strategy against vulvar
carcinoma. Mol Cell Probes. 29:420–426. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
de Melo Maia B, Rodrigues IS, Akagi EM,
Soares do Amaral N, Ling H, Monroig P, Soares FA, Calin GA and
Rocha RM: MiR-223-5p works as an oncomiR in vulvar carcinoma by
TP63 suppression. Oncotarget. 7:49217–49231. 2016.PubMed/NCBI
|
|
14
|
Chen J, Yao D, Li Y, Chen H, He C, Ding N,
Lu Y, Ou T, Zhao S, Li L and Long F: Serum microRNA expression
levels can predict lymph node metastasis in patients with
early-stage cervical squamous cell carcinoma. Int J Mol Med.
32:557–567. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stark MS, Tyagi S, Nancarrow DJ, Boyle GM,
Cook AL, Whiteman DC, Parsons PG, Schmidt C, Sturm RA and Hayward
NK: Characterization of the melanoma miRNAome by deep sequencing.
PLoS One. 5:e96852010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Wang HK, Li Y, Hafner M, Banerjee
NS, Tang S, Briskin D, Meyers C, Chow LT, Xie X, et al: microRNAs
are biomarkers of oncogenic human papillomavirus infections. Proc
Natl Acad Sci USA. 111:pp. 4262–4267. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park S, Eom K, Kim J, Bang H, Wang HY, Ahn
S, Kim G, Jang H, Kim S, Lee D, et al: MiR-9, miR-21 and miR-155 as
potential biomarkers for HPV positive and negative cervical cancer.
BMC Cancer. 17:6582017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wan Y, Vagenas D, Salazar C, Kenny L,
Perry C, Calvopiña D and Punyadeera C: Salivary miRNA panel to
detect HPV-positive and HPV-negative head and neck cancer patients.
Oncotarget. 8:99990–100001. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zeng Y, Zhu J, Shen D, Qin H, Lei Z, Li W,
Huang JA and Liu Z: Repression of Smad4 by miR-205 moderates
TGF-β-induced epithelial-mesenchymal transition in A549 cell lines.
Int J Oncol. 49:700–708. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Voorneveld PW, Kodach LL, Jacobs RJ, Liv
N, Zonnevylle AC, Hoogenboom JP, Biemond I, Verspaget HW, Hommes
DW, de Rooij K, et al: Loss of SMAD4 alters BMP signaling to
promote colorectal cancer cell metastasis via activation of Rho and
ROCK. Gastroenterology. 147:196–208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ormanns S, Haas M, Remold A, Kruger S,
Kruger S, Holdenrieder S, Kirchner T, Heinemann V and Boeck S: The
impact of SMAD4 loss on outcome in patients with advanced
pancreatic cancer treated with systemic chemotherapy. Int J Mol
Sci. 18:E10942017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang Z, Yang J, Di J, Cui M, Xing J, Wu F,
Wu W, Yang H, Zhang C, Yao Z, et al: Downregulated USP3 mRNA
functions as a competitive endogenous RNA of SMAD4 by sponging
miR-224 and promotes metastasis in colorectal cancer. Sci Rep.
7:42812017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee CC, Yang WH, Li CH, Cheng YW, Tsai CH
and Kang JJ: Ligand independent aryl hydrocarbon receptor inhibits
lung cancer cell invasion by degradation of Smad4. Cancer Lett.
376:211–217. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park JW, Jang SH, Park DM, Lim NJ, Deng C,
Kim DY, Green JE and Kim HK: Cooperativity of E-cadherin and Smad4
loss to promote diffuse-type gastric adenocarcinoma and metastasis.
Mol Cancer Res. 12:1088–1099. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Phuah NH, Azmi MN, Awang K and Nagoor NH:
Down-regulation of microRNA-210 confers sensitivity towards
1′S-1′-acetoxychavicol acetate (ACA) in cervical cancer cells by
targeting SMAD4. Mol Cells. 40:291–298. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cheng H, Fertig EJ, Ozawa H, Hatakeyama H,
Howard JD, Perez J, Considine M, Thakar M, Ranaweera R, Krigsfeld G
and Chung CH: Decreased SMAD4 expression is associated with
induction of epithelial-to-mesenchymal transition and cetuximab
resistance in head and neck squamous cell carcinoma. Cancer Biol
Ther. 16:1252–1258. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu N, Yu C, Shi Y, Jiang J and Liu Y:
SMAD4 expression in breast ductal carcinoma correlates with
prognosis. Oncol Lett. 10:1709–1715. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hernanda PY, Chen K, Das AM, Sideras K,
Wang W, Li J, Cao W, Bots SJ, Kodach LL, de Man RA, et al: SMAD4
exerts a tumor-promoting role in hepatocellular carcinoma.
Oncogene. 34:5055–5068. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ling H, Pickard K, Ivan C, Isella C, Ikuo
M, Mitter R, Spizzo R, Bullock M, Braicu C, Pileczki V, et al: The
clinical and biological significance of MIR-224 expression in
colorectal cancer metastasis. Gut. 65:977–989. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cheng D, Zhao S, Tang H, Zhang D, Sun H,
Yu F, Jiang W, Yue B, Wang J, Zhang M, et al: MicroRNA-20a-5p
promotes colorectal cancer invasion and metastasis by
downregulating Smad4. Oncotarget. 7:45199–45213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Haeger SM, Thompson JJ, Kalra S, Cleaver
TG, Merrick D, Wang XJ and Malkoski SP: Smad4 loss promotes lung
cancer formation but increases sensitivity to DNA topoisomerase
inhibitors. Oncogene. 35:577–586. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu L, Nie J, Chen L, Dong G, Du X, Wu X,
Tang Y and Han W: The oncogenic role of microRNA-130a/301a/454 in
human colorectal cancer via targeting Smad4 expression. PLoS One.
8:e555322013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Osugi J, Muto S, Matsumura Y, Higuchi M,
Suzuki H and Gotoh M: Prognostic impact of the high-sensitivity
modified Glasgow prognostic score in patients with resectable
non-small cell lung cancer. J Cancer Res Ther. 12:945–951. 2016.
View Article : Google Scholar : PubMed/NCBI
|