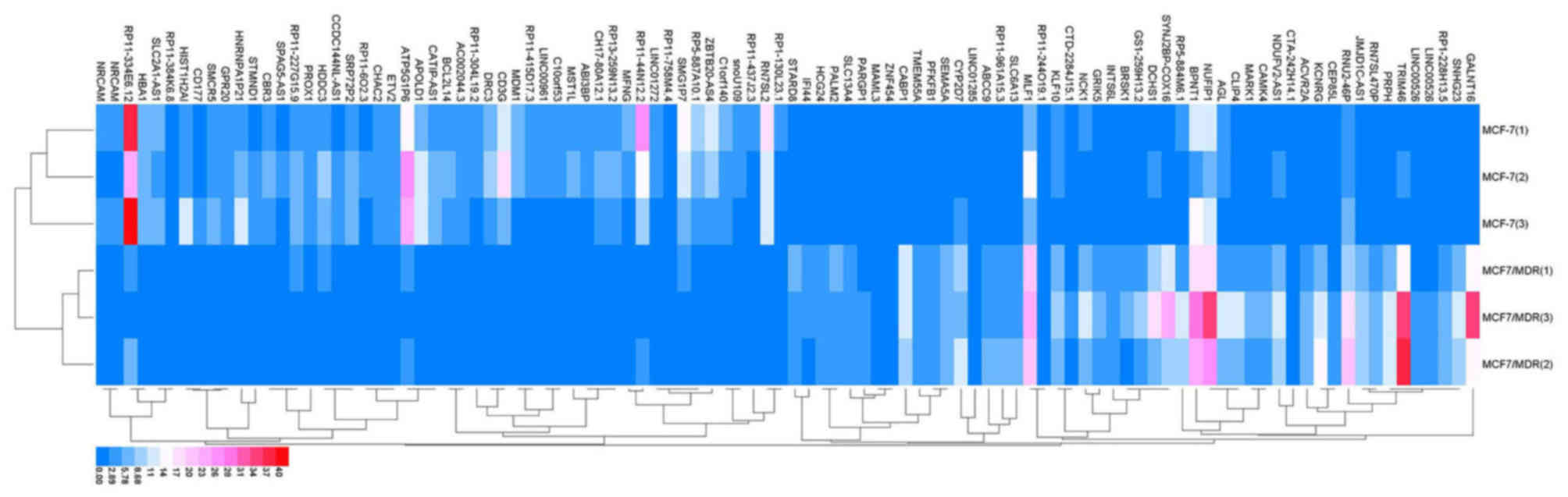
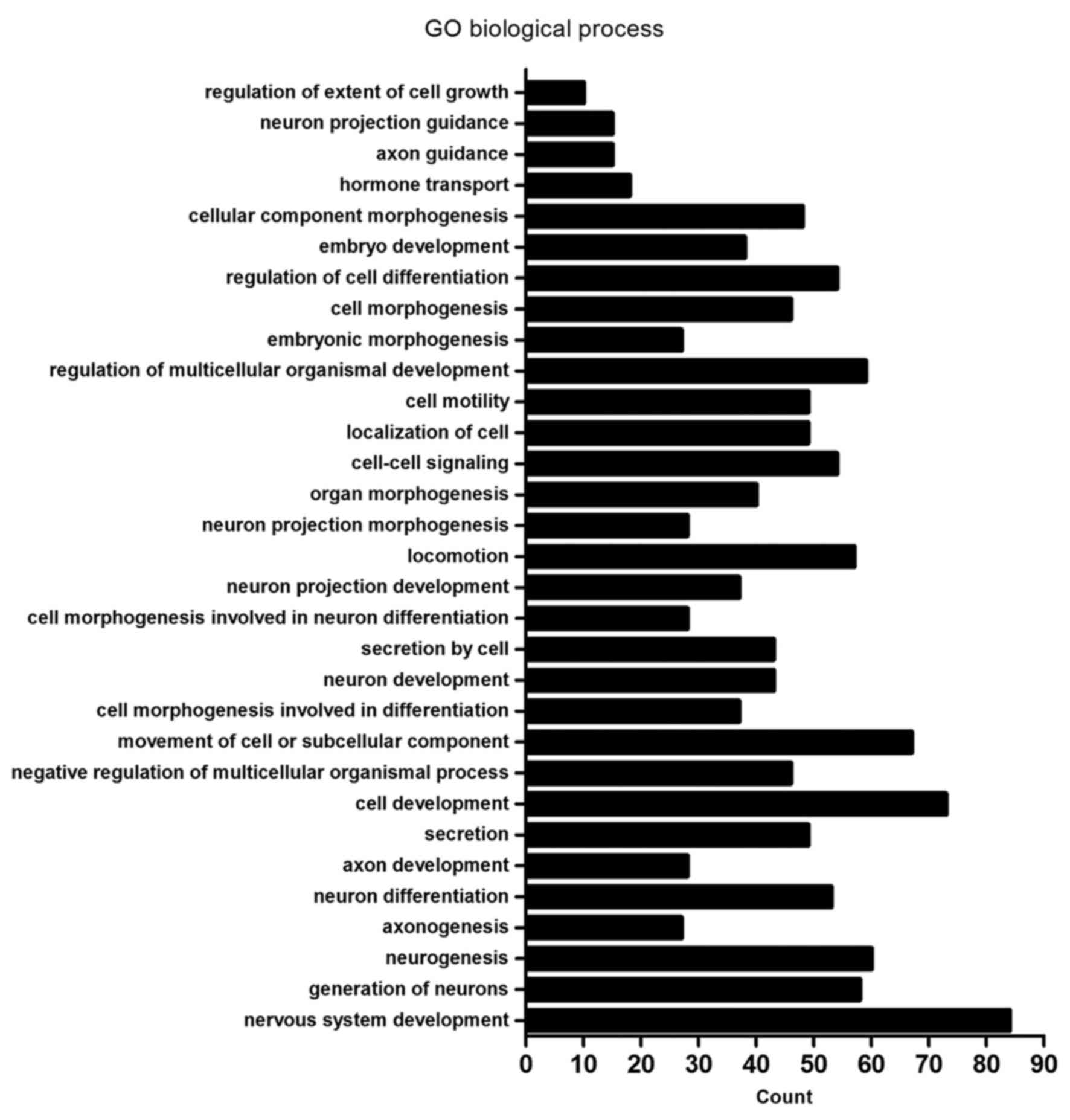
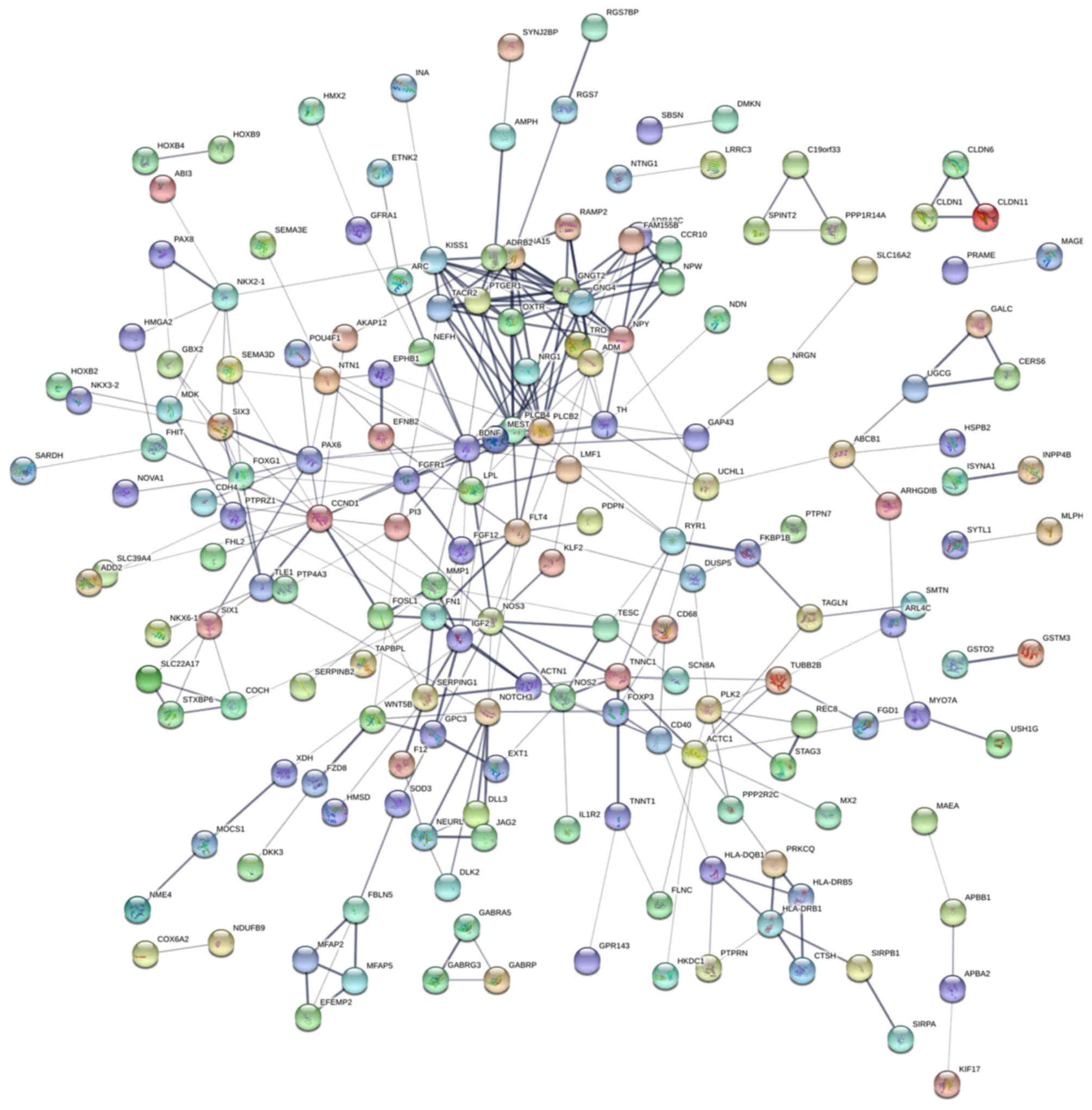
| GO:0007399~nervous
system development | 84 | DLC1, CTHRC1, HMX2,
TUBB2B, MYO7A, USH1G, UCHL1, JAG2, PAX6, ANKRD1, FGF12, RTN1,
ARHGAP4, PCDH1, GSTM3, BDNF, SPINT2, PAX8, SEMA3E, SNPH, SEMA3D,
POU4F1, NRG1, INA, KIF17, CLMN, EFNB1, EFNB2, DLL3, SIX3, GPER1,
HMGA2, PCLO, HOXD9, EPB41L3, PRKCQ, ADM, SIX1, FOXG1, MYRF, NRGN,
EXT1, GAP43, NEURL1, DPF3, FGFR1, WNT5B, NDN, TH, BEX1, MYEF2,
OXTR, CLDN11, FKBP1B, MDK, ITM2C, CDH4, EPHB1, ITM2A, ALCAM,
BCL11A, GBX2, APBA2, NKX2.1, NEFH, ADRA2C, FN1, FZD8, TRNP1,
PTPRZ1, GABRA5, NTNG1, AXL, NTN1, NKX6.1, NOTCH3, SALL2, HOXB2,
NPY, PLK2, H2AFY2, GFRA1, SCN8A, APBB1 | 0.0000335 |
|
GO:0048699~generation of neurons | 58 | CTHRC1, TUBB2B,
MYO7A, USH1G, UCHL1, JAG2, PAX6, ANKRD1, RTN1, ARHGAP4, BDNF, SNPH,
SEMA3E, SEMA3D, POU4F1, NRG1, CLMN, EFNB1, EFNB2, SIX3, DLL3,
GPER1, HMGA2, HOXD9, EPB41L3, PRKCQ, ADM, SIX1, EXT1, GAP43,
NEURL1, FGFR1, WNT5B, NDN, TH, MYEF2, FKBP1B, ITM2C, CDH4, EPHB1,
ALCAM, BCL11A, GBX2, NKX2-1, NEFH, ADRA2C, FN1, FZD8, PTPRZ1,
GABRA5, AXL, NTNG1, NTN1, NKX6-1, NOTCH3, NPY, PLK2, APBB1 | 0.0004872 |
|
GO:0022008~neurogenesis | 60 | CTHRC1, TUBB2B,
MYO7A, USH1G, UCHL1, JAG2, PAX6, ANKRD1, RTN1, ARHGAP4, BDNF, SNPH,
SEMA3E, SEMA3D, POU4F1, NRG1, CLMN, KIF17, EFNB1, EFNB2, DLL3,
SIX3, GPER1, HMGA2, HOXD9, EPB41L3, PRKCQ, ADM, SIX1, MYRF, EXT1,
GAP43, NEURL1, FGFR1, WNT5B, NDN, TH, MYEF2, FKBP1B, ITM2C, CDH4,
EPHB1, ALCAM, BCL11A, GBX2, NKX2-1, NEFH, ADRA2C, FN1, FZD8,
PTPRZ1, GABRA5, AXL, NTNG1, NTN1, NKX6-1, NOTCH3, NPY, PLK2,
APBB1 | 0.0007804 |
|
GO:0007409~axonogenesis | 27 | NDN, UCHL1, PAX6,
CDH4, EPHB1, ARHGAP4, ALCAM, BDNF, BCL11A, SEMA3E, GBX2, SEMA3D,
NEFH, NKX2-1, POU4F1, NRG1, FN1, PTPRZ1, EFNB1, EFNB2, NTNG1, NTN1,
NKX6-1, PRKCQ, EXT1, APBB1, GAP43 | 0.0011163 |
| GO:0030182~neuron
differentiation | 53 | CTHRC1, MYO7A,
USH1G, UCHL1, PAX6, JAG2, ANKRD1, RTN1, ARHGAP4, BDNF, SNPH,
SEMA3E, SEMA3D, POU4F1, NRG1, CLMN, EFNB1, EFNB2, SIX3, HOXD9,
PRKCQ, EPB41L3, ADM, SIX1, EXT1, GAP43, NEURL1, FGFR1, WNT5B, NDN,
TH, MYEF2, ITM2C, CDH4, FKBP1B, EPHB1, ALCAM, BCL11A, GBX2, NKX2-1,
NEFH, ADRA2C, FN1, FZD8, PTPRZ1, GABRA5, NTNG1, NTN1, NKX6-1,
NOTCH3, NPY, PLK2, APBB1 | 0.0013166 |
| GO:0061564~axon
development | 28 | NDN, UCHL1, PAX6,
FKBP1B, CDH4, EPHB1, ARHGAP4, ALCAM, BDNF, BCL11A, SEMA3E, GBX2,
SEMA3D, NEFH, NKX2-1, POU4F1, NRG1, FN1, PTPRZ1, EFNB1, EFNB2,
NTNG1, NTN1, NKX6-1, PRKCQ, EXT1, APBB1, GAP43 | 0.0013888 |
|
GO:0046903~secretion development | 49 | TACR2, ANKRD1,
KISS1, PAX8, SNPH, CHRNA7, NOS2, NRG1, PTPRN2, ACTN1, SERPING1,
GPER1, CD40, PCDH7, HMGA2, PTPRN, PCLO, INHBB, CCND1, ADM, STXBP6,
RAB15, NEURL1, XDH, FGFR1, IL1R2, RAB3B, NKD2, WASH1, HLA-DRB1,
LMF1, OXTR, FKBP1B, AHSG, C1QTNF5, HLA-DRB5, ADRA2C, OLFM2, FN1,
LPL, AXL, IGF2, FOXP3, NKX6-1, ABCB4, FBLN5, APBB1, SYTL1,
SLC17A9 | 0.0016024 |
| GO:0048468~cell
development | 73 | CTHRC1, MAEA,
MYO7A, USH1G, UCHL1, PAX6, ANKRD1, YBX2, ARHGAP4, GSTM3, BDNF,
FRMD6, SPINT2, PAX8, SEMA3E, SEMA3D, POU4F1, NRG1, RAMP2, CLMN,
EFNB1, EFNB2, DLL3, SIX3, SIP A1L3, ACTN1, GPER1, HMGA2, FLNC,
HOXD9, INHBB, EPB41L3, PRKCQ, REC8, ADM, SIX1, EPB41L5, MYRF, RYR1,
EXT1, LRRK1, GAP43, NEURL1, FGFR1, WNT5B, NDN, TH, FHL2, FKBP1B,
ITM2C, CDH4, EPHB1, ALCAM, BCL11A, GBX2, NKX3.2, NKX2.1, NEFH,
ADRA2C, FN1, ACTC1, PTPRZ1, GABRA5, NTNG1, AXL, NTN1, NKX6.1,
NOTCH3, NPY, PLK2, KLF2, APBB1, RFLNB | 0.0021369 |
| GO:0051241~negative
regulation of multicellular organismal process | 46 | XDH, IL1R2, FGFR1,
HLA-DRB1, TACR2, PAX6, OXTR, FKBP1B, ITM2C, AHSG, ARHGAP4, BCL11A,
PAX8, SEMA3E, NKX3-2, SEMA3D, HLA-DRB5, NKX2-1, CHRNA7, NOS3,
ADRA2C, ADAMTS12, NRG1, ARHGDIB, FN1, F12, DLL3, AXL, SIX3, UBE2L6,
SERPING1, FOXP3, HMGA2, NTN1, NKX6-1, NOTCH3, INHBB, ADRB2, TNNT1,
PLK2, ADM, SIX1, SERPINB2, KLF2, TMEM176B, RFLNB | 0.0026439 |
| GO:0006928~movement
of cell or subcellular component | 67 | DLC1, CTHRC1,
TUBB2B, MLPH, TNNC1, MYO7A, UCHL1, JAG2, PAX6, FGF12, AMOTL1, MMP1,
ARHGAP4, BDNF, FRMD6, SPINT2, SEMA3E, SEMA3D, NOS3, CHRNA7, POU4F1,
NRG1, ARC, KIF17, PDPN, EFNB1, ARHGEF5, EFNB2, SIX3, ACTN1, GPER1,
SIRPA, PRKCQ, TNNT1, SIX1, EPB41L5, EXT1, CTSH, GAP43, NEURL1,
FGFR1, WNT5B, APC2, WASH1, NDN, ABI3, PLVAP, MDK, CDH4, EPHB1,
ALCAM, IFT46, GBX2, NKX2-1, NEFH, ADAMTS12, FN1, ARHGDIB, ACTC1,
FLT4, AXL, NTN1, NKX6-1, LAMA1, NPY, PTP4A3, HOXB9 | 0.0056175 |
| GO:0000904~cell
morphogenesis involved in differentiation | 37 | NDN, MYO7A, UCHL1,
PAX6, CDH4, EPHB1, ARHGAP4, ALCAM, BDNF, FRMD6, SPINT2, BCL11A,
PAX8, SEMA3E, GBX2, SEMA3D, NKX2-1, NEFH, POU4F1, NRG1, FN1,
PTPRZ1, EFNB1, EFNB2, NTNG1, AXL, SIPA1L3, ACTN1, HMGA2, NTN1,
NKX6-1, PRKCQ, EPB41L5, EXT1, APBB1, GAP43, RFLNB | 0.0059248 |
| GO:0048666~neuron
development | 43 | FGFR1, CTHRC1, NDN,
MYO7A, USH1G, UCHL1, TH, PAX6, ANKRD1, FKBP1B, ITM2C, CDH4, EPHB1,
ARHGAP4, ALCAM, BDNF, BCL11A, SEMA3E, GBX2, SEMA3D, NKX2-1, NEFH,
POU4F1, NRG1, FN1, PTPRZ1, CLMN, EFNB1, GABRA5, EFNB2, NTNG1, NTN1,
NKX6-1, HOXD9, EPB41L3, PRKCQ, NPY, PLK2, ADM, EXT1, APBB1, GAP43,
NEURL1 | 0.0093174 |
|
GO:0032940~secretion by cell | 43 | IL1R2, FGFR1,
RAB3B, NKD2, HLA-DRB1, WASH1, TACR2, LMF1, OXTR, ANKRD1, FKBP1B,
AHSG, KISS1, C1QTNF5, PAX8, SNPH, HLA-DRB5, CHRNA7, ADRA2C, NOS2,
OLFM2, FN1, LPL, PTPRN2, AXL, ACTN1, SERPING1, IGF2, PCDH7, CD40,
GPER1, PTPRN, FOXP3, HMGA2, PCLO, NKX6-1, INHBB, ADM, STXBP6,
RAB15, SYTL1, APBB1, SLC17A9 | 0.0094679 |
| GO:0048667~cell
morphogenesis involved in neuron differentiation | 28 | NDN, MYO7A, UCHL1,
PAX6, CDH4, EPHB1, ARHGAP4, ALCAM, BDNF, BCL11A, SEMA3E, GBX2,
SEMA3D, NEFH, NKX2-1, POU4F1, NRG1, FN1, PTPRZ1, EFNB1, EFNB2,
NTNG1, NTN1, NKX6-1, PRKCQ, EXT1, APBB1, GAP43 | 0.0107608 |
| GO:0031175~neuron
projection development | 37 | FGFR1, NDN, UCHL1,
PAX6, ANKRD1, FKBP1B, ITM2C, CDH4, EPHB1, ARHGAP4, ALCAM, BDNF,
BCL11A, SEMA3E, GBX2, SEMA3D, NEFH, NKX2-1, POU4F1, NRG1, FN1,
PTPRZ1, CLMN, EFNB1, EFNB2, NTNG1, NTN1, NKX6-1, EPB41L3, PRKCQ,
NPY, PLK2, ADM, EXT1, APBB1, GAP43, NEURL1 | 0.0355076 |
|
GO:0040011~locomotion | 57 | DLC1, CTHRC1,
TUBB2B, JAG2, PAX6, AMOTL1, MMP1, ARHGAP4, BDNF, SPINT2, CCR10,
SEMA3E, SEMA3D, CHRNA7, POU4F1, NOS3, NRG1, FOSL1, ARC, PDPN,
EFNB1, ARHGEF5, EFNB2, SIX3, GPER1, SIRPA, PRKCQ, SIX1, EPB41L5,
EXT1, CTSH, GAP43, NEURL1, FGFR1, WNT5B, WASH1, NDN, APC2, ABI3,
PLVAP, MDK, CDH4, EPHB1, ALCAM, DOCK2, GBX2, NKX2-1, ADAMTS12,
ARHGDIB, FN1, FLT4, AXL, NTN1, NKX6-1, LAMA1, PTP4A3, HOXB9 | 0.0414058 |
| GO:0048812~neuron
projection morphogenesis | 28 | NDN, UCHL1, PAX6,
CDH4, EPHB1, ARHGAP4, ALCAM, BDNF, BCL11A, SEMA3E, GBX2, SEMA3D,
NEFH, NKX2-1, POU4F1, NRG1, FN1, PTPRZ1, EFNB1, EFNB2, NTNG1, NTN1,
NKX6-1, EPB41L3, PRKCQ, EXT1, APBB1, GAP43 | 0.0077004 |
| GO:0009887~organ
morphogenesis | 40 | DLC1, FGFR1, FGD1,
CTHRC1, HMX2, TNNC1, USH1G, MYO7A, TH, JAG2, PAX6, FHL2, ANKRD1,
EPHB1, GPC3, PAX8, GBX2, NKX3-2, NKX2-1, POU4F1, NRG1, ACTC1,
EFNB2, SIX3, TLE1, HMGA2, NTN1, ZNF22, NKX6-1, HOXD9, HOXB4, HOXD8,
HOXB2, ADM, SIX1, RYR1, MFAP2, CTSH, TMEM176B, RFLNB | 0.0264522 |
|
GO:0007267~cell-cell signaling | 54 | CTHRC1, TACR2,
FGF12, SYNGR1, AMOTL1, KISS1, RSPO4, BDNF, PCDH1, GPC3, PAX8, SNPH,
CHRNA7, NOS2, PLCB2, ARC, PTPRN2, EFNB1, EFNB2, SIX3, TLE1, GPER1,
HMGA2, PTPRN, PCLO, INHBB, CCND1, ADRB2, ADM, NRGN, LRRK1, NEURL1,
FGFR1, RAB3B, NKD2, WNT5B, HLA-DRB1, APC2, TH, OXTR, FKBP1B, AMPH,
APBA2, HLA-DRB5, ADRA2C, NOVA1, FZD8, GABRA5, NKX6-1, DKK3, NPY,
PLK2, HOXB9, SYTL1 | 0.0264522 |
|
GO:0051674~localization of cell | 49 | DLC1, CTHRC1,
TUBB2B, PAX6, JAG2, AMOTL1, MMP1, ARHGAP4, SPINT2, SEMA3E, SEMA3D,
CHRNA7, POU4F1, NOS3, NRG1, ARC, PDPN, EFNB1, EFNB2, ARHGEF5, SIX3,
GPER1, SIRPA, PRKCQ, EPB41L5, SIX1, CTSH, NEURL1, FGFR1, WNT5B,
WASH1, NDN, APC2, ABI3, PLVAP, MDK, EPHB1, GBX2, NKX2-1, ADAMTS12,
ARHGDIB, FN1, FLT4, AXL, NTN1, NKX6-1, LAMA1, PTP4A3, HOXB9 | 0.0294439 |
| GO:0048870~cell
motility | 49 | DLC1, CTHRC1,
TUBB2B, PAX6, JAG2, AMOTL1, MMP1, ARHGAP4, SPINT2, SEMA3E, SEMA3D,
CHRNA7, POU4F1, NOS3, NRG1, ARC, PDPN, EFNB1, EFNB2, ARHGEF5, SIX3,
GPER1, SIRPA, PRKCQ, EPB41L5, SIX1, CTSH, NEURL1, FGFR1, WNT5B,
WASH1, NDN, APC2, ABI3, PLVAP, MDK, EPHB1, GBX2, NKX2-1, ADAMTS12,
ARHGDIB, FN1, FLT4, AXL, NTN1, NKX6-1, LAMA1, PTP4A3, HOXB9 | 0.0294439 |
|
GO:2000026~regulation of multicellular
organismal development | 59 | CTHRC1, PAX6,
ANKRD1, ARHGAP4, BDNF, GPC3, PAX8, SEMA3E, SEMA3D, POU4F1, CHRNA7,
NOS3, NRG1, RAMP2, EFNB2, DLL3, SIX3, GPER1, CD40, HMGA2, CCND1,
ADRB2, ADM, SIX1, EPB41L5, MYRF, CTSH, NEURL1, XDH, FGFR1, OXTR,
FKBP1B, ITM2C, CDH4, EPHB1, AHSG, FAM213A, BCL11A, NKX3-2, NKX2-1,
ADRA2C, ADAMTS12, FN1, ARHGDIB, TESC, PTPRZ1, IL1RL2, AXL, FOXP3,
NTN1, NKX6-1, NOTCH3, LAMA1, PLK2, RASSF2, H2AFY2, APBB1, TMEM176B,
RFLNB | 0.0335908 |
|
GO:0048598~embryonic morphogenesis | 27 | DLC1, FGFR1,
CTHRC1, HMX2, USH1G, MYO7A, TH, PAX6, JAG2, GPC3, SPINT2, PAX8,
GBX2, NKX3-2, FN1, SIX3, HMGA2, NTN1, DUSP5, HOXD9, HOXB4, HOXB2,
ADM, SIX1, EPB41L5, MFAP2, EXT1 | 0.0349494 |
| GO:0000902~cell
morphogenesis | 46 | DLC1, FGD1, NDN,
MYO7A, UCHL1, PAX6, CDH4, EPHB1, ALCAM, ARHGAP4, BDNF, FRMD6,
IFT46, SPINT2, BCL11A, PAX8, SEMA3E, GBX2, SEMA3D, NKX2-1, NEFH,
POU4F1, NRG1, FN1, CDC42EP5, COCH, PTPRZ1, PDPN, EFNB1, EFNB2,
NTNG1, AXL, SIPA1L3, ACTN1, HMGA2, NTN1, NKX6-1, EPB41L3, PRKCQ,
EPB41L5, KLF2, EXT1, APBB1, GAP43, RFLNB, NEURL1 | 0.0371107 |
|
GO:0045595~regulation of cell
differentiation | 54 | CTHRC1, PAX6, DLK2,
ANKRD1, ARHGAP4, BDNF, PAX8, SEMA3E, SEMA3D, POU4F1, NRG1, RAMP2,
PRAME, EFNB2, SIX3, DLL3, GPER1, HMGA2, CCND1, ADM, SIX1, EPB41L5,
NEURL1, XDH, FGFR1, WNT5B, APC2, FKBP1B, ITM2C, CDH4, FAM213A,
BCL11A, NKX3-2, NKX2-1, ADRA2C, ADAMTS12, OLFM2, FN1, LPL, TESC,
PTPRZ1, IL1RL2, AXL, FOXP3, NTN1, NKX6-1, NOTCH3, HOXB4, PLK2,
RASSF2, H2AFY2, APBB1, RFLNB, TMEM176B | 0.0381061 |
| GO:0009790~embryo
development | 38 | DLC1, FGFR1,
CTHRC1, HMX2, USH1G, MYO7A, TH, JAG2, PAX6, GPC3, SPINT2, PAX8,
GBX2, NKX3-2, APBA2, NOS3, ETNK2, NRG1, FOSL1, FN1, EFNB1, DLL3,
SIX3, HMGA2, NTN1, HOXD9, DUSP5, HOXB4, SALL2, HOXD8, HOXB2, ADM,
SIX1, EPB41L5, MFAP2, HOXB9, KLF2, EXT1 | 0.0373992 |
| GO:0032989~cellular
component morphogenesis | 48 | DLC1, FGD1, NDN,
MYO7A, UCHL1, PAX6, ANKRD1, CDH4, EPHB1, ALCAM, ARHGAP4, BDNF,
FRMD6, IFT46, SPINT2, BCL11A, PAX8, SEMA3E, GBX2, SEMA3D, NKX2-1,
NEFH, POU4F1, NRG1, FN1, CDC42EP5, COCH, ACTC1, PTPRZ1, PDPN,
EFNB1, EFNB2, NTNG1, AXL, SIPA1L3, ACTN1, HMGA2, NTN1, NKX6-1,
EPB41L3, PRKCQ, EPB41L5, KLF2, EXT1, APBB1, RFLNB, GAP43,
NEURL1 | 0.0375119 |
| GO:0009914~hormone
transport | 18 | FGFR1, HLA-DRB1,
TACR2, PTPRN2, GPER1, HMGA2, PTPRN, PCLO, FKBP1B, NKX6-1, INHBB,
KISS1, SLC16A2, ADM, PAX8, HLA-DRB5, ADRA2C, NOS2 | 0.0415878 |
| GO:0007411~axon
guidance | 15 | EFNB1, EFNB2, PAX6,
CDH4, NTN1, EPHB1, ALCAM, PRKCQ, BDNF, SEMA3E, GBX2, SEMA3D,
NKX2-1, EXT1, GAP43 | 0.0410559 |
| GO:0097485~neuron
projection guidance | 15 | EFNB1, EFNB2, PAX6,
CDH4, NTN1, EPHB1, ALCAM, PRKCQ, BDNF, SEMA3E, GBX2, SEMA3D,
NKX2-1, EXT1, GAP43 | 0.0414269 |
|
GO:0061387~regulation of extent of cell
growth | 10 | ARHGAP4, BDNF,
BCL11A, SEMA3E, SEMA3D, NRG1, NTN1, CDH4, NKX6-1, FN1 | 0.0486247 |