Introduction
Choline kinase (CK) catalyzes the phosphoryl
transfer from ATP to choline, yielding phosphocholine. CK is the
first enzyme in the CDP-choline pathway for the biosynthesis of
phosphatidylcholine, the most abundant phospholipid in the
eukaryotic cell membrane (1). In
humans, CK is encoded by two separate genes, termed CHKA (HGNC no.
1937; referred to as CKα in the present study) and CHKB (HGNC no.
1938; referred to as CKβ in the present study), that produce the
CKα1, CKα2 and CKβ isozymes (2).
Functionally, CKα and CKβ catalyze the same activity, although they
have specific roles in the physiology and development of an
organism (3). Human tumor tissues
usually exhibit increased CKα activity that leads to elevated
levels of its product, phosphocholine (4,5).
Overexpression of CKα has additionally been reported in a variety
of types of human cancer including lung, colorectal and prostate
adenocarcinomas (5–7). CK overexpression additionally
increases the invasiveness and drug resistance of breast cancer
cells (8). CKα inhibitors act as
potent antitumor drugs in vitro and in vivo, as
demonstrated by Rodríguez-González et al (9). Specific inhibition of CKα expression
triggers apoptosis and selectively kills cancer cells, although not
normal cells (10–12). Notably, the possible regulation of
CK gene expression levels in the cells by miRNAs has never been
examined, to the best of the authors' knowledge.
MicroRNAs (miRNAs) are small non-coding regulatory
RNAs with a length of ~20 nucleotides that are involved in diverse
cellular events, including cell development, neuronal cell fate,
cell death and proliferation, and fat storage (13). MiRNAs achieve their functions by
affecting gene expression through sequence-specific interactions
with the 3′-untranslated regions (UTR) or 5′-UTR of target mRNAs
which lead to translational repression and/or destabilization of
the target mRNA (14–17). Dysregulation of miRNA expression is
common in numerous types of cancer (18). In terms of drug discovery,
strategies proposed for miRNA therapeutics include the artificial
introduction of antisense oligonucleotides to inhibit miRNAs
(19) and the design of an
anti-miRNA oligonucleotide that is able to specifically inhibit the
action of miRNAs (20). In
addition, a novel hydrogel technology delivery of an antagomiR to
inhibit an miRNA that promotes tumor growth in mice has been
reported to effectively kill cancer cells (21). Furthermore, the reintroduction of
tumor-suppressive miRNAs has been employed in order to block the
functions of oncogenes (22). The
majority of efforts have been directed at the inhibition of the
activity of CKα by using small molecule inhibitors or RNA
interference (RNAi), and little attention has been given to the
regulation of CK expression by intracellular modulators, including
miRNAs. Therefore, it may be useful to search for miRNAs that
target CKα to discover their potential application in the treatment
of certain types of cancer.
In the present study, two miRNAs, miR-876-5p and
miR-646, predicted to target the CKα gene were studied for their
potential regulation of CKα expression in HepG2 cells. The present
study aimed to test experimentally the effect of miR-876-5p and
miR-646 mimics on CKα gene expression in HepG2 cells. The results
obtained in the present study reveal the involvement of miRNAs in
modulating the expression level of CKα in cancer cells, something
that may be developed into a promising anticancer therapeutic for
the treatment of CKα-overexpressing cancers.
Materials and methods
Cell culture
The HepG2 cell line was obtained from the American
Type Culture Collection (cat. no. HB-8065; Manassas, VA, USA) and
maintained at 37°C, 95% humidity and 5% CO2 in Dulbecco's modified
Eagle's medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% fetal bovine serum (FBS) (Invitrogen;
Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin. The
HepG2 cell line is a hepatoblastoma cell line previously
misidentified as hepatocellular carcinoma (23). The HepG2 cells were seeded in a
24-well plate at a concentration of 1.5×105 cells/well (diluted
with antibiotic-free complete medium) 1 day prior to transfection
with miRNAs.
Prediction of CKα-targeting
miRNAs
TargetScan (www.targetscan.org), DIANA micro T (www.diana.imis.athena-innovation.gr),
microRNA.org (www.microrna.org) and RepTar (reptar.ekmd.huji.ac.il) were used to predict the
miRNAs that potentially bind to the 3′-UTR of CKα mRNA, using the
default settings of the respective programs. The minimum free
energy of the predicted miRNAs was calculated using RNAfold
(rna.tbi.univie.ac.at), mFold (unafold.rna.albany.edu) and KineFold
(kinefold.curie.fr). The site features of the mRNA-miRNA pairings
in terms of G:C, A:U and G:U matches, mismatches and bulges in
miRNAs and mRNAs were analyzed manually to search for high
complementarity at the seed region and the minimum number of
bulges. The potential miRNAs were additionally analyzed based on
the classification of miRNA target sites, according to Brennecke
et al (24), types of
matching to seed region, according to Lewis et al (25)and site contexts, based on Grimson
et al (26).
miRNA transfection
Non-targeting (miRIDIAN microRNA Mimic Negative
Control #1; GE Healthcare Dharmacon, Inc., Lafayette, CO, USA),
GAPDH-targeting (Mimic Housekeeping Positive Control #2; GE
Healthcare Dharmacon, Inc.), miR-876-5p (MISSION®
MicroRNA mimic HMI0929, 5′-UGGAUUUCUUUGUGAAUCACCA-3′; Sigma
Aldrich; Merck KGaA, Darmstadt, Germany) and miR-646
(MISSION® MicroRNA mimic HMI0877,
5′-AAGCAGCUGCCUCUGAGGC-3′; Sigma Aldrich; Merck KGaA) miRNAs were
re-suspended in 1X miRNA buffer (GE Healthcare Dharmacon, Inc.) to
prepare a stock solution of 20 µM. The non-targeting (negative
control) miRNA sequence was based on the Caenorhabditis elegans
cel-miR-67 that was confirmed to exhibit minimal sequence identity
with miRNAs from humans, mice and rats. The GAPDH-targeting miRNA
(referred to as miRNA-GAPDH) targets the 3′UTR of GAPDH mRNA. The
miRNA stock solution was further diluted with RNase-free water to
obtain a 2 µM working solution. Prior to the transfection, the
miRNAs were diluted with serum free Opti-Minimum Essential Medium
(MEM)® I (Thermo Fisher Scientific, Inc.), such that the
final concentrations in the treatment plate were 25, 50 and 100 nM.
In a separate tube, DharmaFECT 2 transfection reagent (GE
Healthcare Life Sciences, Little Chalfont, UK) was additionally
diluted with the serum free Opti-MEM®, according to the
manufacturer's protocol. The diluted miRNAs and transfection
reagent were mixed for 20 min at room temperature to form
transfection complexes. The culture medium from the 24-well plate
was removed and 400 µl complete medium plus 100 µl transfection
complexes were added into each well and cultured at 37°C with 5%
CO2, for a duration of 24 to 48 h. For screening of
miRNA mimics for the downregulation of CKα mRNA levels in the HepG2
cancer cell line, the cells were transfected for 48 h.
Extraction of total cellular RNA
Following the transfection, the medium was removed
and the cells were trypsinized, harvested and washed with PBS prior
to RNA extraction using the RNeasy Mini kit (Qiagen GmbH, Hilden,
Germany), according to the manufacturer's protocol. Any residual
DNA was eliminated by adding RNase-Free DNase I (Qiagen GmbH) onto
the spin column and incubating for 20 min at room temperature. The
concentration of the purified total RNA was measured at 260 nm and
the OD260/280 was determined using a BioPhotometer Plus
(Eppendorf, Hamburg, Germany). The RNA integrity and size
distribution were assessed by running the purified RNA samples on a
1% agarose gel.
Synthesis of cDNA
The first strand cDNA was synthesized from the
extracted RNA using the RevertAid™ H Minus First Strand cDNA
Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.) with a
MyCycler Thermal Cycler (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). A total of 1 µg total RNA was mixed with 0.5 µl each of
oligo(dT)18 (50 µM) and random hexamers (50 µM), with the final
volume made up to 12 µl with distilled water. The mixture was
incubated at 65°C for 5 min. Subsequently, 4 µl 5X reaction buffer,
1 µl RiboLock™ RNase Inhibitor (20 U/µl), 2 µl dNTP mix (10 mM) and
1 µl RevertAid™ H Minus M-MuLV Reverse Transcriptase (200 U/µl) was
added into the mixture, and incubated at 25°C for 5 min and 42°C
for 1 h; the reaction was terminated by a final incubation at 70°C
for 5 min. The synthesized cDNA was stored at −20°C until use.
Quantitative polymerase chain reaction
(qPCR)
The qPCR reactions were performed using an ABI 7500
Fast Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The reactions were performed in 96-well plates
(Axygen; Corning Incorporated, Corning, NY, USA). A relative
quantification method with ubiquitin C (UBC) and tyrosine
3-monooxygenase/tryptophan 5-monooxygenase activation protein ζ
(YWHAZ) as reference genes was used. A negative control without
template DNA was run together with the other samples to detect
possible contamination or non-specific amplification in the
reaction. The PCR reactions with a final volume of 25 µl consisted
of 12.5 µl Power SYBR-Green I Master Mix (Applied Biosystems;
Thermo Fisher Scientific, Inc.), 1.5 µl gene specific primers, 1 µl
1:2 diluted cDNA and water. The cycling parameters were 2 min at
50°C and 10 min at 95°C, followed by 40 cycles of 10 sec at 95°C
and 1 min at 60°C. Following the amplification, PCR specificity was
verified by melting curve analysis with temperatures ranging
between 60 and 95°C with 0.1°C increments. All reactions were run
in triplicate. The reference genes (UBC and YWHAZ) were selected
based on a previous study (27).
The forward primer sequence for UBC was
5′-CTGATCAGCAGAGGTTGATCTT-3′ and its reverse primer sequence was
5′-GTCTTGCCAGTGAGTGTCTT-3′. For YWHAZ, the forward primer sequence
was 5′-TTCTTGATCCCCAATGCTTC-3′ and the reverse primer sequence was
5′-AGTTAAGGGCCAGACCCAGT-3′. The forward and reverse primer
sequences for GAPDH were 5′-CAAGGTCATCCATGACAACTTTG-3′ and
5′-GTCCACCACCCTGTTGCTGTAG-3′, respectively. The forward primer
sequence for total CKα was 5′-TCAGAGCAAACATCCGGAAGT-3′ and the
reverse primer sequence for total CKα was
5′-GGCGTAGTCCATGTACCCAAAT-3′. The forward and reverse primer
sequences for CKα2 specific amplification were
5′-GGCCTTAGCAACATGCTGTTC-3′ and 5′-AGCTTGTTCAGAGCCCTCTTT-3′,
respectively. Relative gene expression levels normalized to the
geometric mean of UBC and YWHAZ Cq values were
determined by the 2−ΔΔCq method (28).
MTT cell viability assay
The MTT cell viability assay was performed by
seeding 1.5×104 HepG2 cells in 100 µl antibiotic-free
complete medium into each well of a 96-well plate and incubating at
37°C with 5% CO2 1 day prior to the transfection with
miR-876-5p mimic. The cells were transfected with 25 nM miR-876-5p
for 48 h with a final volume of 100 µl in each well. The MTT assay
was performed by replacing the medium following transfection with
100 µl fresh culture medium, followed by the addition of 10 µl 12
mM MTT stock solution and incubation at 37°C for 4 h. Following
that, 85 µl medium was removed and 50 µl dimethyl sulfoxide (DMSO)
as the solubilizing agent to dissolve the formazan dye was added to
each well and thoroughly mixed by pipetting, prior to further
incubation at 37°C for 10 min. Finally, the absorbance at 540 nm
was read by using microplate reader (Bio-Rad Laboratories, Inc.)
and the percentage cell viability was calculated based on the
average absorbance values for the samples and control.
Scanning electron microscopy
The morphological alterations in HepG2 cells
following treatment with either DMSO or the desired miRNAs for 48 h
were observed under a scanning electron microscope (Quanta 200;
FEI; Thermo Fisher Scientific, Inc.). Following transfection, cells
were grown on a plastic cover slip (Thermo Fisher Scientific, Inc.)
and were fixed with McDowell-Trump fixative solution [1% (v/v)
glutaraldehyde and 4% (v/v) formaldehyde in 0.1 M phosphate buffer;
pH 7.2] (29) at 4°C for 24 h. The
following day, the samples were washed with 0.1 M PBS and incubated
in 1% osmium tetroxide at room temperature for 1–2 h for secondary
fixation. The samples were dehydrated via sequential incubation of
15 min each in 50, 75, 95 and 100% ethanol. The dehydrated samples
were immersed in hexamethyldisilazane (HMDS): acetone (1:1 ratio)
for 15 min and in 100% HMDS three times, for 15 min each, prior to
being left overnight in a desiccator. The dried samples were
mounted onto the sample stub prior to being coated with gold and
viewed under the scanning electron microscope.
Statistical analysis
All data were analyzed using Student's t-test and
one-way analysis of variance with the Bonferroni post hoc test.
P<0.05 was considered to indicate a statistically significant
difference, and all analyses were performed using SPSS software
version 22.0 (IBM Corp., Armonk, NY, USA). All data are presented
as the mean ± standard error of mean from three independent
experiments.
Results
Selection of miRNAs for downregulation
of CKα gene expression
A total of 54 non-repeating miRNAs were predicted to
target CKα mRNA by the online programs used in the present study.
Of these, 22 miRNAs were shortlisted based on their sequence
conservation among different species. Subsequently, the miRNAs were
selected based on favorable minimum free energy (≤-1.0 kcal/mol),
miRNA-mRNA binding site features at the seed region,
complementarity (either the stronger 5′dominant or the weaker
3′compensatory), types of matching to the seed region (with site
efficacy following this order:
8mer>7mer-m8>7mer-A1>6mer>Offset 6 mer) and other site
contexts that improve efficacy (extra Watson-Crick base pairing at
nucleotides 12–17 of the miRNA, and target sites that are located
≥15 nucleotides downstream of the stop codon, near either end of
the 3′-UTR).
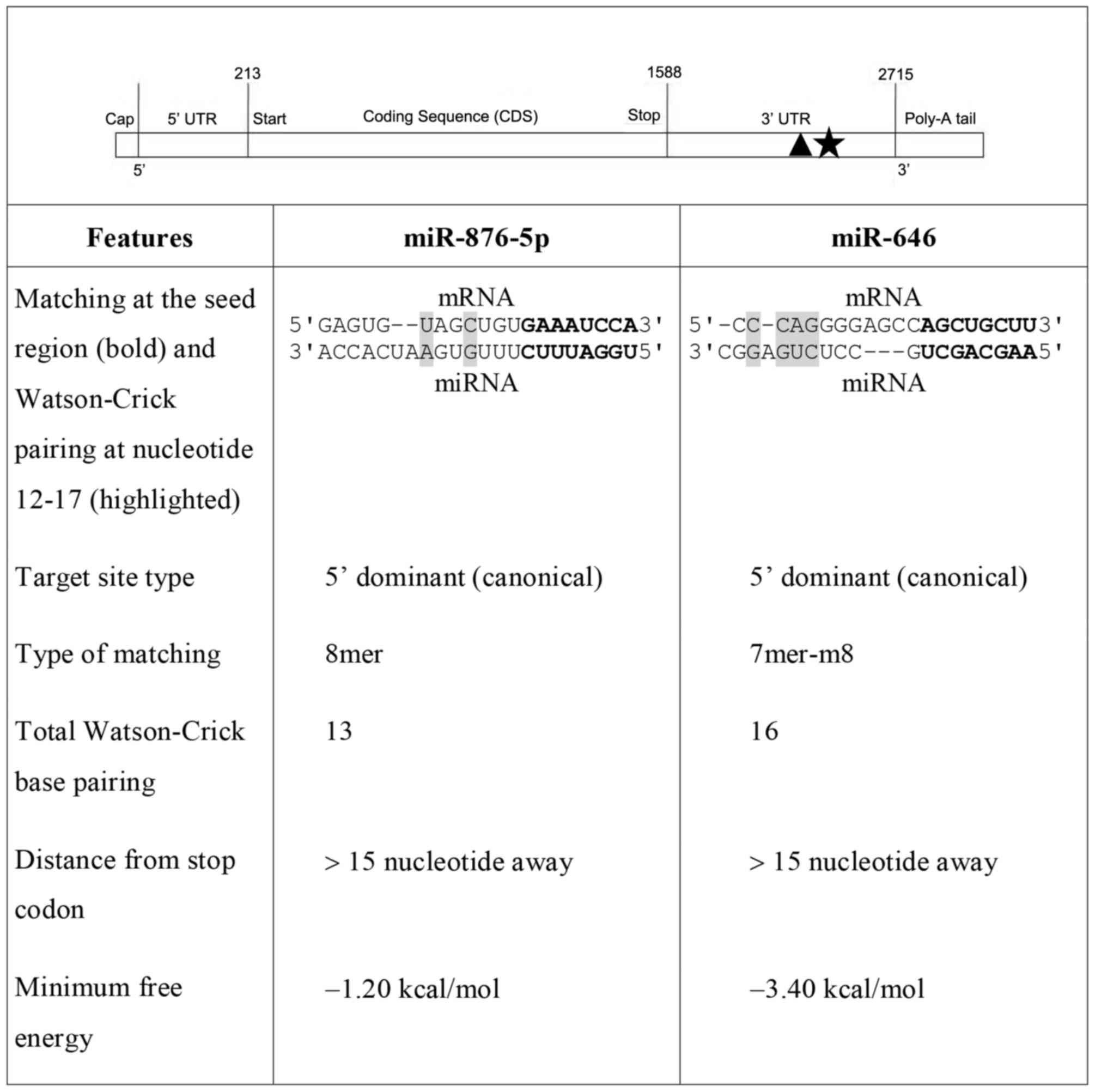
Based on the selection criteria described above, two
miRNA candidates (miR-876-5p and miR-646) were selected for
experimental validation. The principal features of these two miRNAs
and the locations of their target sites at the 3′-UTR of CKα mRNA
are presented in Fig. 1. In
addition to exhibiting other favorable characteristics, miR-876-5p
was selected based on its 8mer site type and two Watson-Crick base
pairings at nucleotides 12–17. Although miR-646 only has a 7mer-m8
site type, it was selected for the four Watson-Crick base pairings
at nucleotides 12–17 and the high GC base pairing throughout the
whole miRNA sequence with its target site. In summary, the two
miRNAs possess favorable seed types, pairings at nucleotides 12–17,
distances from the stop codon, minimum free energy, and the absence
of GU wobble pairs and mismatches at the seed region. The two
miRNAs additionally possess the 5′dominant canonical target site
type, which is more effective than the 5′dominant seed and
3′compensatory types.
Screening of miRNA mimics for the
downregulation of CKα mRNA levels in the HepG2 cancer cell
line
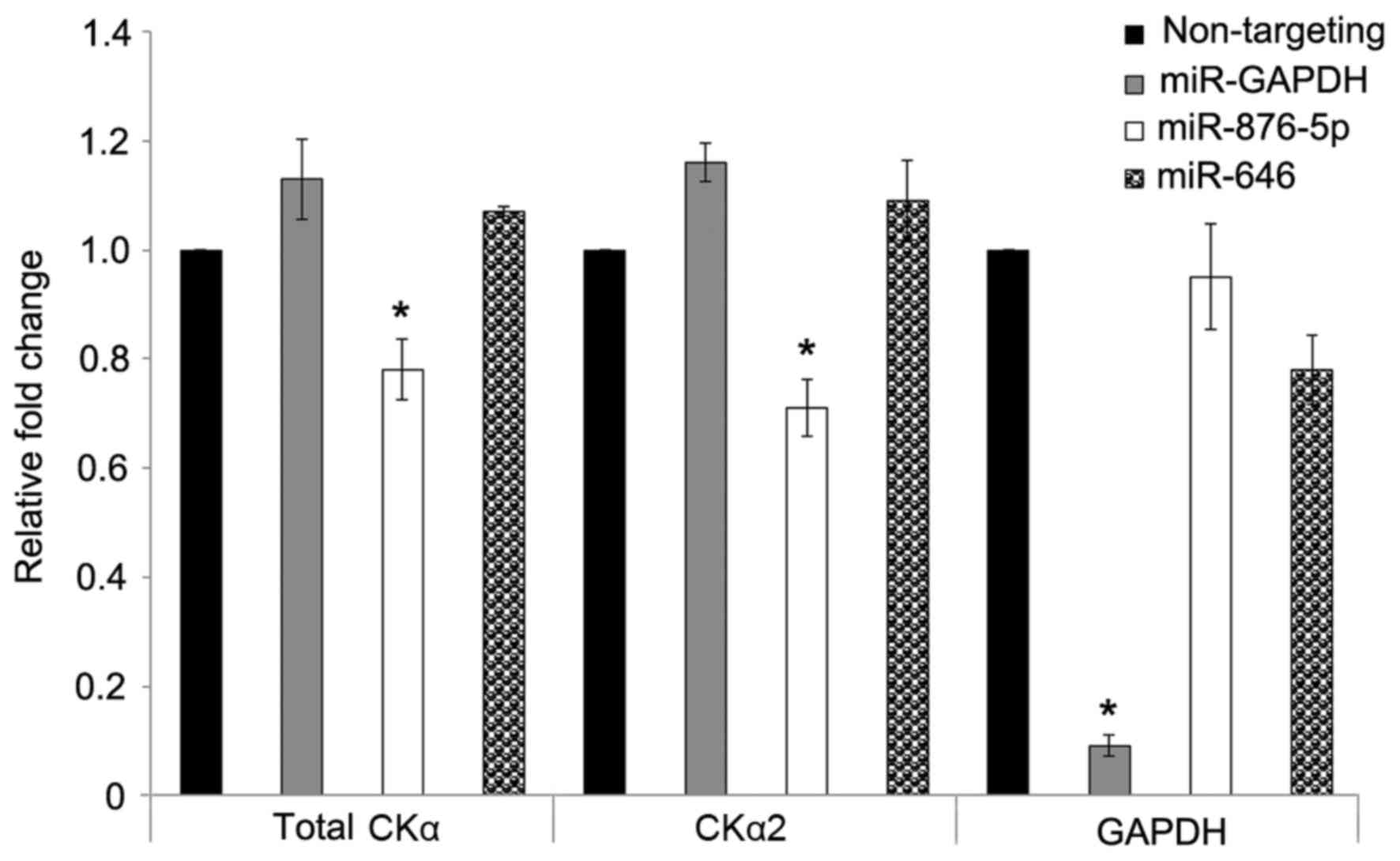
The levels of CKα mRNA were determined by reverse
transcription-qPCR following transfection of HepG2 cells with 25 nM
different miRNA mimics for 48 h. Fig.
2 illustrates that miR-876-5p mimic significantly (P<0.05)
downregulated the expression of total CKα and CKα2 by ~0.8 and
0.7-fold, respectively. miR-646 did not significantly affect the
expression levels of total CKα and CKα2 mRNA. The successful
transfection of HepG2 cells with the miRNAs was confirmed by the
GAPDH-targeting miRNA (positive control miRNA), which resulted in
strong downregulation of GAPDH to ~10% compared with the negative
control (90% downregulation). The results additionally demonstrated
that CKα2 was the predominant CKα isoform, since the levels of
downregulation by miR-876-5p for total CKα and CKα2 were very
similar. Due to the significant downregulation of CKα mRNA by
miR-876-5p, the effects of transfection concentration and duration
were subsequently determined in the present study.
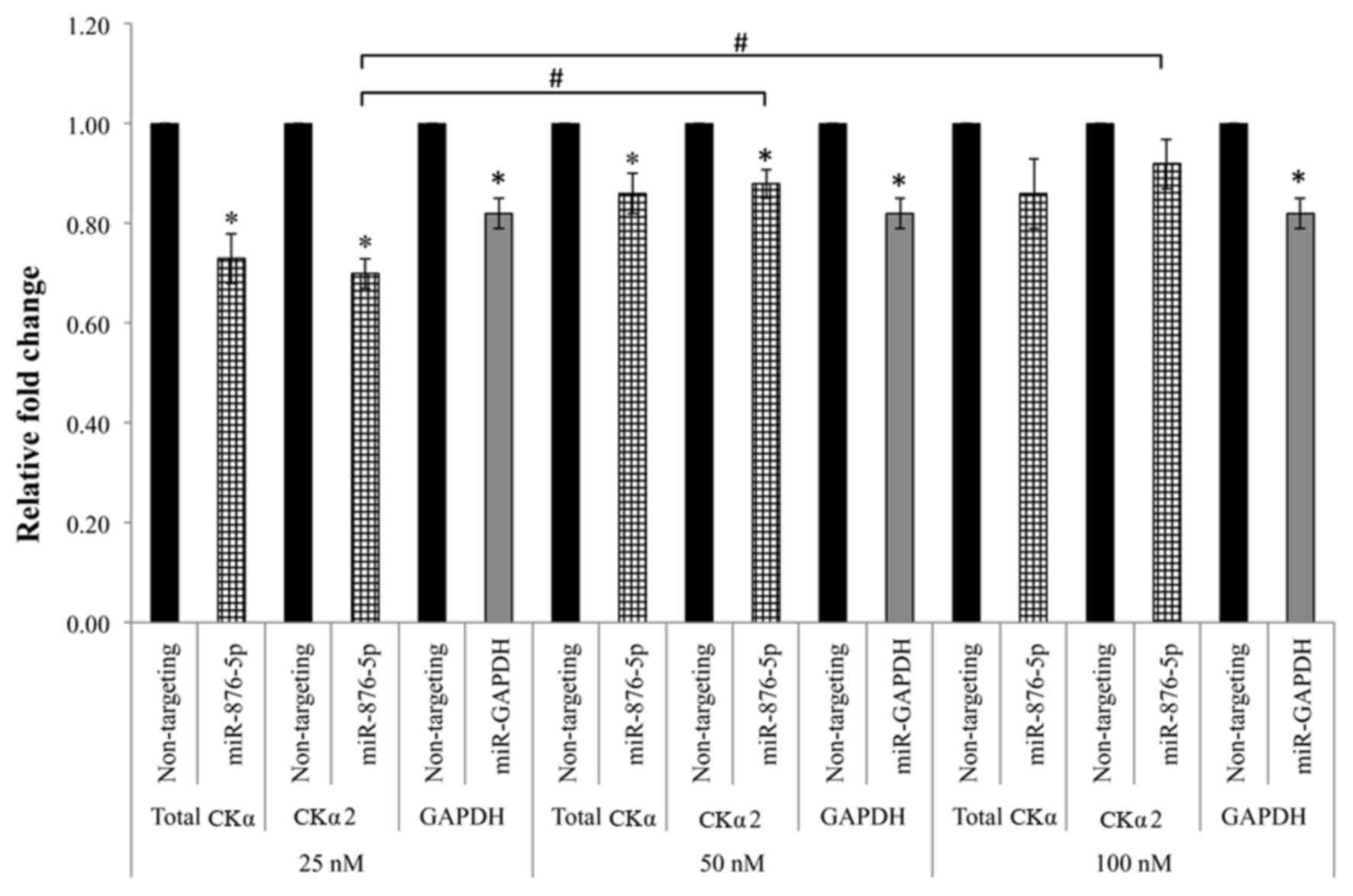
Effect of miR-876-5p concentration on
the downregulation of CKα mRNA expression
A total of three concentrations of miR-876-5p were
tested at the 48 h transfection duration. According to the results
presented in Fig. 3, the total CKα
and CKα2 mRNA expression levels were significantly downregulated to
~0.7 fold (P<0.05) compared with the negative control when the
cells were transfected with 25 nM miR-876-5p. The total CKα and
CKα2 mRNA expression levels were significantly downregulated
(P<0.05) to 86% and 88%, respectively, compared with the
negative control following the transfection with 50 nM miR-876-5p
mimic. Transfection with 100 nM miR-876-5p mimic did not
significantly downregulate the total CKα and CKα2 mRNA expression
levels compared with the negative control (P>0.05). Comparison
between the three concentrations demonstrated that there were
significant differences between CKα2 levels in cells treated with
25 and 50 nM, and 25 and 100 nM (P<0.05). Notably, the
downregulation caused by miR-876-5p may be underestimated for all
the concentrations tested, as the level of GAPDH mRNA was not
effectively downregulated by the positive control miRNA as in
previous experiments. However, the overall results demonstrated
that the concentration of 25 nM miR-876-5p mimic was able to
produce the most marked downregulation of CKα gene expression.
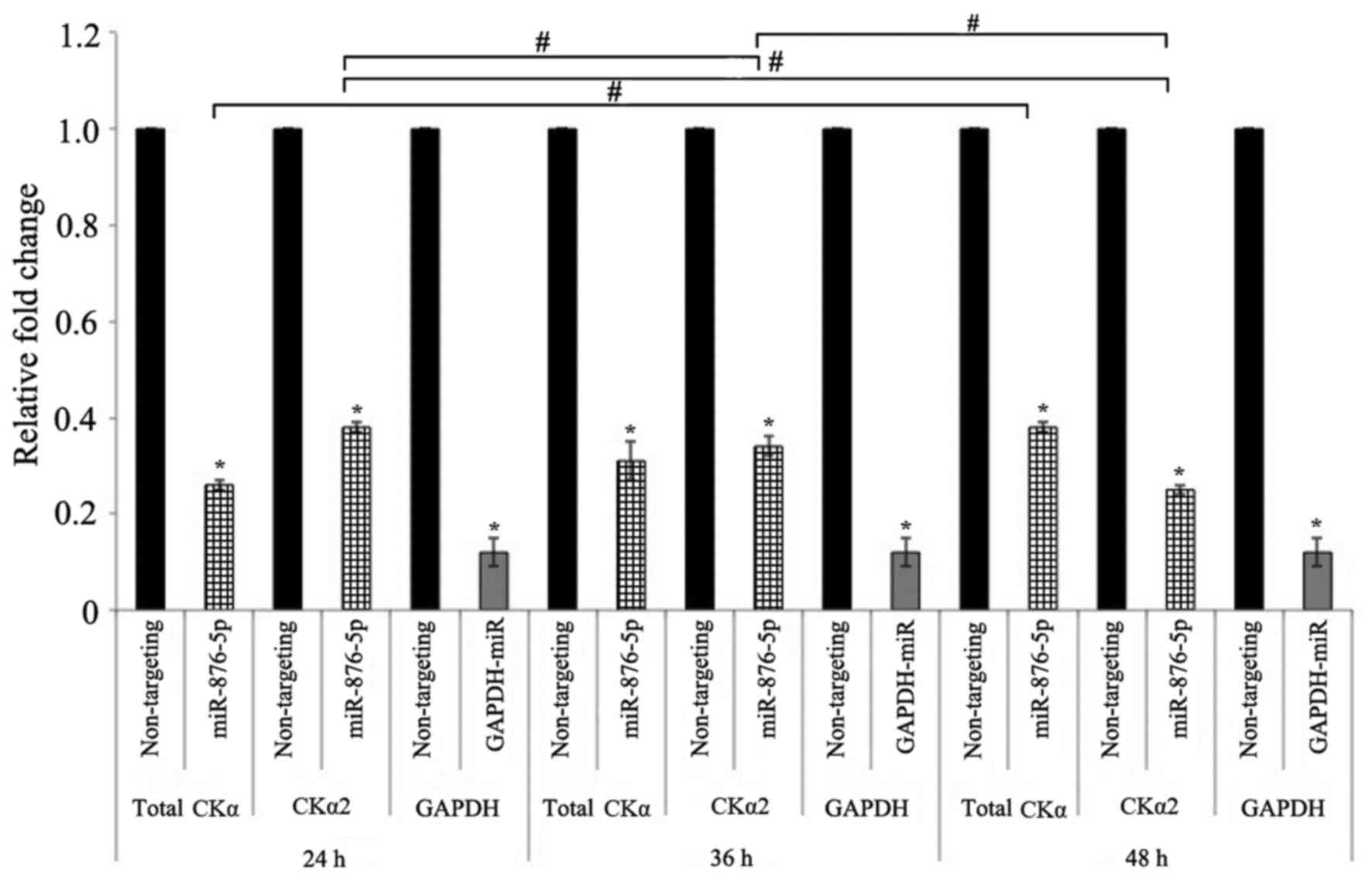
Effect of miR-876-5p transfection
duration on the downregulation of CKα mRNA expression
HepG2 cells were transfected with 25 nM miR-876-5p
mimic at three different transfection durations. Fig. 4 illustrates that the expression
levels of total CKα mRNA were significantly downregulated to 26, 31
and 38% compared with the negative control following transfection
for 24, 36 and 48 h, respectively. The expression levels of CKα2
mRNA were additionally downregulated to 38, 34 and 25% compared
with the negative control at the 24, 36 and 48 h transfection
durations, respectively. All the downregulations were significant
when compared with the negative control (P<0.05). Comparison
between the three transfection durations demonstrated that there
were significant differences in CKα2 expression levels between all
the durations tested, while total CKα expression levels were only
significantly different between cells transfected for 24 and 48 h
(P<0.05). The results were more reliable compared with the
experiments for the optimization of miR-876-5p concentration, as
the GAPDH mRNA levels were markedly downregulated by the positive
control miRNA at all transfection durations tested. Since human
CKα2 is more active than the CKα1 isoform (11) and the strongest downregulation of
CKα2 mRNA expression levels in the HepG2 cell line was obtained
with 48 h transfection of 25 nM miR-876-5p mimic, these parameters
were selected for subsequent experiments. Taken together, the
results affirm the ability of miR-876-5p to downregulate CKα gene
expression.
Effect of miR-876-5p on HepG2 cell
viability and cellular morphology
The viability of HepG2 cells following transfection
with miR-876-5p was determined by MTT cell proliferation assay. As
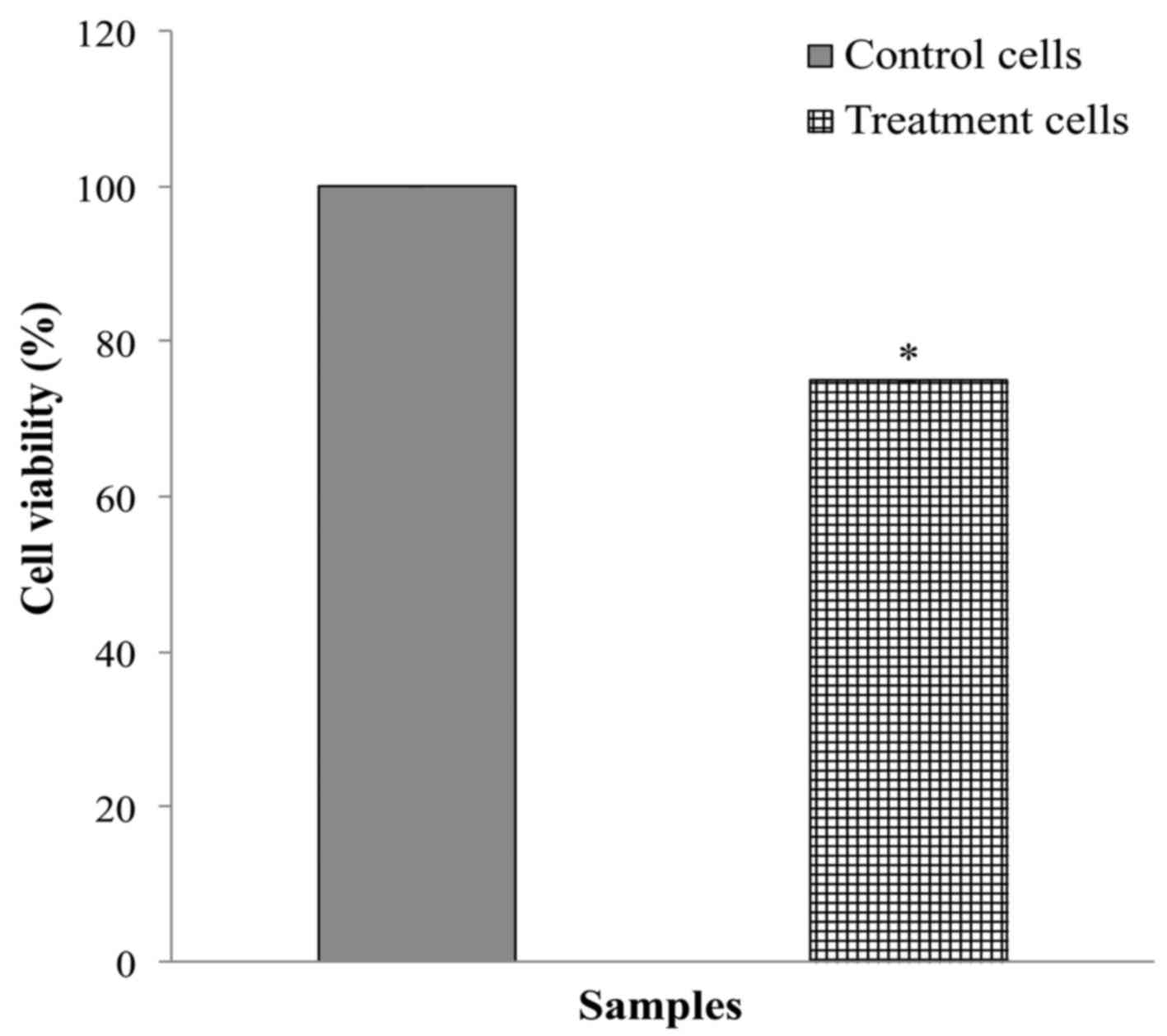
presented in Fig. 5, the viability
of HepG2 cells transfected with 25 nM miR-876-5p mimic for 48 h was
~25% lower compared with cells transfected with the non-targeting
miRNA. There was a significant difference (P<0.05) between the
downregulation levels of miR-876-5p and the negative control
miRNA.
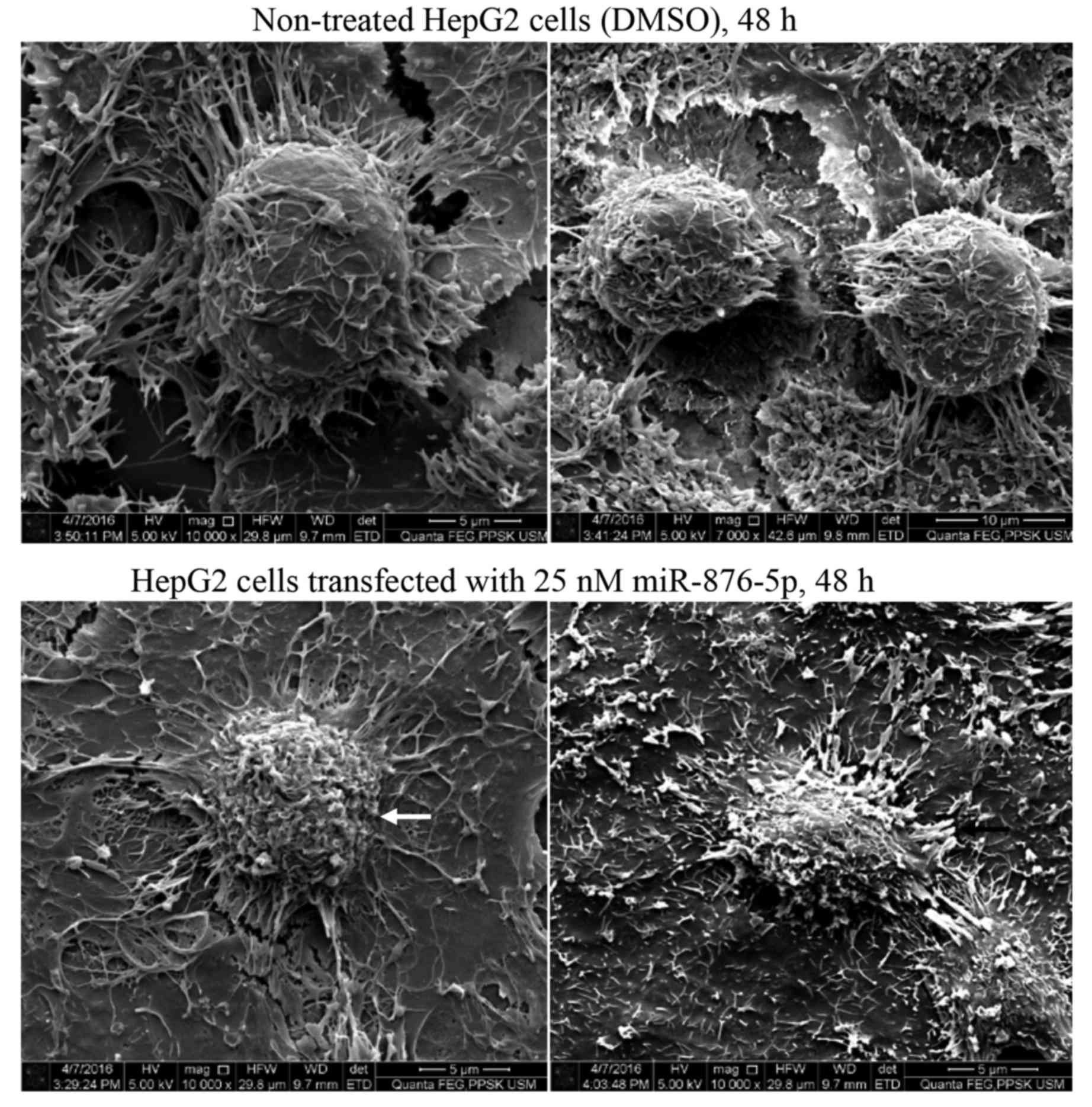
Scanning electron microscopy was used to investigate
the effect of miR-876-5p transfection on HepG2 cellular morphology,
particularly the effect on the cell membrane structure. Fig. 6 presents representative images of
non-treated and miR-876-5p-treated HepG2 cells. The majority of the
non-treated cells (upper panels) appeared healthy, exhibiting a
round shape with filopodia structures at the edges of their
surfaces. By contrast, the cells transfected with miR-876-5p
exhibited signs of apoptosis, including cell shrinkage, membrane
blebbing and loss of filopodia on their membranes. Since the
non-treated cells were not exposed to the liposomal transfection
reagent, it must be noted that the apoptosis-indicating morphology
observed in miR-876-5p-transfected cells may be due to the
transfection reagent.
Discussion
Gene expression may be negatively regulated by
non-coding, single-stranded RNAs of ~22 nucleotides, termed miRNAs,
which cause mRNA cleavage or translational repression (30). In the present study, miRNAs that
potentially bind to the 3′-UTR of CKα mRNA were predicted by four
online programs that used dissimilar algorithms and approaches.
TargetScan, RepTar, microRNA.org
and DIANA micro T flexibly allow users to search for potential
miRNAs, according to target gene name (31). A total of eight features were taken
into consideration for selecting the two best miRNAs: i) Minimum
free energy; ii) mismatches at the seed region; iii) G:U wobble
pair; iv) miRNA target site; v) miRNA site type; vi) Watson-Crick
pairing at nucleotides 12–17; vii) distance from stop codon; and
viii) its position at the 3′ UTR of mRNA.
Human hsa-miR-876-5p (MiRBase accession no.
MIMAT0004924, http://www.mirbase.org) with the
sequence 5′-UGGAUUUCUUUGUGAAUCACCA-3′ originates from the stem-loop
(MiRBase accession no. MI0005542) encoded by a microRNA gene (NCBI
accession no. NR_030597), located on chromosome 9. Recently, the
serum level of miR-876-5p has been identified to be elevated by 9.5
fold in response to severe human enterovirus 71 infections
(32). Knockdown of miR-876-5p
reduced viral load in cultured cells and rendered EV71-related
symptoms less severe in infected mice (32). It has been reported that miR-876-5p
is expressed in primary human myocytes, and that oxytocin may
modulate the expression of this miRNA (33). miR-876-5p appears to be one of the
most downregulated miRNAs in lymph nodes metastases (34), while higher expression of
miR-876-5p is correlated with longer overall survival rates of
hepatocellular carcinoma (35),
rendering this miRNA a potential target for liver cancer therapy
(36). Liu et al (35) demonstrated that miR-876-5p was one
of the top eight miRNAs targeting nima related kinase 11 for
downregulation, leading to drug resistance in ovarian cancer. Human
hsa-miR-646 (MiRBase accession no. MIMAT0003316) with the sequence
5′-AAGCAGCUGCCUCUGAGGC-3′ originates from the stem-loop (MiRBase
accession no. MI0003661) and is encoded by a microRNA gene (NCBI
accession no. NR_030376) located on chromosome 20. miR-646 was
initially recognized in a study of miRNAs expressed in human
cerebral cortical gray and white matter (37). It was reported to be downregulated
in numerous types of cancer, and to be of importance as a tumor
suppressor (38). Accelerated
osteosarcoma tumor progression according to Tumor, Node, Metastasis
classification, and a large tumor diameter were the outcomes of
decreased expression of miR-646 (38). Lower expression of miR-646 was
associated with the metastasis of osteosarcoma, and it was
identified that this miRNA downregulated the expression of
fibroblast growth factor 2 to inhibit osteosarcoma metastasis
(37). miR-646 additionally
downregulated nin one binding protein, and inhibited the growth and
proliferation of renal cancer cells. Downregulation of this miRNA
resulted in the metastasis of renal carcinoma (39). However, the correlation between
miR-876-5p and miR-646, and human choline kinase gene expression,
is yet to be reported, to the best of the authors' knowledge.
In the present study, the use of as low as 25 nM
miRNA was sufficient to downregulate the expression of the target
gene. This concentration is acceptable as it has been reported that
miRNA concentrations >100 nM may decrease cell viability due to
apparent toxicity (40). Higher
concentrations may cause off-target or nonspecific effects, as
demonstrated by Borawski et al (41). At a very low concentration of
miRNA, the target gene expression may not be efficiently repressed
and subtle downregulation of target gene expression may be
difficult to measure (42).
The transfection duration requires optimization in
order to determine the time point following transfection where
target gene downregulation is the strongest. A prolonged
transfection duration may affect cell survival and may not be
suitable. A significant downregulation of the target gene mRNA
expression level may be readily observed at 24 h post-transfection
and the effect may become stronger with longer transfection
duration, with the suppression of protein expression usually
detected at 12 to 24 h following mRNA downregulation, depending on
the half-life of the protein (43).
Total CKα mRNA was downregulated to ~26% of the
negative control with 25 nM miR-876-5p at 24 h, while CKα2 mRNA was
downregulated to ~25% of the negative control with the same amount
of miR-876-5p at 48 h post-transfection. The magnitude of CKα mRNA
downregulation exhibited by miR-876-5p was relatively large since,
according to Mukherji et al (44), typical miRNA gene repression is
relatively small when measured in a whole population of cultured
cells. The results of the present study support the use of
bioinformatics prediction of miRNAs targeting a specific gene of
interest as a promising approach to search for novel miRNA
modulators of the target gene. The reason for miR-646 not exerting
any significant downregulation of CKα is unclear. The effect of
miR-646 may require other transfection conditions. However, it may
be that miR-646 does not bind to the 3′-UTR of CKα mRNA, although
it was one of the best predicted candidates. It must be emphasized
that further experiments are required to confirm the downregulation
of CKα by miR-876-5p and to address certain limitations of the
present study. These limitations include the lack of proof of a
direct interaction between miR-876-5p and the 3′-UTR of CKα mRNA,
and this requires further investigation with a luciferase reporter
assay. Western blot analysis is require to confirm the alterations
in CKα protein expression levels following treatment with
miR-876-5p. An additional limitation of the present study was the
variation in transfection efficiency, as illustrated by the
unusually low GAPDH downregulation in the experiments to determine
the optimal miR-876-5p concentration compared to the results of the
miRNA screening and determination of optimal miR-876-5p
transfection duration. The experiments ought to be repeated to
obtain more accurate results for the determination of the optimum
miR-876-5p transfection concentration.
The results of the cell viability assay and scanning
electron microscopy additionally suggested that miRNA-876-5p
induced suppression of CKα gene expression, caused decreased cell
viability and induced apoptotic cell death. However, further
experiments are required to verify these results. Future studies
may include cell proliferation and apoptosis assays at different
time points, and the negative control cells may be transfected with
non-targeting miRNA for direct comparison with
miR-876-5p-transfected cells under the scanning electron
microscope. Previously, knockdown of CKα expression by RNAi has
been demonstrated to promote cancer cell death (11). Thus, miR-876-5p downregulation may
generate a similar effect on cancer cells. The present study is the
first, to the best of our knowledge, to report on the miRNA
regulation of CKα gene expression, and the miR-876-5p identified
here may be developed into a promising anticancer therapeutic
agent.
In conclusion, the present study demonstrated that,
out of the two tested miRNA candidates, miR-876-5p was able to
significantly decrease the expression level of CKα mRNA. At
concentrations as low as 25 nM, miR-876-5p mimic was able to exert
the strongest downregulation of CKα gene expression in HepG2 cells.
Additionally, HepG2 cells transfected with miR-876-5p exhibited
decreased viability compared with the non-treated cells, in
addition to certain signs of apoptosis. All of these observations
indicated the role of miR-876-5p in modulating CKα gene expression
and its direct involvement in cancer cell proliferation. Further
studies are required to examine the expression pattern of
miR-876-5p in different tumors, and to investigate the association
between the level of this miRNA with tumor type or invasiveness.
The results of the present study may be further confirmed by
looking at the effect of treating appropriate cell lines with
anti-miR-876-5p oligonucleotides on CKα gene expression. In
addition, it is also important to demonstrate that the effect
observed in the present study was due to direct binding of
miR-876-5p to the 5′-UTR of CKα mRNA by luciferase reporter assay.
The effect of this miRNA on the CKα protein expression level
requires investigation.
Acknowledgements
The authors wish to acknowledge the laboratory staff
of the School of Health Sciences for their technical
assistance.
Funding
The present study was supported by the Fundamental
Research Grant Scheme (grant no. 203/PPSK/6171171) and Universiti
Sains Malaysia Research University (grant no. 1001/PPSK/812161).
SMAK was a postgraduate supported by the Universiti Sains Malaysia
Fellowship Scheme.
Availability of data and materials
The data generated and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
SMAK performed the experiments, analyzed the data
and wrote the manuscript. LLF designed the experiments, analyzed
the data and wrote the manuscript. WCST conceived and designed the
experiments, analyzed the data and was the major contributor in
writing the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lykidis A, Wang J, Karim MA and Jackowski
S: Overexpression of a mammalian ethanolamine-specific kinase
accelerates the CDP-ethanolamine pathway. J Biol Chem.
276:2174–2179. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Malito E, Sekulic N, Too WC, Konrad M and
Lavie A: Elucidation of human choline kinase crystal structures in
complex with the products ADP or phosphocholine. J Mol Biol.
364:136–151. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aoyama C, Ohtani A and Ishidate K:
Expression and characterization of the active molecular forms of
choline/ethanolamine kinase-alpha and -beta in mouse tissues,
including carbon tetrachloride-induced liver. Biochem J.
363:777–784. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gallego-Ortega D, Ramirez de Molina A,
Ramos MA, Valdes-Mora F, Barderas MG, Sarmentero-Estrada J and
Lacal JC: Differential role of human choline kinase alpha and beta
enzymes in lipid metabolism: Implications in cancer onset and
treatment. PLoS One. 4:e78192009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ramírez de Molina A, Gutiérrez R, Ramos
MA, Silva JM, Silva J, Bonilla F, Sánchez JJ and Lacal JC:
Increased choline kinase activity in human breast carcinomas:
Clinical evidence for a potential novel antitumor strategy.
Oncogene. 21:4317–4322. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nakagami K, Uchida T, Ohwada S, Koibuchi
Y, Suda Y, Sekine T and Morishita Y: Increased choline kinase
activity and elevated phosphocholine levels in human colon cancer.
Jpn J Cancer Res. 90:419–424. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ramírez de Molina A, Rodríguez-González A,
Gutiérrez R, Martínez-Piñeiro L, Sánchez J, Bonilla F, Rosell R and
Lacal J: Overexpression of choline kinase is a frequent feature in
human tumor-derived cell lines and in lung, prostate, and
colorectal human cancers. Biochem Biophys Res Commun. 296:580–583.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shah T, Wildes F, Penet MF, Winnard PT Jr,
Glunde K, Artemov D, Ackerstaff E, Gimi B, Kakkad S, Raman V and
Bhujwalla ZM: Choline kinase overexpression increases invasiveness
and drug resistance of human breast cancer cells. NMR Biomed.
23:633–642. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rodríguez-González A, Ramirez de Molina A,
Fernández F and Lacal JC: Choline kinase inhibition induces the
increase in ceramides resulting in a highly specific and selective
cytotoxic antitumoral strategy as a potential mechanism of action.
Oncogene. 23:8247–8259. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bañez-Coronel M, Ramírez de Molina A,
Rodríguez-González A, Sarmentero J, Ramos MA, García-Cabezas MA,
García-Oroz L and Lacal JC: Choline kinase alpha depletion
selectively kills tumoral cells. Curr Cancer Drug Targets.
8:709–719. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gruber J, See Too WC, Wong MT, Lavie A,
McSorley T and Konrad M: Balance of human choline kinase isoforms
is critical for cell cycle regulation: Implications for the
development of choline kinase-targeted cancer therapy. FEBS J.
279:1915–1928. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
MacKeigan JP, Murphy LO and Blenis J:
Sensitized RNAi screen of human kinases and phosphatases identifies
new regulators of apoptosis and chemoresistance. Nat Cell Biol.
7:591–600. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Almeida MI, Reis RM and Calin GA: MicroRNA
history: Discovery, recent applications, and next frontiers. Mutat
Res. 717:1–8. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vella MC and Slack FJ: C. elegans
microRNAs. WormBook. 1–9. 2005.PubMed/NCBI
|
|
17
|
Williams AH, Liu N, van Rooij E and Olson
EN: MicroRNA control of muscle development and disease. Curr Opin
Cell Biol. 21:461–469. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Broderick JA and Zamore PD: MicroRNA
therapeutics. Gene Ther. 18:1104–1110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stenvang J, Petri A, Lindow M, Obad S and
Kauppinen S: Inhibition of microRNA function by antimiR
oligonucleotides. Silence. 3:12012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Conde J, Oliva N, Atilano M, Song HS and
Artzi N: Self-assembled RNA-triple-helix hydrogel scaffold for
microRNA modulation in the tumour microenvironment. Nat Mater.
15:353–363. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jansson MD and Lund AH: MicroRNA and
cancer. Mol Oncol. 6:590–610. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
24
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:e852005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chua SL, See Too WC, Khoo BY and Few LL:
UBC and YWHAZ as suitable reference genes for accurate
normalisation of gene expression using MCF7, HCT116 and HepG2 cell
lines. Cytotechnology. 63:645–654. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McDowell EM and Trump BF: Histologic
fixatives suitable for diagnostic light and electron microscopy.
Arch Pathol Lab Med. 100:405–414. 1976.PubMed/NCBI
|
|
30
|
Bandres E, Agirre X, Ramirez N, Zarate R
and Garcia-Foncillas J: MicroRNAs as cancer players: Potential
clinical and biological effects. DNA Cell Biol. 26:273–282. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Peterson SM, Thompson JA, Ufkin ML,
Sathyanarayana P, Liaw L and Congdon CB: Common features of
microRNA target prediction tools. Front Genet. 5:232014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang RY, Weng KF, Huang YC and Chen CJ:
Elevated expression of circulating miR876-5p is a specific response
to severe EV71 infections. Sci Rep. 6:241492016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cook JR, MacIntyre DA, Samara E, Kim SH,
Singh N, Johnson MR, Bennett PR and Terzidou V: Exogenous oxytocin
modulates human myometrial microRNAs. Am J Obstet Gynecol.
213:65.e1–e9. 2015. View Article : Google Scholar
|
|
34
|
Saiselet M, Gacquer D, Spinette A, Craciun
L, Decaussin-Petrucci M, Andry G, Detours V and Maenhaut C: New
global analysis of the microRNA transcriptome of primary tumors and
lymph node metastases of papillary thyroid cancer. BMC Genomics.
16:8282015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu X, Gao Y, Lu Y, Zhang J, Li L and Yin
F: Downregulation of NEK11 is associated with drug resistance in
ovarian cancer. Int J Oncol. 45:1266–1274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang YH, Lin KH, Chen HC, Chang ML, Hsu
CW, Lai MW, Chen TC, Lee WC, Tseng YH and Yeh CT: Identification of
postoperative prognostic microRNA predictors in hepatocellular
carcinoma. PLoS One. 7:e371882012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun XH, Geng XL, Zhang J and Zhang C:
miRNA-646 suppresses osteosarcoma cell metastasis by downregulating
fibroblast growth factor 2 (FGF2). Tumour Biol. 36:2127–2134. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Azam AT, Bahador R, Hesarikia H, Shakeri M
and Yeganeh A: Downregulation of microRNA-217 and microRNA-646 acts
as potential predictor biomarkers in progression, metastasis, and
unfavorable prognosis of human osteosarcoma. Tumour Biol.
37:5769–5773. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Z, Wu G, Sher RB, Khavandgar Z,
Hermansson M, Cox GA, Doschak MR, Murshed M, Beier F and Vance DE:
Choline kinase beta is required for normal endochondral bone
formation. Biochim Biophys Acta. 1840:2112–2122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bollin F, Dechavanne V and Chevalet L:
Design of experiment in CHO and HEK transient transfection
condition optimization. Protein Expr Purif. 78:61–68. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Borawski J, Lindeman A, Buxton F, Labow M
and Gaither LA: Optimization procedure for small interfering RNA
transfection in a 384-well format. J Biomol Screen. 12:546–559.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jin HY, Gonzalez-Martin A, Miletic AV, Lai
M, Knight S, Sabouri-Ghomi M, Head SR, Macauley MS, Rickert RC and
Xiao C: Transfection of microRNA mimics should be used with
caution. Front Genet. 6:3402015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hengstermann A, D'Silva MA, Kuballa P,
Butz K, Hoppe-Seyler F and Scheffner M: Growth suppression induced
by downregulation of E6-AP expression in human
papillomavirus-positive cancer cell lines depends on p53. J Virol.
79:9296–9300. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mukherji S, Ebert MS, Zheng GX, Tsang JS,
Sharp PA and van Oudenaarden A: MicroRNAs can generate thresholds
in target gene expression. Nat Genet. 43:854–859. 2011. View Article : Google Scholar : PubMed/NCBI
|