Introduction
Melanoma is a type of cancer that originates from
cells containing pigment (1).
Although melanoma is less common compared with squamous cell
carcinomas and cutaneous basal cell carcinomas, melanoma is
considered to be one of the most common causes of cancer-associated
mortalities in young adults and children (2). The highest incidence of melanoma has
been observed in Australia and New Zealand, followed by North
America and Northern Europe, while the incidence of this disease is
relatively low in Latin America, Africa and Asia (3). In addition, melanoma was also
revealed to be more common in men than in women (4). With proper treatment, the 5-year
survival rate of patients only with local tumors may reach 98%.
However, once metastasis has occurred, the 5-year survival rate may
be as low as 17% (5). Similar to
other tumors, the development and progression of melanoma is a
complex process with multiple signaling pathways involved (6,7).
Therefore, an in-depth analysis of the signal transduction
regulation involved in melanoma will facilitate the development of
novel treatment strategies for this disease.
Long non-coding RNA (lncRNA), is a group of
noncoding RNAs with a length >200 nucleotides (8). A previous study has demonstrated that
the onset and development of different human cancers requires the
involvement of various lncRNAs (9). As a lncRNA, H19 has been demonstrated
to serve a role as an oncogene in various human malignant tumors
(10,11). However, based on current knowledge,
the functionality of H19 in melanoma remains unclear.
In the present study, the expression of H19 in the
tumor tissues and adjacent healthy tissues of 59 patients with
melanoma was detected. The effects of H19 on proliferation,
migration and invasion of two melanoma cells lines were
investigated. In addition, the interactions between H19 and the
nuclear factor (NF)-κB signaling pathway were also explored. The
results of the present study may benefit future studies on the
pathogenesis of melanoma and the clinical treatment of this
disease.
Materials and methods
Patients
A total of 49 patients with melanoma were recruited
to the present study in the Tianjin Medical University Cancer
Institute and Hospital (Tianjin, China) from October 2013 to August
2017. Patients diagnosed with other severe diseases and mental
health problems were excluded. Patients were diagnosed via imaging
examinations and pathological tests. The patients were then divided
into 3 tumor stage groups according to tumor thickness. These
groups were characterized by the following diagnostic criteria:
Stage I, tumor thickness <1.0 mm; stage II, tumor thickness
between 1.0 and 4.0 mm; and stage III, tumor thickness >4.0 mm.
Patients in stage I included 7 males and 8 females, with an age
range of 21 to 68 years and an average age of 43±11.6 years;
patients in stage II included 8 males and 9 females, with an age
range of 27 to 72 years and an average age of 48±13.1 years;
patients in stage III included 8 males and 7 females, with an age
range of 24 to 73 years and an average age of 46±11.0 years. No
significant differences in age, sex and other basic information
were found among different stages. All patients received surgical
resection, and tumor tissues and surrounding healthy tissue (within
1 cm around tumor) were collected during the operation. The present
study was approved by the Ethics Committee of Tianjin Medical
University Cancer Institute and Hospital, and all patients provided
written informed consent.
Cell lines and cell culture
Human melanoma cell lines C32 and SK-MEL-28, and the
normal skin cell line CCD-1059Sk were all purchased from American
Type Culture Collection (Manassas, VA, USA). Cell culture was
performed according to manufacturer's protocol in Eagle's minimum
Essential medium (EMEM; American Type Culture Collection)
containing 10% fetal bovine serum (FBS, Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Cells were harvested during the logarithmic
growth phase for subsequent experiments.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tumor tissues, adjacent
healthy tissues and cells using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA). Tissues were ground in
liquid nitrogen before the addition of TRIzol reagent. RNA quality
was determined using a NanoDrop™ 2000 Spectrophotometer
(Thermo Fisher Scientific, Inc.), and only RNA samples with a
A260/A280 ratio between 1.8 and 2.0 were used to synthesize cDNA
via reverse transcription using SuperScript III Reverse
Transcriptase (Thermo Fisher Scientific, Inc.). Reaction conditions
for reverse transcription were: 65°C for 5 min, 50°C for 50 min and
85°C for 5 min. A qPCR reaction system was prepared using cDNA and
SYBR®-Green Real-Time PCR Master mix (Thermo Fisher
Scientific, Inc.). Following primers were used in the PCR
reactions: 5′-TGAGCTCTCAGGAGGGAGGATGG-3′ (forward) and
5′-TTGTCACGTCCACCGGACCTG-3′ (reverse) for H19;
5′-GACCTCTATGCCAACACAGT-3′ (forward) and 5′-AGTACTTGCGCTCAGGAGGA-3′
(reverse) for β-actin. The qPCR thermocycling conditions were as
follows: 95°C for 42 sec, followed by 40 cycles of 95°C for 10 sec
and 60°C for 35 sec. Data were analyzed using the 2−ΔΔCq
method (12), and the relative
expression levels of H19 were normalized to those of the endogenous
control β-actin.
Establishment of H19 small interfering
(si)RNA silencing cell lines
H19 siRNA silencing cell lines were constructed
using commercial siRNAs. Silencer™ Select Negative
Control No. 1 siRNA (cat. no. 4390843; Thermo Fisher Scientific,
Inc.) and H19 siRNA (cat. no. 1299001; Thermo Fisher Scientific,
Inc.) were used. Prior to transfection, cells were cultured EMEM
containing 10% FBS overnight to reach 80–90% confluence.
Lipofectamine 2000™ reagent (cat. no. 11668-019;
Invitrogen, Thermo Fisher Scientific, Inc.) was used to transfect
40 nM siRNA into 5×105 cells by incubation with the
cells at 37°C in a CO2 incubator for 4 h according to
the manufacturer's protocol, followed by incubation in RPMI-1640
medium (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) at
37°C for 48 h prior to subsequent experimentation.
Cell proliferation assay
Cell proliferation ability was measured using a Cell
Counting kit-8 (CCK-8; Sigma-Aldrich; Merck KGaA) according to the
manufacturer's protocol. Briefly, 100 µl cell suspension containing
2×104 cells was transferred to each well of a 96-well
plate. CCK-8 solution (10 µl) was added into each well for cell
culture for 24, 48, 72 and 96 h. Following incubation at 37°C for a
further 4 h, the optical density values were measured at a
wavelength of 450 nm using a microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Wound-healing assay
Cells were seeded into a 24-well plate at
1×105 cells per well. Once cell adhesion was reached
(following 48 h), a pipette tip was used to scratch the cells.
Cells were washed with PBS and cultured in an incubator (37°C, 5%
CO2) for 24 h to reach ~70–80% confluence. Cell
migration was observed under an inverted microscope (Olympus
Corporation, Tokyo, Japan), 10 fields of view were randomly
selected and images were obtained. ImagePro Plus version 5.0
software (National Institutes of Health, Bethesda, MD, USA) was
used to analyze all images.
Cell invasion assay
Transwell cell invasion assay (BD Biosciences,
Franklin Lakes, NJ, USA) was performed to measure the cell invasion
ability. The upper chamber was pre-coated with Matrigel (cat. no.
356234; EMD Millipore, Billerica, MA, USA) and filled with
serum-free RPMI-1640 medium (Thermo Fisher Scientific, Inc.)
containing 5×104 cells. RPMI-1640 medium containing 20%
fetal calf serum (Sigma-Aldrich, Merck KGaA) was used to fill the
lower chamber. Following incubation at 37°C for 24 h, membranes
were collected and stained with 0.5% crystal violet (Sigma-Aldrich;
Merck KGaA) at room temperature for 20 min. A total of 10 fields of
vision were randomly selected and stained cells were counted under
an optical microscope (Olympus Corporation).
Western blotting
Following protein extraction using
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.), the concentration of protein samples was measured using a
Bicinchoninic Acid assay. Then, 10% SDS-PAGE gel electrophoresis
was performed using 30 µg of protein from each sample, followed by
transfer to a polyvinylidene difluoride membrane under 25 V for 1
h. Blocking was performed using 5% skimmed milk at room temperature
for 1 h. Following washing with TBST (0.1% Tween-20), membranes
were then incubated with the corresponding primary antibodies
including rabbit anti-phosphorylated (p)-phosphoinositide 3-kinase
(PI3K) antibody (1:2,000; cat. no. ab182651; Abcam, Cambridge, UK),
anti-PI3K antibody (1:2,000; cat. no. ab5451; Abcam),
anti-p-protein kinase B (AKT) antibody (1:2,000; cat. no. ab18206;
Abcam), anti-AKT antibody (1:2,000; cat. no. ab126811; Abcam),
anti-inhibitor of nuclear factor κβ (IκBα) antibody (1:1,000; cat.
no. ab76429; Abcam), anti-NF-κB p65 antibody (1:1,000; cat. no.
ab16502; Abcam), anti-NF-κB p50 antibody (1:1,000; cat. no.
ab32360; Abcam) and anti-β-actin primary antibody (1:1,000; cat.
no. ab8226; Abcam) overnight at 4°C. Following washing with PBS,
membranes were further incubated with anti-rabbit IgG-horseradish
peroxidase secondary antibody (1:1,000; cat. no. MBS435036;
MyBioSource, San Diego, CA, USA) at room temperature for 1 h.
Membranes were then washed with TBST and signals were detected
using an enhanced chemiluminescence substrate (Sigma-Aldrich; Merck
KGaA) method. Relative expression levels of each protein were
normalized to the endogenous control, β-actin, using ImageJ 1.8.0
software (National Institutes of Health).
Statistical analysis
SPSS software version 19.0 (IBM Corp., Armonk, NY,
USA) was used for all statistical analyses. Each experiment was
performed three times, and results are presented as the mean ±
standard deviation. Data between two groups were analyzed by
Student's t-test and multiple comparisons were analyzed by one way
analysis of variance with a least significant difference post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
H19 is upregulated in melanoma tumor
tissue compared with adjacent healthy tissue
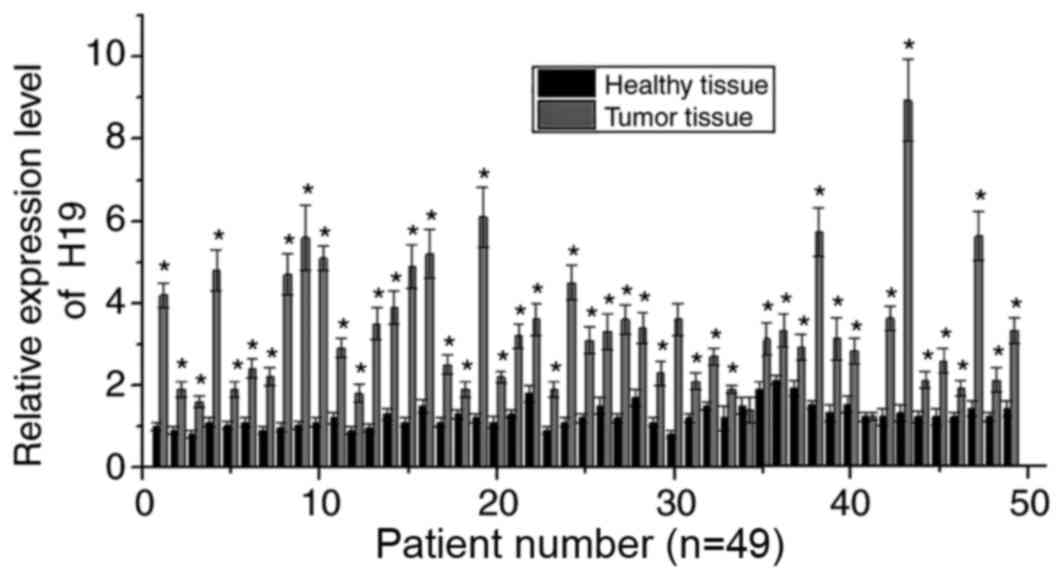
RT-qPCR was performed to detect the mRNA expression
of H19 in the tumor and adjacent healthy tissues of 49 patients
with melanoma. As shown in Fig. 1,
compared with adjacent healthy tissues, the mRNA expression levels
of H19 were significantly increased in the cancerous tissues of 47
of 49 patients (P<0.05), suggesting the potential involvement of
H19 in the development of melanoma.
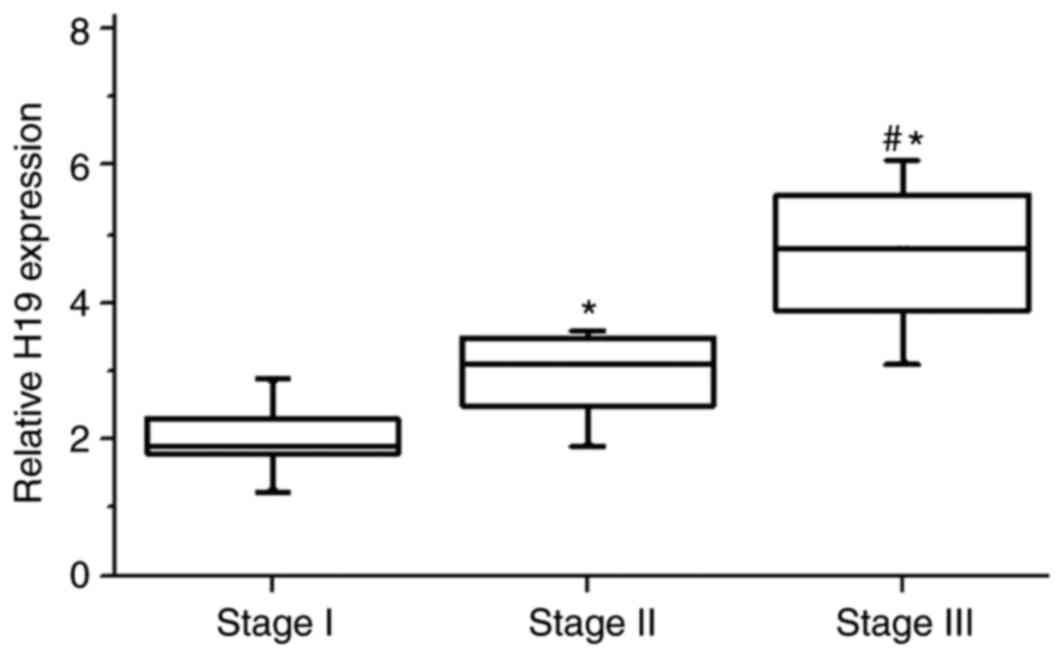
Expression of H19 increases with the
development of melanoma
Patients were divided into different stages
according to their tumor thickness. As shown in Fig. 2, the expression levels of H19 RNA
in tumor tissues were significantly higher in patients with stage
II than in patients with stage I (P<0.05). Similarly, the
expression levels of H19 RNA in tumor tissues were also
significantly higher in patients with stage III compared with
patients with stage II (P<0.05). These results suggested that
the expression of H19 was greater with increasing stages of
melanoma.
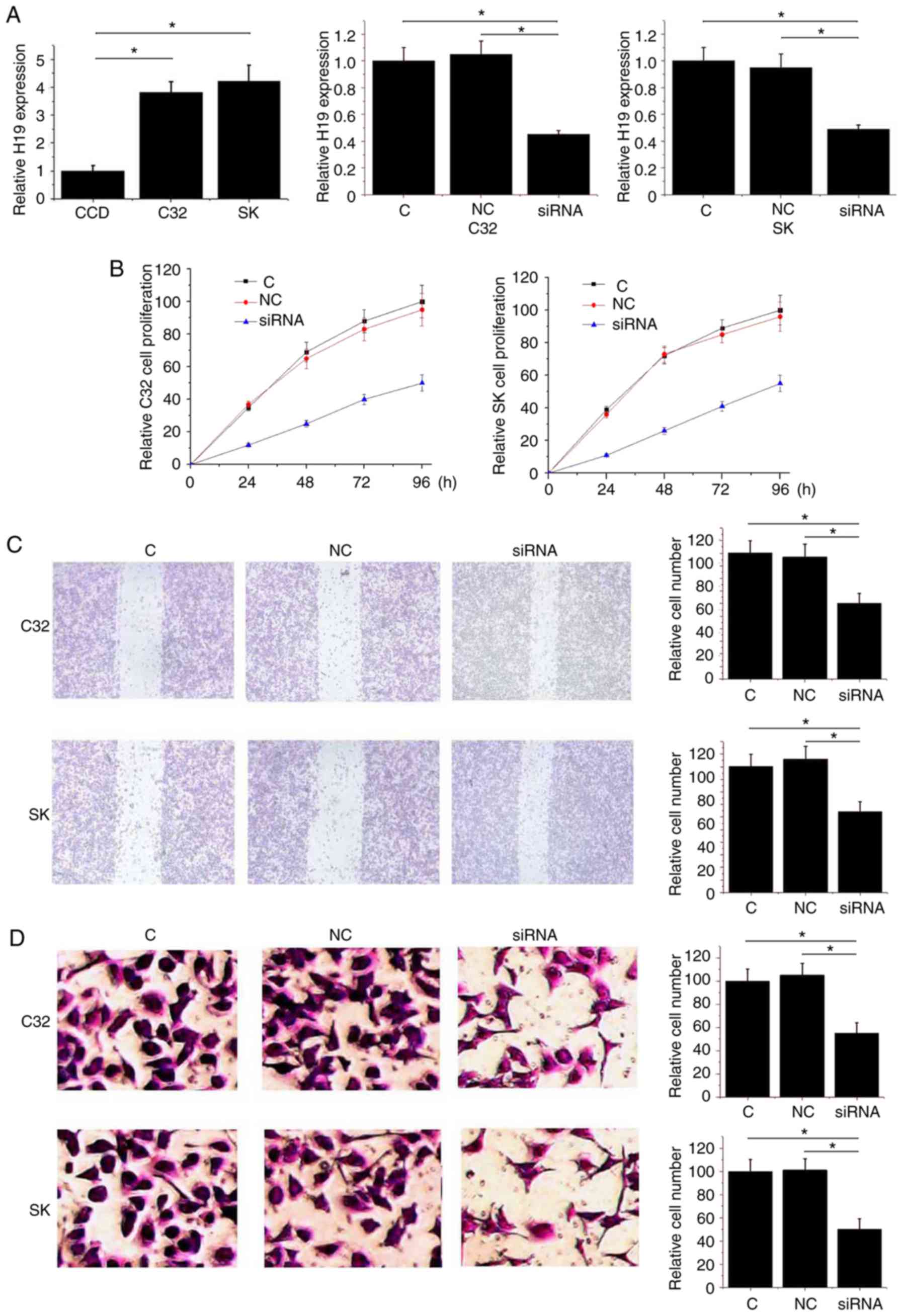
H19 knockdown inhibits the
proliferation, migration and invasion of melanoma cells
As shown in Fig.
3A, compared with the normal skin cell line CCD-1059Sk,
expression levels of H19 RNA were significantly increased in the
human melanoma cell line C32 and SK-MEL-28 (P<0.05). Following
siRNA transfection, the expression levels of H19 RNA were
significantly reduced in the C32 and SK-MEL-28 cell lines
(P<0.05; Fig. 3A). As shown in
Fig. 3B, downregulation of H19
significantly reduced the proliferation ability of C32 and
SK-MEL-28 cells (P<0.05). Similarly, downregulation of H19 also
significantly reduced the migration and invasion ability of C32 and
SK-MEL-28 (P<0.05; Fig. 3C and
D). The data suggested that H19 knockdown inhibited the
proliferation, migration and invasion of melanoma cells.
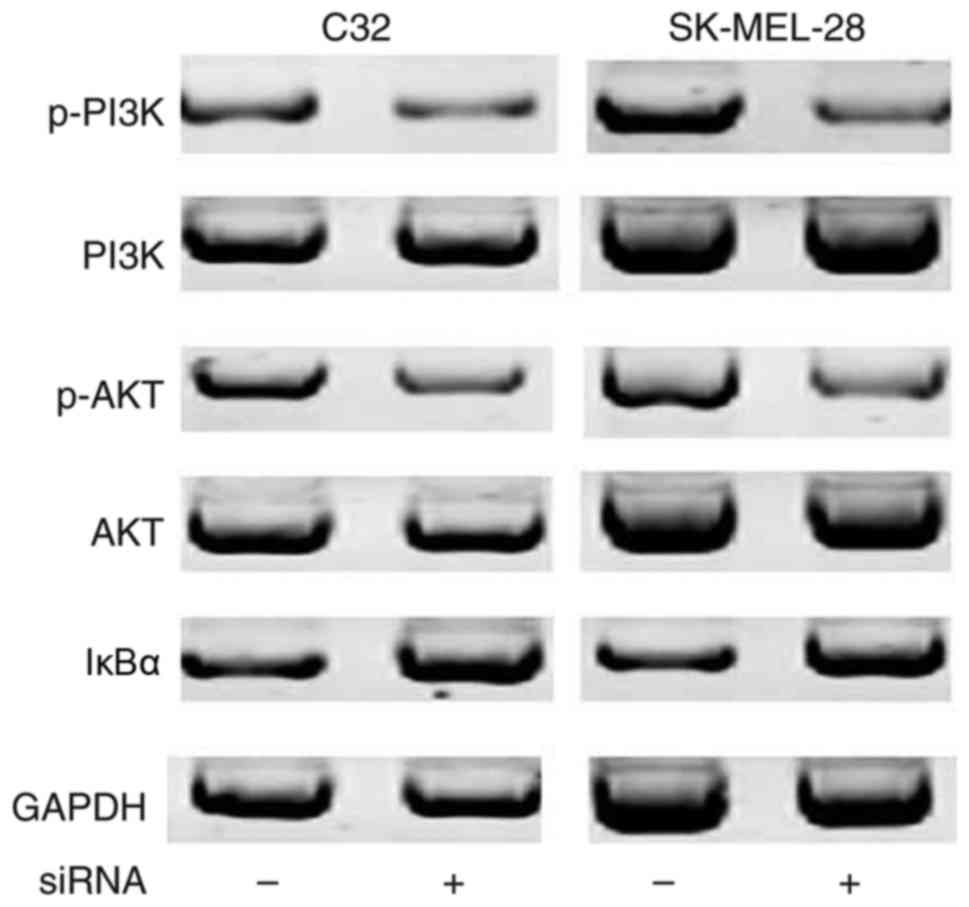
H19 siRNA silencing inhibits the
activation of the NF-κB signaling pathway
It has been well established that the NF-κB
signaling pathway may be activated by the PI3K/Akt signaling
pathway via the degradation of IκBα. As shown in Fig. 4, no differences in the expression
levels of PI3K and Akt were observed between control cells and
cells transfected with siRNA. Compared with cells without siRNA
transfection, the expression levels of p-PI3K and p-Akt were
decreased in cells transfected with siRNA, indicating inactivation
of the PI3K/Akt signaling pathway following H19 knockdown. In
addition, compared with cells without siRNA transfection,
expression levels of the IκBα protein were increased in cells with
siRNA transfection, indicating reduced degradation of IκBα
following H19 knockdown. These data suggested that H19 knockdown
inactivated the PI3K/Akt signaling pathway, which in turn inhibited
the activation of the NF-κB signaling pathway.
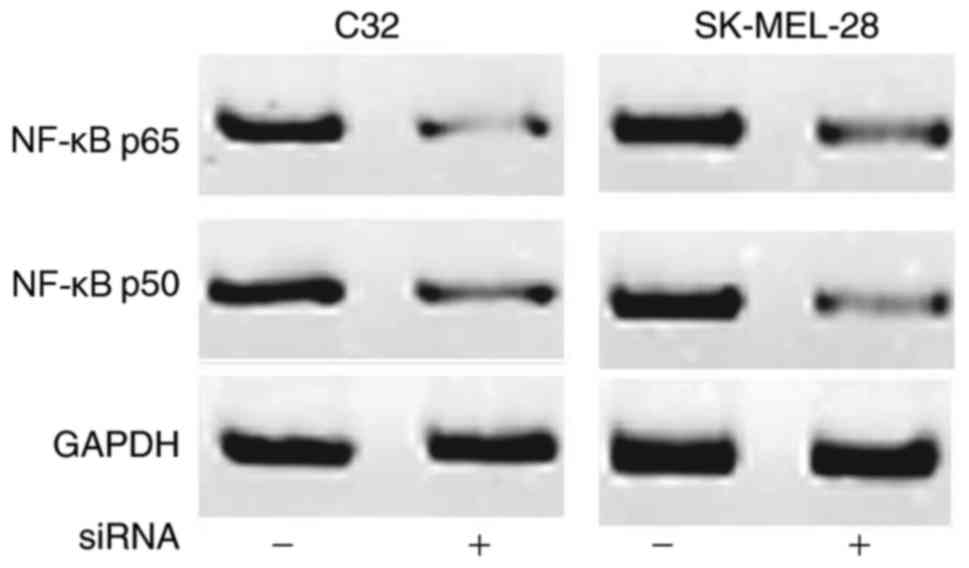
H19 inhibits the expression of NF-κB
p65 and p50
The expression of NF-κB p65 and p50 may be regulated
by certain lncRNAs. Therefore, the effect of H19 knockdown on the
expression levels of NF-κB p65 and p50 were investigated. As shown
in Fig. 5, compared with the cells
without siRNA transfection, expression levels of NF-κB p65 and p50
were decreased in cells following siRNA transfection. These data
suggested that H19 knockdown inhibited the NF-κB signaling pathway
by downregulating the expression levels of NF-κB p65 and p50.
Discussion
Previous studies have demonstrated that the
development of melanoma is accompanied by alterations in the
expression levels of various lncRNAs (13–15).
The expression levels of lncRNA urothelial cancer associated 1 and
metastasis associated lung adenocarcinoma transcript 1 have been
reported to be significantly increased along with the development
of melanoma, and the expression of these two lncRNAs were
significantly associated with the treatment outcomes and prognosis
of this disease (14). By
contrast, lncRNA growth arrest specific 5 (GAS5) may serve a role
as an anticancer factor in melanoma by regulating gelatinase A and
B, and the overexpression of lncRNA GAS5 significantly inhibited
the development of this disease (16). As an oncogene, H19 was upregulated
in patients with different types of cancers (17–19).
However, the expression pattern of H19 in patients with melanoma
remains unknown. In the present study, the expression levels of H19
were observed to be significantly higher in tumor tissues than in
the adjacent healthy tissues of 47 out of 49 patients with
melanoma. In addition, the expression of H19 was also revealed to
be significantly increased along with the stage development of
melanoma. These data suggested that H19 may be involved in the
development of melanoma.
Numerous studies have shown that lncRNA H19
participates in different human cancers primarily by promoting
tumor metastasis. In a study of gastric cancer, Zhou et al
(20) reported that H19 negatively
regulated the expression of microRNA (miR)-141, which in turn
promoted the proliferation and invasion of cancer cells. In another
study, H19 was demonstrated to serve pivotal roles in regulating
the invasion of glioma cells, and this function of H19 was
associated with miR-675 derived from H19 (21). A previous study on
cholangiocarcinoma reported that the expression levels of H19 were
upregulated by oxidative stress, and the increased expression level
of H19 promoted the migration and invasion of tumor cells by
targeting interleukin-6 and C-X-C chemokine receptor type 4
(22). In the present study, the
effects of H19 on the development of melanoma was investigated
using H19 knockdown via siRNA transfection. Proliferation, invasion
and migration abilities of melanoma cells were revealed to be
significantly reduced following siRNA transfection. These data
suggested that H19 served a role as an oncogene in the development
of melanoma, and the downregulation of H19 may inhibit the
progression of tumors by reducing the invasion of migration
abilities of tumor cells.
As a key factor in cancerogenesis, the NF-κB
signaling pathway has important functions in almost every step of
the initiation, development and progression of tumorigenesis
(23). It has been reported that
the NF-κB signaling pathway interacts with H19 to perform its
biological functions (24). It is
also well known that IκBα degradation mediated by the activation of
the PI3K/Akt signaling pathway may lead to the activation of the
NF-κB signaling pathway (25,26).
In the present study, no obvioust differences in the expression
levels of PI3K and Akt were identified between melanoma cells with
and without H19 knockdown. However, phosphorylation levels of PI3K
and Akt were decreased, and the expression levels of IκBα were
increased in melanoma cells with H19 knockdown than in cells
without H19 knockdown. These data suggested that H19 knockdown may
inactivate the PI3K/Akt signaling pathway, which in turn may lead
to the inactivation of the NF-κB signaling pathway.
In conclusion, H19 was upregulated in melanoma tumor
tissue. Downregulation of H19 inhibited the proliferation,
migration and invasion of melanoma cells, and the function of H19
in melanoma may be achieved by the inhibition of the NF-κB
signaling pathway via the inactivation of the PI3K/Akt signaling
pathway. However, the present study may be limited by the small
sample size. Thus, further studies with larger sample sizes are
required to further confirm these conclusions.
References
|
1
|
Hodis E, Watson IR, Kryukov GV, Arold ST,
Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C,
et al: A landscape of driver mutations in melanoma. Cell.
150:251–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allen DC: Malignant melanomaHistopathology
Reporting. Springer; London: pp. 207–216. 2013, View Article : Google Scholar
|
|
3
|
Stewart BW and Wild CP: World Cancer
Report 2014. IARC Publications; Lyon: 2017
|
|
4
|
Azoury SC and Lange JR: Epidemiology, risk
factors, prevention, and early detection of melanoma. Surg Clin
North Am. 94(945–62): vii2014. View Article : Google Scholar
|
|
5
|
Topalian SL, Sznol M, McDermott DF, Kluger
HM, Carvajal RD, Sharfman WH, Brahmer JR, Lawrence DP, Atkins MB,
Powderly JD, et al: Survival, durable tumor remission, and
long-term safety in patients with advanced melanoma receiving
nivolumab. J Clin Oncol. 32:1020–1030. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Atefi M, Avramis E, Lassen A, Wong DJ,
Robert L, Foulad D, Cerniglia M, Titz B, Chodon T, Graeber TG, et
al: Effects of MAPK and PI3K pathways on PD-L1 expression in
melanoma. Clin Cancer Res. 20:3446–3457. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pal HC, Sharma S, Strickland LR, Katiyar
SK, Ballestas ME, Athar M, Elmets CA and Afaq F: Fisetin inhibits
human melanoma cell invasion through promotion of mesenchymal to
epithelial transition and by targeting MAPK and NFκB signaling
pathways. PLoS One. 9:e863382014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perkel JM: Visiting ‘noncodarnia’.
Biotechniques. 54(301): 303–304. 2013.
|
|
9
|
Ning S, Zhang J, Wang P, Zhi H, Wang J,
Liu Y, Gao Y, Guo M, Yue M, Wang L and Li X: Lnc2Cancer: A manually
curated database of experimentally supported lncRNAs associated
with various human cancers. Nucleic Acids Res. 44:D980–D985. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. FEBS J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo M, Li Z, Wang W, Zeng Y, Liu Z and Qiu
J: Long non-coding RNA H19 increases bladder cancer metastasis by
associating with EZH2 and inhibiting E-cadherin expression. Cancer
Lett. 333:213–221. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khaitan D, Dinger ME, Mazar J, Crawford J,
Smith MA, Mattick JS and Perera RJ: The melanoma-upregulated long
noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer
Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tian Y, Zhang X, Hao Y, Fang Z and He Y:
Potential roles of abnormally expressed long noncoding RNA UCA1 and
Malat-1 in metastasis of melanoma. Melanoma Res. 24:335–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang L, Zhang W, Su B and Yu B: Long
noncoding RNA HOTAIR is associated with motility, invasion, and
metastatic potential of metastatic melanoma. Biomed Res Int.
2013:2510982013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen L, Yang H, Xiao Y, Tang X, Li Y, Han
Q, Fu J, Yang Y and Zhu Y: LncRNA GAS5 is a critical regulator of
metastasis phenotype of melanoma cells and inhibits tumor growth in
vivo. Onco Targets Ther. 9:4075–4087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF and Waye MM: The lncRNA
H19 promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu M, Chen Q, Liu X, Sun Q, Zhao X, Deng
R, Wang Y, Huang J, Xu M, Yan J and Yu J: lncRNA H19/miR-675 axis
represses prostate cancer metastasis by targeting TGFBI. FEBS J.
281:3766–3775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou X, Ye F, Yin C, Zhuang Y, Yue G and
Zhang G: The interaction between MiR-141 and lncRNA-H19 in
regulating cell proliferation and migration in gastric cancer. Cell
Physiol Biochem. 36:1440–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi Y, Wang Y, Luan W, Wang P, Tao T,
Zhang J, Qian J, Liu N and You Y: Long non-coding RNA H19 promotes
glioma cell invasion by deriving miR-675. PLoS One. 9:e862952014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang WT, Ye H, Wei PP, Han BW, He B, Chen
ZH and Chen YQ: LncRNAs H19 and HULC, activated by oxidative
stress, promote cell migration and invasion in cholangiocarcinoma
through a ceRNA manner. J Hematol Oncol. 9:1172016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hoesel B and Schmid JA: The complexity of
NF-κB signaling in inflammation and cancer. Mol Cancer. 12:862013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan JX: LncRNA H19 promotes
atherosclerosis by regulating MAPK and NF-kB signaling pathway. Eur
Rev Med Pharmacol Sci. 21:322–328. 2017.PubMed/NCBI
|
|
25
|
Hyam SR, Lee IA, Gu W, Kim KA, Jeong JJ,
Jang SE, Han MJ and Kim DH: Arctigenin ameliorates inflammation in
vitro and in vivo by inhibiting the PI3K/AKT pathway and polarizing
M1 macrophages to M2-like macrophages. Eur J Pharmacol. 708:21–29.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han W, Xiong Y, Li Y, Fang W, Ma Y, Liu L,
Li F and Zhu X: Anti-arthritic effects of clematichinenoside (AR-6)
on PI3K/Akt signaling pathway and TNF-α associated with
collagen-induced arthritis. Pharm Biol. 51:13–22. 2013. View Article : Google Scholar : PubMed/NCBI
|