Introduction
Osteosarcoma, a primary bone malignancy, most
commonly occurs during childhood and adolescence (1–3). In
~20% of patients, pulmonary metastasis is clinically detected,
which is the major cause of OS-associated mortality (4). At present, the standard treatment
strategy for patients with OS usually consists of surgery resection
and multi-drug chemotherapy, which has improved the cure rates of
patients with OS by ≥50% since the 1970s (5,6).
However, chemotherapy is associated with severe side effects and
there are currently no efficient biomarkers that allow the dosage
of chemotherapeutic drugs to be adapted to restrict dose-associated
toxicity. Therefore, the identification of novel and effective
therapeutic approaches to further improve the treatment for
patients with OS is required.
MicroRNAs (miRNAs/miRs), endogenous noncoding RNAs,
are 18–24 nucleotides in length and are involved in the regulation
of the expression of two-thirds of all human genes through binding
to mRNA 3′-untranslated regions (3′-UTRs) (7–9).
miRNAs are not only implicated in the maintenance of normal
cellular processes, but also act as tumor suppressors or promotors
by targeting specific genes. For example, it was demonstrated that
miR-21 knockdown inhibited the cell proliferation of OS cell lines,
while elevated miR-21 expression accelerated the proliferation of
OS cells and increased their resistance to cisplatin (10). miR-22 has been reported to be
downregulated in several tumor types, including lung, liver and
colorectal tumors, as well as in OS (11–13).
Xia et al (11)
demonstrated that miR-22 significantly attenuated OS, prostate
cancer, cervical cancer and lung cancer cell proliferation and
invasion. However, the mechanism by which miR-22 exerts these
antitumor effects and the association with chemotherapy regimens in
OS treatment remains unclear.
S100 calcium-binding protein A11 (S100A11), which is
also termed calgizzarin or S100C, belongs to the S100 protein
family, which are 10–12 kDa in molecular weight and able to bind
calcium by EF-hand motifs (14).
S100A11 is expressed ubiquitously in tissues and exhibits various
cellular functions, including cancer progression (15,16).
Previous studies have demonstrated that increased S100A11
expression is associated with tumor metastasis and a poor prognosis
in pancreatic, lung and colon cancers (17–19).
The current study aimed to investigate the role of
miR-22 in the carcinogenesis and progression of OS, and to
determine whether modulation of miR-22 expression may affect the
susceptibility of OS cells to the standard chemotherapy regimens in
OS treatment.
Materials and methods
Patients and specimens
A total of 4 male and 3 female patients with OS
(aged 12–22 years), and a control group consisting of 7 healthy
volunteers (4 male and 3 female, aged 11–22 years), were recruited
from February 2016 to February 2017 at the Second Affiliated
Hospital of Xi'an Jiaotong University (Xi'an, China). Prior to the
present study, none of the 7 patients with OS had received surgery,
preoperative chemotherapy or radiotherapy. Patient information is
presented in Table I. The ages of
patients and healthy volunteers were not significantly different.
OS cases were definite diagnoses based on accepted
clinicopathological and radiological criteria. The study was
authorized by the Ethics Committee of The Second Affiliated
Hospital of Xi'an Jiaotong University (Xi'an, China). All patients
and volunteers were anonymous and gave written informed consent.
Blood samples were serially collected from the venous blood of
patients and volunteers, which was centrifuged (4°C, 600 × g, for
10 min) and plasma was shipped on dry ice to a central repository
and stored at −80°C until further biochemical analysis.
 | Table I.Clinical information for patients
with OS included in the current study. |
Table I.
Clinical information for patients
with OS included in the current study.
| Sex | Age, years | Pathological
type |
|---|
| Female | 12 | Parosteal OS |
| Male | 16 | Conventional
OS |
| Male | 15 | Chondroblastic
OS |
| Female | 20 | Conventional
OS |
| Female | 17 | Conventional
OS |
| Male | 22 | Osteoblastoma
OS |
| Male | 18 | Telangiectatic
OS |
Cell culture
The MG-63 human osteosarcoma and hFOB 1.19 normal
osteoblast cell lines were purchased from the Cell Bank of Type
Culture Collection of the Chinese Academy of Sciences (Shanghai,
China). Cells were cultured at 37°C in 5% CO2 in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal
bovine serum (FBS; Bioind, Kibbutz Beit-Haemek, Israel), 100 U/ml
penicillin/streptomycin mixed solution and 2 mM glutamine (Beyotime
Institute of Biotechnology, Haimen, China).
Cell transfection
Hsa-miR-22 overexpression plasmid (pGCMV/EGFP) and
the negative control (NC, empty pGCMV/EGFP plasmid) were supplied
by Shanghai GenePharma Co., Ltd. (Shanghai, China). The sequence of
the miR-22 used within the plasmid is 5′-ACGGUACCCCGGCUGGGUGUU-3,
the scrambled sequence was used within the NC group is
5′-ACGGUACCCCGGCUAGGGUGUC-3. The human S100A11 gene was constructed
into a pcDNA3.1+HA empty vector (Invitrogen; Thermo Fisher
Scientific, Inc.) and the NC of the pcDNA3.1+HA-S100A11 group was
the pcDNA3.1+HA-empty group. Untransfected MG-63 cells were
employed as a blank control group. For transfection, MG-63 cells
were seeded in a 24-well plate at a density of 1×105/ml
for 24 h and were subsequently transfected with 100 nM plasmids
using Lipofectamine® 2000 (DNA/Lipofectamine®
2000=1/2.5; Invitrogen; Thermo Fisher Scientific, Inc.) and
incubated for 6 h at 37°C. After 6 h post-transfection, the medium
was changed to 2% FBS-DMEM with blasticidin (12 µg/ml) for 15 days
at 37°C. Cells where stable transfection was verified were stored
in liquid nitrogen for further experiments. Reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) and
western blotting were used to determine stable transfection as
described below.
Cell Counting Kit (CCK)-8 assay
The CCK-8 was supplied by Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany). To analyze cell proliferation, MG-63 stable
transfection cells (miR-22 and miR-22 + pcDNA3.1+HA-S100A11) were
seeded at a density of 8×103 cells/well in 96-well
plates and incubated for 12, 24, 48 or 72 h at 37°C. The optical
density (OD) value was measured at 480 nm using a Bio-Rad 2550 EIA
Reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
To analyze cell viability, MG-63 stable transfection
cells (miR-22 and miR-22 + pcDNA3.1+HA-S100A11) were seeded at a
density of 1×105 cells/well in 6-well plates and
incubated for 24 h at 37°C. Subsequently, 0, 0.1, 0.2 and 0.3 µM
cisplatin (20) (Sigma-Aldrich;
Merck KGaA) were added to the cells. After 48 h incubation at 37°C
with the CCK-8 reagent, the OD values were measured and relative
cell viability was obtained by setting corresponding untreated
groups as 100%. Half maximal inhibitory concentration
(IC50) values were calculated using GraphPad Prism 6.0
software (GraphPad Software, Inc., La Jolla, CA, USA).
Transwell migration assay
A Transwell migration assay was used to determine
cell migration ability. A total of ~103 MG-63 stable
transfection cells (miR-22 and miR-22 + pcDNA3.1+HA-S100A11) were
cultured with DMEM medium in the upper chamber of 24-well plates,
and the lower chamber was filled with 20% FBS-DMEM medium as a
chemoattractant. After incubation for 48 h at 37°C, cells that
migrated to the lower side of the inserts were fixed with 4%
paraformaldehyde at room temperature for 20 min and stained with
0.1% crystal violet (Beyotime Institute of Biotechnology) at room
temperature for 20 min, and then the cells from five independent,
randomly chosen visual fields were counted under a light microscope
(magnification, ×100; Nikon Corporation, Japan) for quantification
of cells.
RT-qPCR
Isolation of the total RNA from serum or cells was
performed using an RNeasy mini-kit (Qiagen GmbH, Hilden, Germany)
and single-strand cDNA was reverse-transcribed by using
ThermoScript One Step RT-PCR kit (Thermo Fisher Scientific, Inc.)
following the manufacturer's instructions (Invitrogen; Thermo
Fisher Scientific, Inc.). qPCR was performed on a real-time
quantified PCR system with a Rotor-Gene SYBR-Green PCR kit (400)
(Qiagen China Co., Ltd., Shanghai, China) on an ABI PRISM 7500
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with the following steps: Step 1: Predenaturation at 95°C for
5 min; step 2: 40 cycles of denaturation at 95°C for 10 sec, then
annealing and extension at 60°C for 34 sec. The experiments were
repeated three times. The relative miRNA or mRNA expression levels
were determined using the 2−ΔΔCq method (21) and normalized to U6 and GAPDH,
respectively. The reverse transcription primer for miR-22 was
5′-CCAGCTAAAGCTGCCAGTTGAAGAACTG-3′. The qPCR primers were as
follows: miR-22, 5′-AAGCUGCCAGUUGAAGAACUGU-3′ (forward) and
5′-AGUUCUUCAACUGGCAGCUUUU-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′
(forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse); S100A11,
5′-TCTCCAAGACAGAGTTCCTAAGC-3′ (forward) and
5′-TCATGCGGTCAAGGACAC-3′ (reverse); and GAPDH,
5′-GAAACCAGATCTCCACCGCA-3′ (forward) and 5′-GCGCCCAATACGACCAAATC-3′
(reverse).
Western blot analysis
Cells were harvested and extracted using
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology). Protein concentration was determined via a
Bradford's assay. Denatured cell lysates (30 µg) were resolved by
8% SDS-PAGE, transferred to nitrocellulose membranes. The membrane
was blocked with the blocking buffer for 30 min at room
temperature, and then incubated overnight with antibodies against
marker of proliferation Ki-67 (1:1,000; ab15580, Abcam, Cambridge,
UK), proliferating cell nuclear antigen (PCNA; 1:1,000; ab29;
Abcam), matrix metallopeptidase MMP2 (1:1,000; ab37150; Abcam) and
GAPDH (1:1,000, ab8245; Abcam). Tris-buffered saline with 0.05%
Tween-20 was used to wash the membranes three times for 10 min.
Subsequently, the membranes were incubated with a secondary
antibody, rabbit horseradish peroxidase-conjugated anti-goat
immunoglobulin G (1:2,000; sc2771; Santa Cruz Biotechnology Inc.,
Dallas, TX, USA) for 1 h at room temperature. A chemiluminescence
system (ECL Plus; Odyssey Infrared Imaging system; LI-COR
Biosciences, Lincoln, NE, USA) was used for visualization. The
bands were scanned on a gel imaging and analysis system and
analyzed by Quantity One 4.4 (Bio-Rad Laboratories, Inc.).
ELISA
A Quantikine ELISA kit (KA2413; R&D Systems,
Inc., Minneapolis, MN, USA) was used to detect the expression of
S100A11 protein. Twenty-four hours post-transfection, the cell
supernatants of different MG-63 cell treatment groups or serum from
patients were collected and analyzed according to the
manufacturer's protocol.
Reporter gene transfection and
luciferase assays
TargetScan (http://www.targetscan.org/vert_71/) and miRanda
(http://34.236.212.39/microrna/home.do) databases were
used to predict the potential miR-22 binding sites of S100A11. In
the luciferase reporter assay, 1×106 MG-63 cells/well
that were stably transfected with miR-22 were further transfected
with 30 ng wild-type or mutant 3′-UTR of S100A11 via
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, cells were collected and luciferase
activity was measured by a Dual-Luciferase Reporter Assay System
(Promega Corporation, Madison, WI, USA) according to the
manufacturer's protocol.
The S100A11 3′-UTR was cloned into a luciferase
pMIR-REPORT vector (Shanghai GeneChem Co., Ltd., Shanghai, China)
and mutations were introduced in potential miR-22 binding sites.
Values were double normalized to firefly luciferase activity.
PRL-SV40 vector carrying the Renilla luciferase gene
(Promega Corporation) was used as an internal control for
transfection efficiency. The sequences included: S100A11 3′-UTR,
5′-UCUGAGUUCUUGAAGCAUUUCAA-3′, hsa-miR-22,
3′-GAUCACCAGGAUUUGUAAAGUG-5′ and S100A11 3′-UTR,
5′-UCUGAGUUCUUGAAGAUCGAUCA-3′.
Statistical analysis
Experiments were performed at least three
independent times and data are presented as the mean ± standard
deviation. Statistical significance was evaluated by one-way
analysis of variance followed by a Tukey's post hoc test or
two-tailed Student's t-tests using SPSS 20.0 software (IBM Corp.,
Armonk, NY, USA) and GraphPad Prism 6.0 software. P<0.05 was
considered to indicate a statistically significant difference.
Results
Endogenous expression of miR-22 in
patients with OS and an OS cell line
The expression of miR-22 from the serum of patients
with OS and healthy controls, and in vitro cultured OS and
normal osteoblast cells, was investigated by RT-qPCR. As
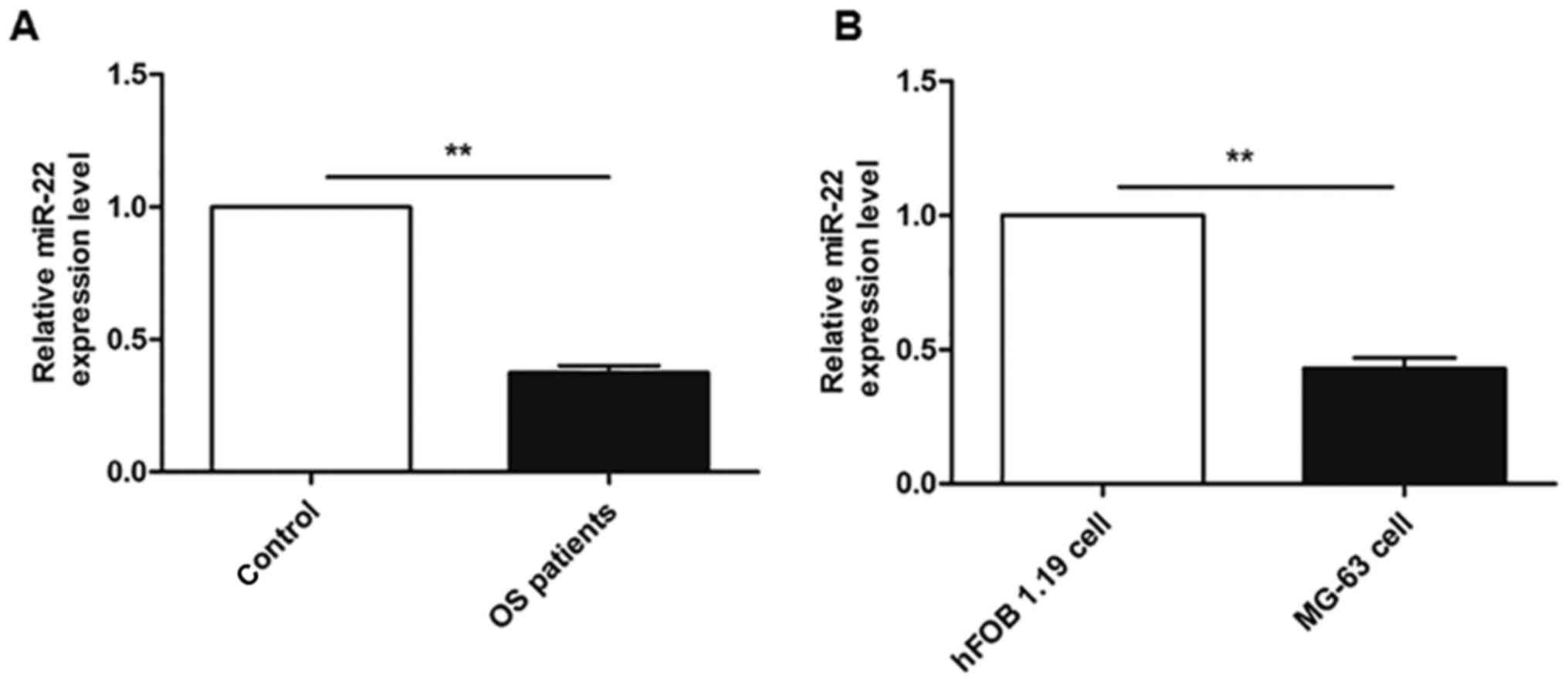
demonstrated in Fig. 1A, the serum
expression of miR-22 was lower in samples from patients with OS
compared with healthy volunteers (P<0.05). Furthermore, Fig. 1B also indicated that the miR-22
levels were ~two fold lower in the MG-63 OS cell line compared with
the hFOB 1.19 osteoblast cell line (P<0.01).
miR-22 overexpression inhibits the
proliferation and migration of MG-63 Cells
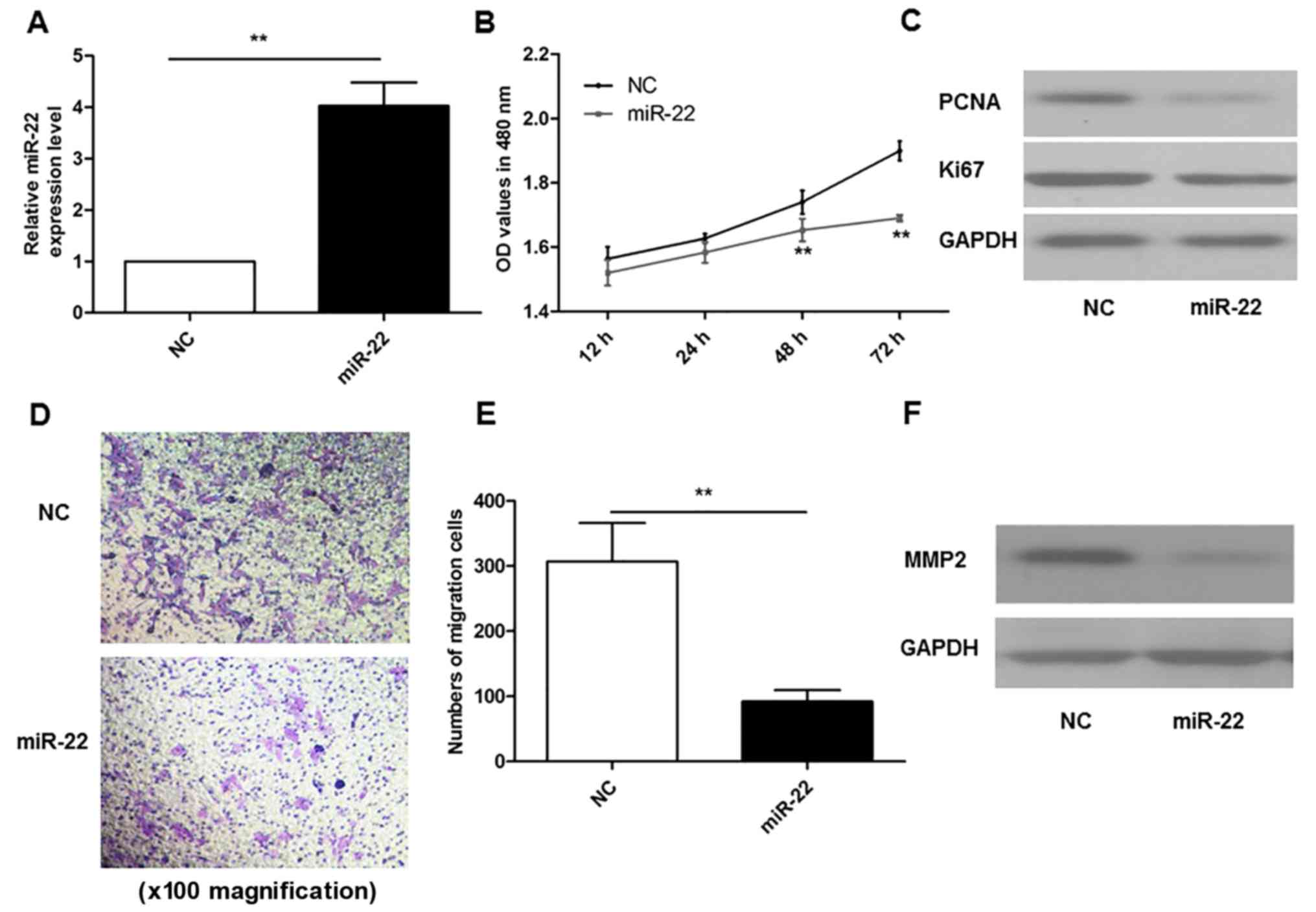
As demonstrated in Fig.
2A, MG-63 cells were successfully transfected with miR-22
overexpression plasmids, and qPCR results indicated that the levels
of miR-22 were ~4-fold higher than those in the NC group
(P<0.01). A CCK-8 assay was performed to investigate cell
proliferation. The results demonstrated that overexpression of
miR-22 markedly inhibited the proliferation of MG-63 cells at 48
and 72 h, compared with the NC group (P<0.01; Fig. 2B). In addition, miR-22
overexpression also reduced the protein expression levels of Ki67
and PCNA compared with the NC group (Fig. 2C).
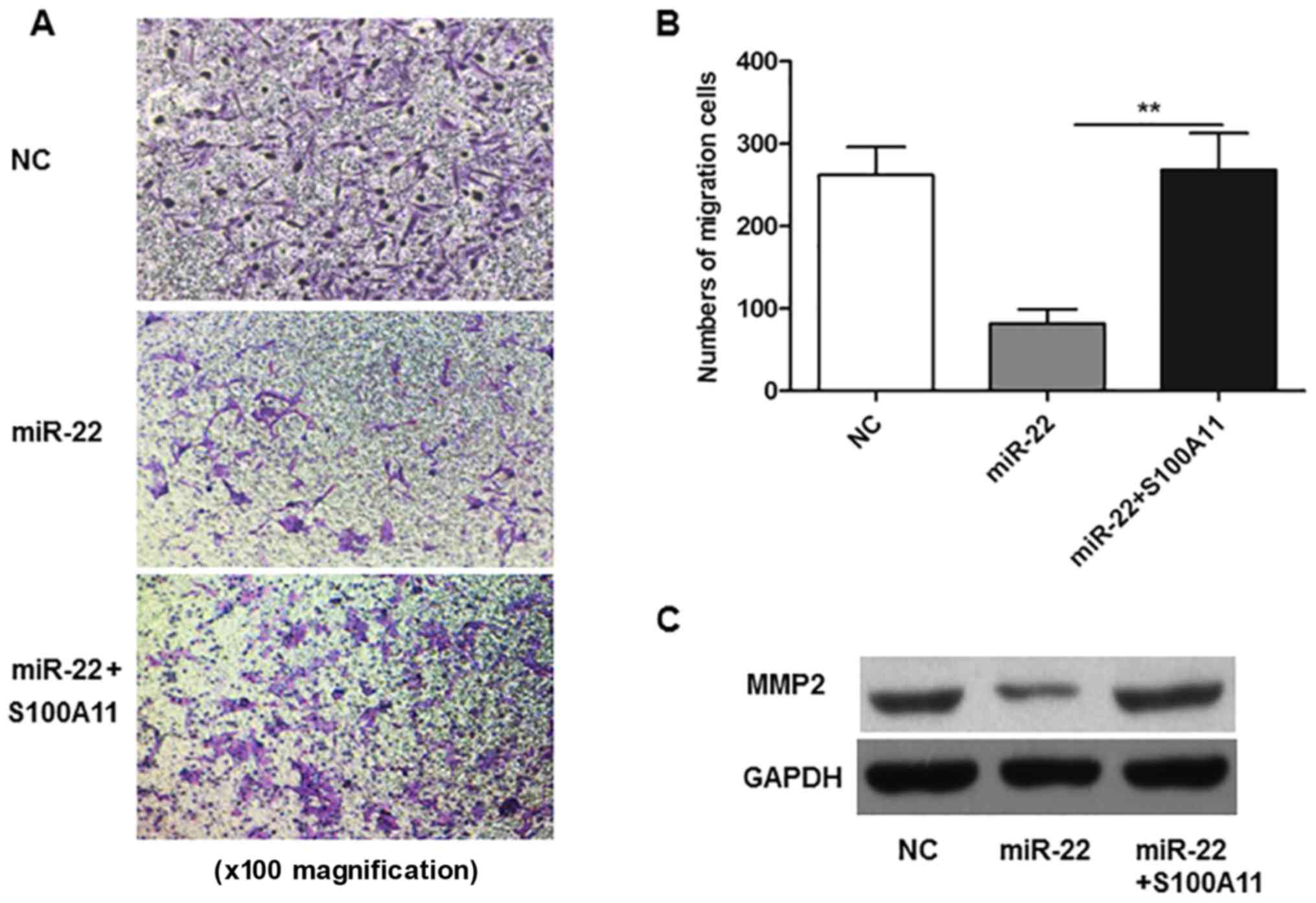
A Transwell migration assay was performed to
determine the effects of miR-22 on cell migration ability. The
results demonstrated that overexpression of miR-22 resulted in the
inhibition of MG-63 cell migration, compared with the NC group
(P<0.01; Fig. 2D and E). In
accordance with migration results, miR-22 overexpression also
reduced the protein expression levels of MMP2 compared with the NC
group (Fig. 2F).
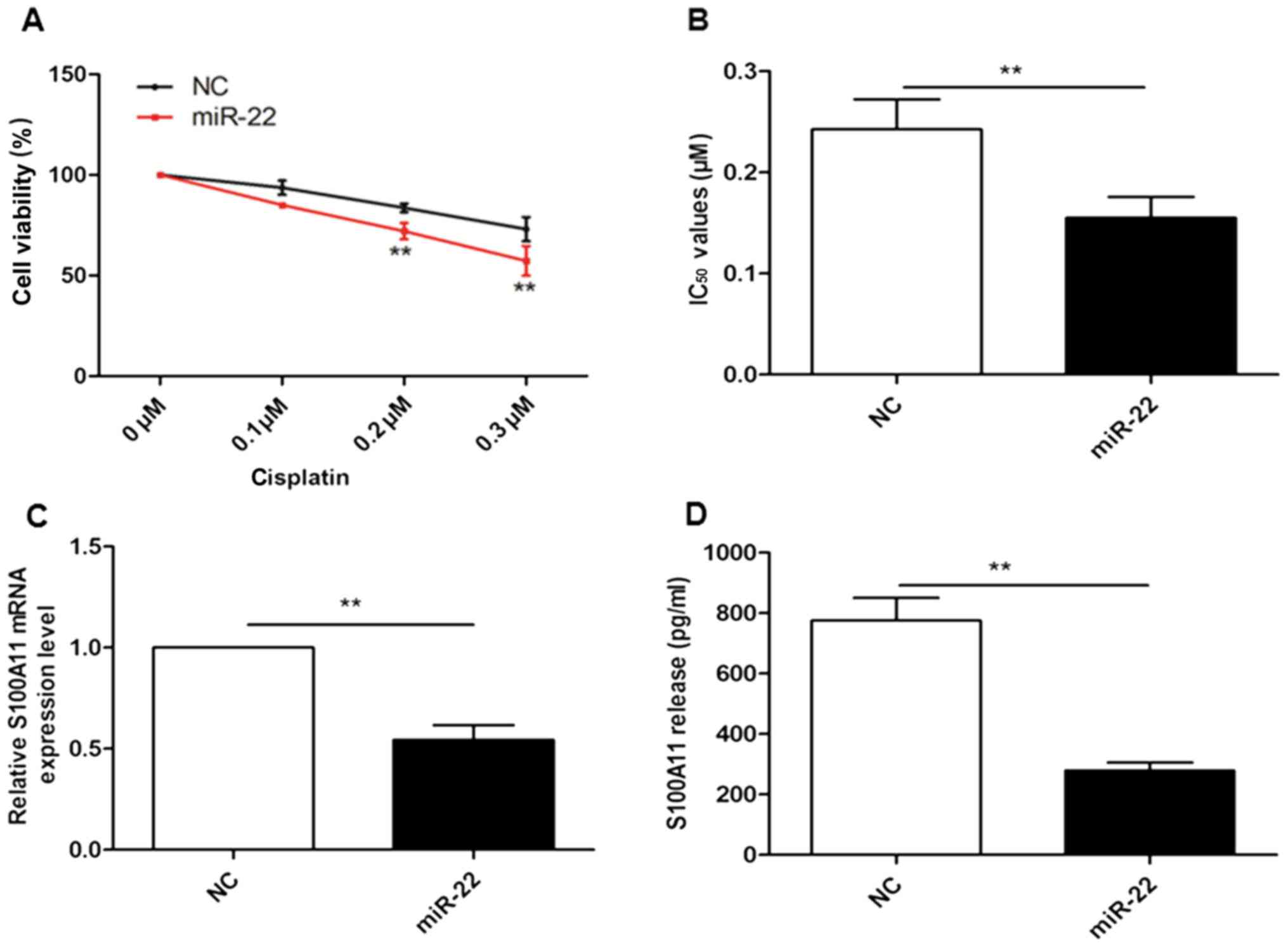
miR-22 overexpression sensitizes MG-63
cells to cisplatin treatment and reduces the expression of
S100A11
As cisplatin is frequently used clinically for OS
treatment (22), the current study
assessed whether modulated miR-22 levels in MG-63 cells may affect
the sensitivity of cells to cisplatin chemotherapy. The results in
Fig. 3A and B demonstrate that the
cell viability of MG-63 cells that overexpress miR-22 was decreased
in a dose-dependent manner for cisplatin treatment; overexpression
of miR-22 increased the sensitivity of MG-63 to cisplatin and
resulted in a significant reduction in the calculated
IC50 value, compared with the NC group (P<0.01). The
mRNA expression of S100A11 in transfected cells, and the levels of
S100A11 in the cell culture medium, was also investigated. The
results in Fig. 3C and D
demonstrated that reduced mRNA levels of S100A11, and reduced
release of S100A11 protein into the culture medium, were observed
in MG-63 cells with miR-22 overexpression, compared with the NC
group (P<0.01).
S100A11 is a direct target of
miR-22
The present study employed bioinformatics tools to
predict the putative miRNAs targeting S100A11, and identified that
miR-22 possesses the conserved binding sites (Fig. 4A). Subsequently, a luciferase
reporter gene assay was performed to verify whether miR-22 directly
binds to the 3′-UTR of S100A11 in OS. The results in Fig. 4B indicate that co-transfection of
wild-type S100A11 3′-UTR with miR-22 overexpression reduced the
relative luciferase activity by ~50%, compared with NC
(miR-NC)+wild-type 3′-UTR of S100A11 group. Furthermore,
co-transfection of cells with the mutant 3′-UTR of S100A11 and
miR-22 overexpression reversed this suppressive effect of miR-22
(P<0.01). Additionally, the results in Fig. 4C and D indicate that the release of
S100A11 into the serum or cell culture medium of patients with OS
or MG-63 cells was significantly increased compared with healthy
volunteers or hFOB 1.19 normal osteoblast cells, respectively
(P<0.01).
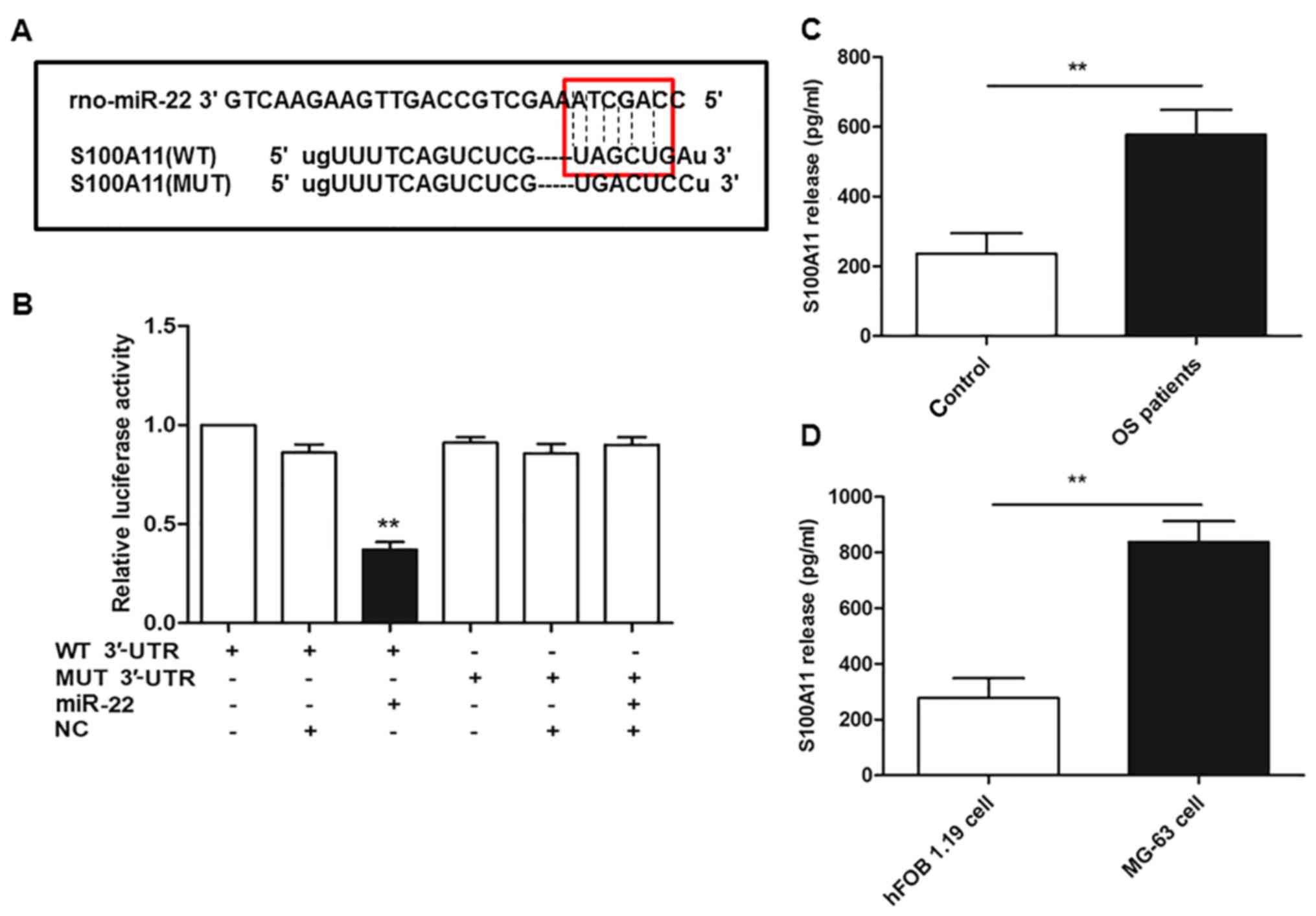 | Figure 4.S100A11 is a direct target of miR-22.
(A) Sequences of WT S100A11 3′-UTR and MUT S100A11 3′-UTR, and the
ability of miR-22 to bind to the WT sequence. (B) Relative
luciferase activity in MG-63 cells following co-transfection of
miR-22 or NC and the WT or MUT S100A11 3′-UTR. Levels of S100A11 in
(C) the serum of patients with OS and healthy volunteers, and (D)
the cell culture medium of MG-63 OS cells and hFOB 1.19 normal
osteoblasts, were determined by ELISA. For part B, **P<0.01 vs.
MG-63 cells co-transfected with NC and the WT 3′UTR of S100A11; for
parts C and D, **P<0.01, as indicated. S100A11, S100
calcium-binding protein A11; miR, microRNA; WT, wild-type; UTR,
untranslated region; MUT, mutant; NC, negative control; OS,
osteosarcoma. |
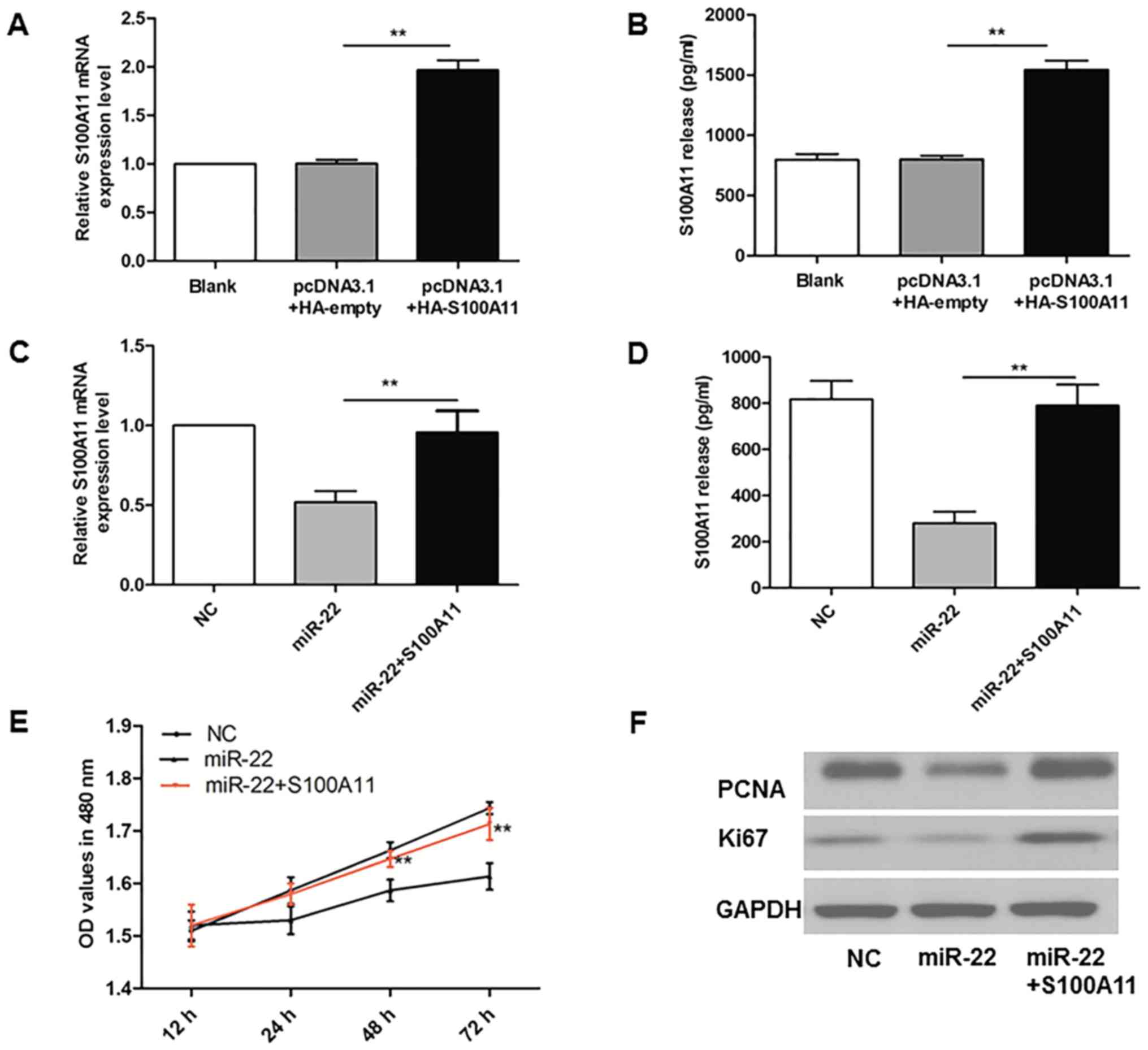
Successful overexpression of S100A11
in MG-63 cells
As demonstrated in Fig.
5A and B, MG-63 cells were successfully transfected with
pcDNA3.1+HA-S100A11, and RT-qPCR and ELISA results demonstrated
that the levels of S100A11 were ~2-fold of those in the
pcDNA3.1+HA-empty group (P<0.01).
S100A11 reverses the reduction in
proliferation induced by overexpression of miR-22 in MG-63
cells
To investigate the functional mediator role of
S100A11 for miR-22 in OS cells, the present study overexpressed
S100A11 in MG-63 cells in combination with miR-22 overexpression.
The results in Fig. 5C and D
demonstrate that the levels of S100A11 mRNA in MG-63 cells, and
S100A11 levels in the cell culture medium, were increased in the
miR-22 overexpression + S100A11 overexpression group compared with
cells where only miR-22 was overexpressed (P<0.01). As indicated
in Fig. 5E, miR-22 overexpression
led to reduced MG-63 cell proliferation compared with the NC group,
while simultaneous S100A11 overexpression reversed the
anti-proliferative effect of miR-22 (P<0.01). In addition,
miR-22-induced reductions in the protein expression of PCNA and
Ki67 were reversed in co-transfected cells (Fig. 5F).
S100A11 reverses miR-22-induced
reductions in the migratory ability of MG-63 cells
Results in Fig. 6A and
B demonstrate that S100A11 overexpression reversed the
inhibitory effect of miR-22 on MG-63 cell migration (P<0.01). In
addition, the expression of MMP2 in co-transfected cells was
increased compared with cells that only overexpressed miR-22
(Fig. 6C).
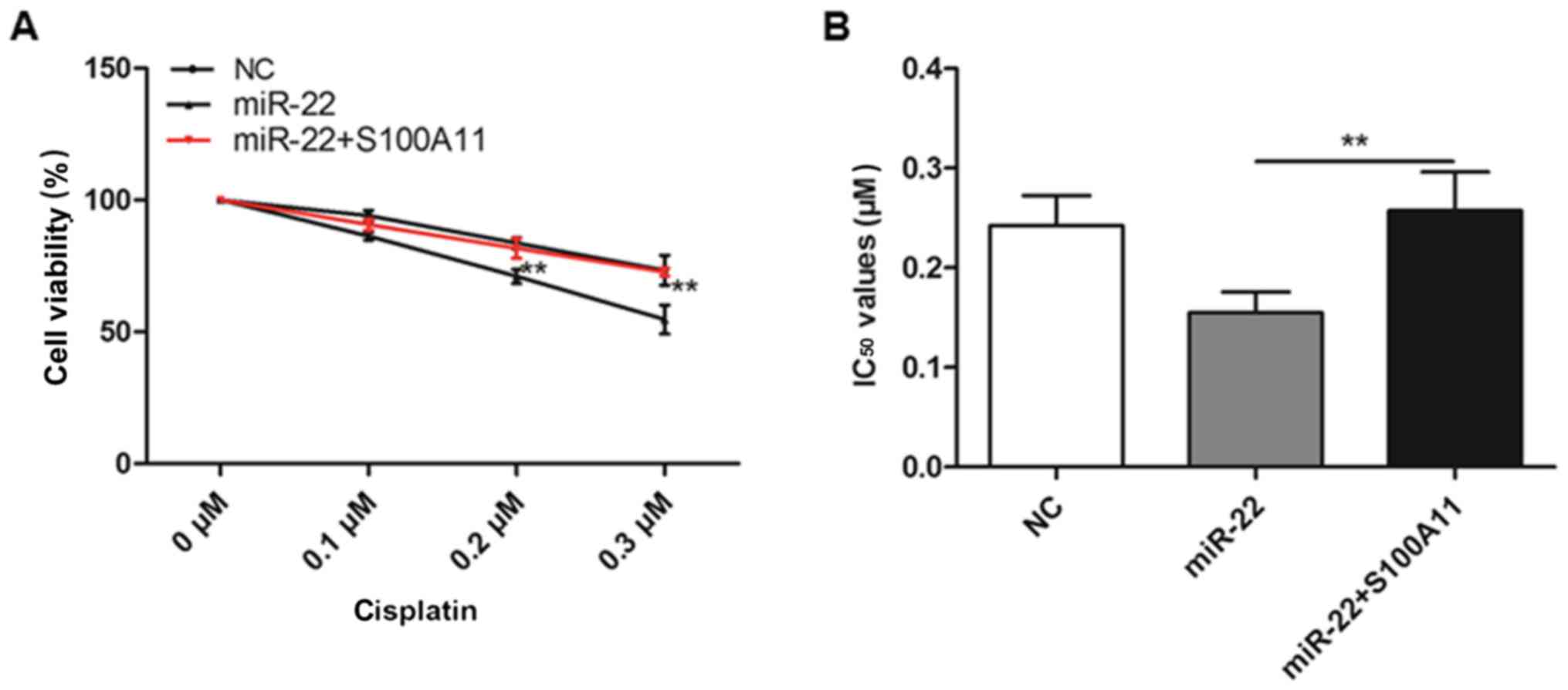
S100A11 desensitizes MG-63 cells to
cisplatin treatment
As demonstrated in Fig.
7, the cell viability of MG-63 cells that were co-transfected
with miR-22 and S100A11 decreased in a dose-dependent manner for
cisplatin treatment; S100A11 desensitized MG-63 cells to cisplatin
and resulted in a significant increase in the calculated
IC50 value, compared with cells that only overexpressed
miR-22 (P<0.01).
Discussion
The present study observed that the expression
levels of miR-22 were significantly reduced in patients with OS and
the MG-63 OS cell line compared with healthy volunteers and the
hFOB 1.19 normal osteoblast cell line, respectively. In addition,
the expression of miR-22 was inversely associated with S100A11
levels, which was significantly upregulated in patients with OS and
MG-63 cells, compared with the respective controls. In vitro
results demonstrated that overexpression of miR-22 inhibited the
proliferation and migratory ability, and increased the sensitivity
to cisplatin treatment, in MG-63 cells. Notably, the present study
identified S100A11 as a direct target gene of miR-22 in MG-63
cells. Further experiments demonstrated that the alterations in
proliferation, migration and chemosensitivity were partially
abolished by overexpression of the S100A11 gene. Therefore, the
current study revealed that S100A11 is a specific target gene for
miR-22 and that this miRNA may function as a tumor suppressor in OS
through enhancing the chemosensitivity of cells to cisplatin.
OS often occurs in children and adolescents
(23); the 5-year survival rate
for OS remains <70% for OS cases that are most resistant to
therapies (24). miRNAs have been
implicated in the development of chemosensitivity and
chemoresistance, which may aid the understanding of the molecular
basis of this aggressive bone cancer (25,26).
Recently, scientists have reported that several miRNAs may be
involved in the sensitivity of cancers to chemotherapeutic agents.
Vanas et al (10)
demonstrated that reduced miR-21 expression inhibited the
proliferation of OS cells lines, while elevated miR-21 expression
increased the proliferation and overexpression of miR-21 increased
the resistance of cells to cisplatin treatment. Furthermore, Geng
et al (20) reported an
inverse association between miR-224 and Rac family small GTPase 1
(Rac1); miR-224 inhibited the proliferation of OS cell lines, as
well as their metastasis and sensitivity to cisplatin treatment,
through direct targeting of Rac1. In addition, miR-138 was also
reported to be involved in cisplatin resistance. Research
demonstrated that miR-138 was reduced in OS tissue and restoring
miR-138 expression markedly inhibited cell proliferation and
invasion, and increased sensitivity to cisplatin (27). These findings confirmed that miRNAs
affect the sensitivity of cancers to chemotherapeutic agents.
However, to the best of our knowledge, no previous studies have
described the effect of miR-22 in the modulation of sensitivity to
cisplatin in OS. The present study, the present study demonstrated
for the first time that miR-22 may be a promising therapeutic
target and may have potential for use in combination with
chemotherapeutic agents.
S100A11, a member of the S100 protein family, is
widely expressed in human tissues. Increased levels of S100A11 have
been reported in various cancer types and was closely associated
with tumor progression (28,29).
S100A11 has been proposed to exhibit pleiotropic effects in various
biological contexts, including in nuclear and cytoplasmic
compartments, and S100A11 has been reported to act as a growth
inhibitor in normal keratinocytes (30,31).
Furthermore, when the protein was secreted in dimerized form and
bound to receptor for advanced glycation end products, S100A11
induced the growth stimulation in normal keratinocytes and various
cancer cells (32). At present,
S100A11 in OS has not been fully characterized, and the present
study aimed to investigate the role of this protein in OS
progression. The results of the current study indicated that
S100A11 was highly upregulated in the serum of patients with OS and
the culture medium of the MG-63 OS cell line compared with
respective controls, and further research identified S100A11 as a
specific target gene of miR-22. These novel findings may aid the
understanding of the extracellular role of dimerized S100A11 in the
OS cell microenvironment and its association with cancer
progression.
In conclusion, the present study demonstrated that
levels of miR-22 were significantly reduced in patients with and
the MG-63 OS cell line, while the expression of S100A11 was
inversely associated with miR-22 levels. Overexpression of miR-22
inhibited the proliferation and migration, and increased the
sensitivity to cisplatin treatment, in MG-63 cells. However,
overexpression of S100A11 partially abolished the alterations in
proliferation, migration and chemosensitivity that were induced by
miR-22. Furthermore, the present study identified S100A11 as a
direct target gene of miR-22 in MG-63 cells. The current study
demonstrated for the first time that miR-22 may be a promising
therapeutic target and may have potential in combination with
chemotherapeutic agents for OS.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XH designed the experiment, XZho conducted the
experimental and statistical work and wrote the paper. DN, XZha and
ZG helped XZho conduct the cell experiments.
Ethics approval and consent to
participate
The present study was authorized by the Ethics
Committee of The Second Affiliated Hospital of Xi'an Jiaotong
University (Xi'an, China). All patients and volunteers were
anonymous and provided written informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Messerschmitt PJ, Garcia RM, Abdul-Karim
FW, Greenfield EM and Getty PJ: Osteosarcoma. J Am Acad OrthopSurg.
17:515–527. 2009. View Article : Google Scholar
|
|
2
|
Bousquet M, Noirot C, Accadbled F, Sales
de Gauzy J, Castex MP, Brousset P and Gomez-Brouchet A: Whole-exome
sequencing in osteosarcoma reveals important heterogeneity of
genetic alterations. Ann Oncol. 27:738–744. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mirabello L, Troisi RJ and Savage SA:
International osteosarcoma incidence patterns in children and
adolescents, middle ages and elderly persons. Int J Cancer.
125:229–234. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Miller BJ, Cram P, Lynch CF and Buckwalter
JA: Risk factors for metastatic disease at presentation with
osteosarcoma: An analysis of the SEER database. J Bone Joint Surg
Am. 95:e892013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ferrari S and Palmerini E: Adjuvant and
neoadjuvant combination chemotherapy for osteogenic sarcoma. Curr
Opin Oncol. 19:341–346. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Allison DC, Carney SC, Ahlmann ER,
Hendifar A, Chawla S, Fedenko A, Angeles C and Menendez LR: A
meta-analysis of osteosarcoma outcomes in the modern medical era.
Sarcoma. 2012:7048722012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee RC and Ambros V: An extensive class of
small RNAs in Caenorhabditis elegans. Science. 294:862–864. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Irwandi RA and Vacharaksa A: The role of
microRNA in periodontal tissue: A review of the literature. Arch
Oral Biol. 72:66–74. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vanas V, Haigl B, Stockhammer V and
Sutterlüty-Fall H: MicroRNA-21 increases proliferation and
cisplatin sensitivity of osteosarcoma-derived cells. PLoS One.
11:e01610232016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia SS, Zhang GJ, Liu ZL, Tian HP, He Y,
Meng CY, Li LF, Wang ZW and Zhou T: MicroRNA-22 suppresses the
growth, migration and invasion of colorectal cancer cells through a
Sp1 negative feedback loop. Oncotarget. 8:36266–36278. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Qiao DD, Yang J, Lei XF, Mi GL, Li SL, Li
K, Xu CQ and Yang HL: Expression of microRNA-122 and microRNA-22 in
HBV-related liver cancer and the correlation with clinical
features. Eur Rev Med Pharmacol Sci. 21:742–747. 2017.PubMed/NCBI
|
|
13
|
Chen J, Wu FX, Luo HL, Liu JJ, Luo T, Bai
T, Li LQ and Fan XH: Berberine upregulates miR-22-3p to suppress
hepatocellular carcinoma cell proliferation by targeting Sp1. Am J
Transl Res. 8:4932–4941. 2016.PubMed/NCBI
|
|
14
|
Shankar J, Messenberg A, Chan J, Underhill
TM, Foster LJ and Nabi IR: Pseudopodial actin dynamics control
epithelial-mesenchymal transition in metastatic cancer cells.
Cancer Res. 70:3780–3790. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jaiswal JK, Lauritzen SP, Scheffer L,
Sakaguchi M, Bunkenborg J, Simon SM, Kallunki T, Jäättelä M and
Nylandsted J: S100A11 is required for efficient plasma membrane
repair and survival of invasive cancer cells. Nat Commun.
5:37952014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Woo T, Okudela K, Mitsui H, Tajiri M, Rino
Y, Ohashi K and Masuda M: Up-Regulation of S100A11 in lung
adenocarcinoma-its potential relationship with cancer progression.
PLoS One. 10:e01426422015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao MB, Jiang F, Ni WK, Chen BY, Lu CH,
Li XY and Ni RZ: High expression of S100A11 in pancreatic
adenocarcinoma is an unfavorable prognostic marker. Med Oncol.
29:1886–1891. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tian T, Hao J, Xu A, Hao J, Luo C, Liu C,
Huang L, Xiao X and He D: Determination of metastasis-associated
proteins in non-small cell lung cancer by comparative proteomic
analysis. Cancer Sci. 98:1265–1274. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Melle C, Ernst G, Schimmel B, Bleul A and
von Eggeling F: Colon-derived liver metastasis, colorectal
carcinoma, and hepatocellular carcinoma can be discriminated by the
Ca2+-binding proteins S100A6 and S100A11. PLoS One.
3:e37672008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Geng S, Gu L, Ju F, Zhang H, Wang Y, Tang
H, Bi Z and Yang C: MicroRNA-224 promotes the sensitivity of
osteosarcoma cells to cisplatin by targeting Rac1. J Cell Mol Med.
20:1611–1619. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gorlick R: Current concepts on the
molecular biology of osteosarcoma. Cancer Treat. Res. 152:467–478.
2009.
|
|
23
|
Bacci G, Briccoli A, Rocca M, Ferrari S,
Donati D, Longhi A, Bertoni F, Bacchini P, Giacomini S, Forni C, et
al: Neoadjuvant chemotherapy for osteosarcoma of the extremities
with metastases at presentation: Recent experience at the Rizzoli
Institute in 57 patients treated with cisplatin, doxorubicin, and a
high dose of methotrexate and ifosfamide. Ann Oncol. 14:1126–1134.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu X, Li Z, Yu J, Chan MT and Wu WK:
MicroRNAs predict and modulate responses to chemotherapy in
colorectal cancer. Cell Prolif. 48:503–510. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song B, Wang Y, Xi Y, Kudo K, Bruheim S,
Botchkina GI, Gavin E, Wan Y, Formentini A, Kornmann M, et al:
Mechanism of chemoresistance mediated by miR-140 in human
osteosarcoma and colon cancer cells. Oncogene. 28:4065–4074. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martinez-Sanchez A, Dudek KA and Murphy
CL: Regulation of human chondrocyte function through direct
inhibition of cartilage master regulator SOX9 by microRNA-145
(miRNA-145). J Biol Chem. 287:916–924. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhu Z, Tang J, Wang J, Duan G, Zhou L and
Zhou X: MiR-138 acts as a tumor suppressor by targeting EZH2
and enhances cisplatin-induced apoptosis in osteosarcoma cells.
PLoS One. 11:e01500262016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hao J, Wang K, Yue Y, Tian T, Xu A, Hao J,
Xiao X and He D: Selective expression of S100A11 in lung cancer and
its role in regulating proliferation of adenocarcinomas cells. Mol
Cell Biochem. 359:323–332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang LN, Tong SW, Hu HD, Ye F, Li SL, Ren
H, Zhang DZ, Xiang R and Yang YX: Quantitative proteome analysis of
ovarian cancer tissues using a iTRAQ approach. J Cell Biochem.
113:3762–3772. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sakaguchi M, Murata H, Sonegawa H,
Sakaguchi Y, Futami J, Kitazoe M, Yamada H and Huh NH: Truncation
of Annexin A1 is a regulatory lever for linking epidermal growth
factor signaling with cytosolic phospholipase A2 in
normal and malignant squamous epithelial cells. J Biol Chem.
282:35679–35686. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sakaguchi M, Miyazaki M, Takaishi M,
Sakaguchi Y, Makino E, Kataoka N, Yamada H, Namba M and Huh NH:
S100C/A11 is a key mediator of Ca2+-induced growth
inhibition of human epidermal keratinocytes. J Cell Biol.
163:825–835. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sakaguchi M, Sonegawa H, Murata H, Kitazoe
M, Futami J, Kataoka K, Yamada H and Huh NH: S100A11, an dual
mediator for growth regulation of human keratinocytes. Mol Biol
Cell. 19:78–85. 2008. View Article : Google Scholar : PubMed/NCBI
|