Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies in the world (1). Although great progress has been made
in the diagnosis of HCC, the outcome for HCC still remains very
poor (1). Therefore, the
identification of novel therapeutic targets in HCC is
important.
The aberrant activation of Wnt/β-catenin signaling
is frequently observed in hepatocellular carcinoma (HCC) (1,2).
Previous studies have demonstrated that activation of Wnt/β-catenin
signaling promotes the growth, migration, invasion and metastasis
of HCC cells (3,4). Therefore, improving understanding of
Wnt/β-catenin signaling regulation may improve treatment of
HCC.
The expression of β-catenin, a key molecule involved
in the Wnt/β-catenin signaling pathway, is tightly regulated in
quiescent cells (5). Serine and
threonine residues in the N-terminal of β-catenin are
phosphorylated sequentially by cyclin-dependent kinase inhibitor
and glycogen synthase kinase 3β, which leads to the degradation of
β-catenin via the ubiquitin-proteasome system (5). The disassociation of the β-catenin
destruction complex following the binding of Wnt ligands to the
low-density lipoprotein receptor related protein 5/6 receptor
causes the accumulation of β-catenin in the cytoplasm and the
subsequent translocation of β-catenin to the nucleus. In the
nucleus, β-catenin forms a complex with transcription factor 4
(TCF7L24) and regulates the expression of multiple genes, including
c-Myc, cyclin D1 and Snail (6).
Improved understanding of β-catenin/TCF7L2 complex regulation may
help in the development of a novel therapeutic strategy to treat
HCC.
Transcription termination factor (TTF)-1 interacting
protein 5 (TIP5) is a component of the chromatin remodeling complex
(7,8). Human TIP5 is a 211 kDa protein
containing several protein and DNA interaction domains (8). Previous studies have revealed that
TIP5 mediates the epigenetic regulation of recombinant RNA genes
(9,10). The function of TIP5 in cancer is
currently poorly understood. TIP5 is involved in epigenetic
alterations in prostate cancer and its overexpression predicts
disease recurrence (11). However,
the role that TIP5 serves in the progression of HCC remains
unknown.
In the present study, it was determined whether TIP5
was a binding partner of TCF7L2. In addition, the functions of TIP5
in the progression of HCC were investigated.
Materials and methods
Cell culture and patient samples
Normal human liver L02 and HCC 7404, MHCC97, Hep3B
and Huh7 cell lines were obtained from the American Type Culture
Collection (Manassas, VA, USA). Cells were cultured in Dulbecco's
Modified Eagle medium (DMEM, Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(Thermo Fisher Scientific, Inc.), penicillin (100 U/ml) and
streptomycin (100 µg/ml). Cell cultures were maintained in a
humidified incubator containing 5% CO2 at 37°C for
passages 1–10. Patients with HCC (n=53; 23 females, 30 males; age,
47–72) between January 2015-December 2016 were enrolled in the
present study. The collection of samples from patients was approved
by the Institutional Ethics Committee of Cangnan People's Hospital
(Wenzhou, China)and written informed consent was received from all
patients in the present study. HCC samples were collected
immediately following hepatectomy. The pathology was confirmed by
the pathologist.
TIP5 knockdown in HCC cell lines
The lentivirus to knock down the expression of TIP5
in 7404 and Huh7 cells was purchased from GeneChem (Shanghai,
China). Cells were incubated with the viral particles for 24 h
(MOI=1) at 37°C and the green fluorescent protein (GFP)-positive
cells were sorted by flow cytometry at 4°C and analyzed with WinMDI
software (version 2.9; the Scripps Research Institute, La Jolla,
CA, USA). The GFP was included in the purchased lentivirus. The
following sequences were used: Small hairpin (sh) TIP5 1#,
5′-AAGAGTTCTGGGCCAACGG-3′; sh TIP5 2#, 5′-AAGAGCAGCCAGAACTGA-3′;
and shcontrol (con), 5′-AAGATTTCGGGGGGCCCAA-3′. Experiments were
subsequently performed one week post-transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) and cDNA was synthesized using the
TaqMan microRNA reverse transcription kit (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) according to the manufacturer's
instructions. qPCR was performed with SYBR-Green (Takara
Biotechnology Co., Ltd., Dalian, China) in triplicates in a total
volume of 8 µl (4 µl master mix and 4 µl cDNA) using a 7900
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with the following thermocycling conditions: 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. The
relative expression of target genes was normalized to 18S (Applied
Biosystems, Thermo Fisher Scientific, Inc.) and were calculated
using the 2−ΔΔCq method (4). The primer sequences used were as
follows: TIP5 forward, 5′-ACTGTATCTCACACTACTAC-3′ and reverse,
5′-GAAGGTTAGTGTTATGACTT-3′. 18S forward, 5′-GAGAAACGGCTACCACATCC-3′
and reverse, 5′-CACCAGACTTGCCCTCCA-3′.
Protein extraction and western blot
analysis
HCC cells (normal human liver cell line L02 and HCC
cell lines 7404, MHCC97, Hep3B and Huh7) were collected and
incubated in radioimmunoprecipitation assaylys is buffer (Thermo
Fisher Scientific, Inc.) containing protease inhibitor
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 30 min for
protein extraction. Subsequently, cell lysates were centrifuged at
120,000 × g for 20 min at 4°C. The concentration of the protein was
determined using Bradford reagent. Protein (10 µg/lane) was loaded
and subjected to 10% SDS-PAGE, and were transferred to a
polyvinylidene fluoride membrane. Samples were blocked in 5%
non-fat milk with 1X Tris-buffered saline with Tween-20 for 1 h.
Subsequently, membranes were incubated with primary antibodies
against TIP5 (cat. no. 195278; Abcam, Cambridge, UK; 1:1,000
dilution), Snail (cat. no. 3879; CST Biological Reagents Co., Ltd.,
Shanghai, China; 1:1,000 dilution), vimentin (cat. no. 5741; CST
Biological Reagents Co., Ltd.; 1:1,000 dilution), N-cadherin (cat.
no. 13116; CST Biological Reagents Co., Ltd.; 1:1,000 dilution),
c-Myc (cat. no. 13987; CST Biological Reagents Co., Ltd.; 1:1,000
dilution) and GAPDH antibody (cat. no. 5174; CST Biological
Reagents Co., Ltd.; 1:5,000 dilution) overnight at 4°C. Secondary
anti-rabbit antibody conjugated with peroxidase (cat. no. 7074; CST
Biological Reagents Co., Ltd.; 1:1,000 dilution) was applied to
membranes for 1 h at room temperature. Following immunoblotting,
bands were visualized using an enhanced chemiluminescent kit
(Thermo Fisher Scientific, Inc.).
MTT assay
Huh7 and 7404 cells were counted with a
hemocytometerand placed in 96-well plates (1,000 cells/well) in
triplicate and incubated with fresh DMEM medium (containing 10%
FBS) at 37°C for 0, 1, 3, 5 or 7 days. MTT (Dojindo Molecular
Technologies Inc. Rockville, MD, USA) Cells were incubated with MTT
(200 µl/ml medium) for 4 h and subsequently cultured for 0, 1, 3, 5
and 7 days to assess cell proliferation. Cell proliferation curves
were determined once absorbance at 540 nm was measured using a
microplate reader.
Plasmid construction and cell
transfection
The TIP5 sequence was amplified from the cDNA of HCC
tissues and generated using PCR with KOD DNA polymerase (Takara
Biotechnology Co., Ltd.) and the following thermocycling
conditions: 95°C for 30 sec, followed by 40 cycles of 95°C for 5
sec, 60°C for 30 sec and 68°C for 90 sec. TIP5 was subsequently
subcloned into the pcDNA3.1 vector (Takara Biotechnology Co.,
Ltd.). The forward primer sequence for TIP5 is
5′-ATGGAGGCAAACGACCATTTT-3′; the reverse primer sequence was
5′-TCACAAGATTGGCCTGTTTTCC-3′. TIP5 (0.5 µg) or dominant negative
(DN) β-catenin plasmids (0.5 µg; provided by Dr Michael Levin;
Rockefeller University, New York, NY, USA) were transfected into
7404 or Huh7 cells using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Following 48 h culture, transfected
cells were selected with G418. After 2 weeks, G418-resistant cells
were pooled and the expression of myc-TIP5 was verified using
anti-Myc antibody (1:1,000, cat. no. sc-4084; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) in western blot analysis
(overnight at 4°C).
Cell migration assays
Cells (7404 and Huh7) were seeded on the top layer
of the Boyden chamber with 50 µl DMEM supplemented with 1% FBS. 152
µl DMEM supplemented with 10% FBS was used as a chemoattractant in
the bottom chamber. Cells (1×105) were incubated at 37°C
for 12 h. Non-migratory cells in the top chambers were removed with
cotton swabs. The migrated cells in the lower membrane surface were
fixed in 100% methanol for 5 min, air-dried, stained with eosin for
5 min and then counted under a light microscope (magnification,
×10). Cell morphology alterations were assessed under the
microscope.
Immunohistochemistry
Tissues were fixed with 4% paraformaldehyde
overnight at 4°C. Xylene and ethanol was used to deparaffinize and
rehydrate the 5 µm paraffin-embedded HCC tissue. Endogenous
peroxidase activity was blocked with 0.35%
H2O2 solution. Antigen retrieval was
performed at 100°C using a microwave and tissues were subsequently
washed with PBS. Non-specific binding was blocked using 1% bovine
serum albumin (Sangon Biotech Co., Ltd., Shanghai, China)solution
at room temperature for 1 h. Sections were stained with TIP5
antibody (1:100) at 4°C overnight and incubated with secondary
antibody at room temperature for 1 h (DAKO; Agilent Technologies,
Inc., Santa Clara, CA, USA). Slides were subsequently developed
with 3,3′-diaminobenzidine at room temperature for 3 min and
counterstained with hematoxylin. The slides were examined under a
light microscope (magnification, ×10).
Glutathione S-transferase (GST)
pull-down assay
The GST pull-down was performed as previously
described (5). The coding sequence
of TCF7L2 from 7404 cells was cloned into the vector pGEX-4T-1 to
obtain the GST tag. The GST fusion protein was purified by
sepharose 4B beads, according to the manufacturer's instructions
(GE Healthcare Life Sciences, Shanghai, China) (5). Cells (7404) were grown to 80%
confluence and were harvested with lysis buffer (50 mM Tris-HCl,
0.5% Triton X-100) and centrifuged (12,000 × g at 4°C) for 10 min.
Cell lysates were incubated with 5 µg GST or GST-TCF7L24 protein
overnight at 4°C. Subsequently, Sepharose 4B beads (GE Healthcare
Life Sciences) were added to capture the GST fusion protein. The
pulled down protein (1 µg/well) was separated using 10% SDS-PAGE
and western blot was performed.
Immunoprecipitation
Cells (7404) were cultured until 70% confluence was
reached and were harvested with lysis buffer, and centrifuged
(12,000 × g) at 4°C for 20 min. The supernatant (500 µl) of cell
lysate was incubated with the following primary antibodies:
Anti-myc (1:1,000), anti-hemagglutin in tag (cat. no. H9658;
Sigma-Aldrich; Merck KGaA, 1:5,000 dilution) and anti-TCF4 (cat.
no. 2569; CST Biological Reagents Co., Ltd; 1:500 dilution) and
incubated overnight at 4°C. Subsequently, protein A beads were
added to the cell supernatant and incubated for a further 4 h at
4°C. Beads were washed with lysis buffer three times and the
immuneprecipitated proteins (1 µg/well) were eluted with loading
buffer (CST Biological Reagents Co., Ltd.; 1:1,000 dilution) and
separated by 10% SDS-PAGE. The bands were visualized with an
enhanced chemiluminescence kit (Thermo Fisher Scientific,
Inc.).
Mass spectrometry
Mass spectrometry was performed by the commercial
services of the Institute of Biophysics, Chinese Academy of Science
(Beijing, China).
Topflash reporter assay
Cells (7404) were grown to 80% confluence and
reporter assays were performed using 0.1 µg Topflash (Takara
Biotechnology Co., Ltd.), 0.5 µg TIP5 shRNA vector (pLKO.1) and
0.05 µg TK Renilla luciferase. The reporter was constructed
using the tandem repeats of the TCF binding elements, as described
previously (12). The plasmids
were transfected using Lipofectamine® 2000. After 48 h,
cells were treated with Wnt3a (100 ng/ml) protein for 8 h. Cell
lysates were prepared and reporter activity was measured using the
dual-luciferase reporter assay system (Promega Corp., Madison, MA,
USA) and normalized to Renilla luciferase activity to
confirm transfection efficiency.
Soft agar assay
Control and TIP5 shRNA cells (7404; 1×104
cells/well) were seeded in 1 ml 0.5% noble agar in complete DMEM
medium overlaying 2 ml 1% agar in the same medium. Following 2
weeks culture at 37°C, cell colonies were examined under a light
microscope (magnification, ×10).
Statistical analysis
Results were expressed as the mean ± standard
deviation as indicated. Differences between values were
statistically analyzed by SPSS (version 15.0; SPSS, Inc., Chicago,
IL, USA) using a Student's t-test or one-way analysis of variance
followed by Dunnett's test for multiple comparisons. P<0.05 was
considered to indicate a statistically significant difference.
Results
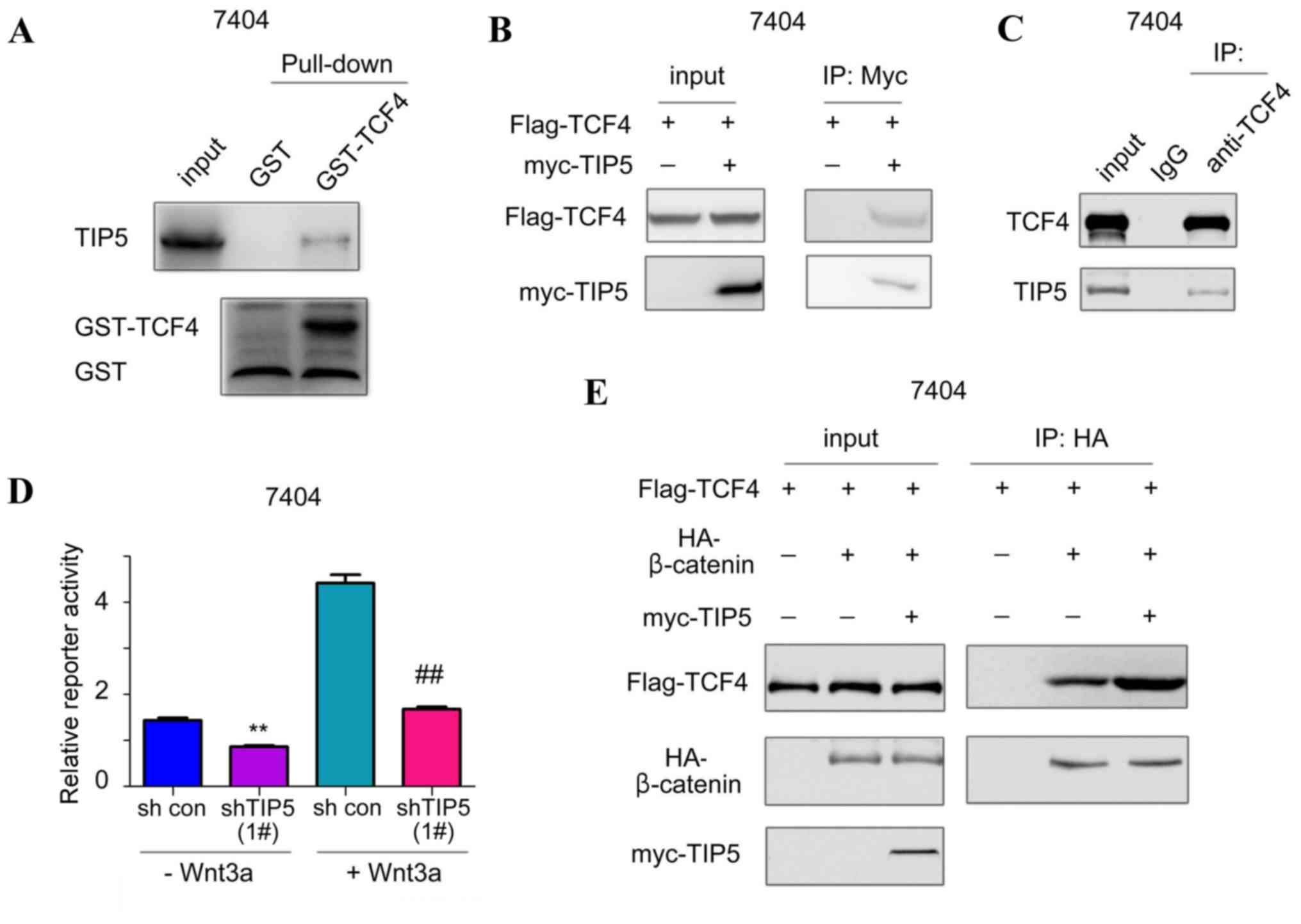
TIP5 interacts with TCF7L24 in HCC
cell lines
To screen novel regulators for β-catenin/TCF7L2
signaling, 7404 cell lysate was immunoprecipitated with
anti-TCF7L24 antibody and the TCF7L24-binding protein was
identified using mass spectrometry (data not shown). TIP5, a
nuclear transcriptional activator, was identified to be a potential
candidate for TCF7L24 binding. To further investigate the
interaction between TCF7L24 and TIP5, a GST pull-down assay was
performed. The fusion protein GST-TCF7L24 was purified and
incubated with the 7404 cell lysates. As indicated in Fig. 1A, the endogenous TIP5 in 7404 cells
was pulled down by the GST-TCF7L24 fusion protein, suggesting an
interaction occurred between TCF7L24 and TIP5. In addition, the
ectopically expressed TIP5 (myc-TIP5) and TCF7L24 (Flag-TCF7L24)
formed a complex in 7404 cells (Fig
1B). Furthermore, endogenous TIP5 and TCF7L24 bound to each
other in the immunoprecipitation assay (Fig. 1C). These data suggest that TIP5 and
TCF7L24 interact within 7404 HCC cells. The effects of TIP5 on the
activity of β-catenin/TCF7L2 signaling were assessed using a
Topflash reporter assay. It was indicated that TIP5 knockdown
significantly impaired the Topflash reporter activity both at the
basal level and following the treatment with Wnt3a (Fig. 1D). Consistent with these
observations, overexpression of TIP5 enhanced the interaction
between β-catenin and TCF7L24 (Fig.
1E). Taken together, these data suggest that TIP5 interacts
with TCF7L24 and activates β-catenin/TCF7L2 signaling.
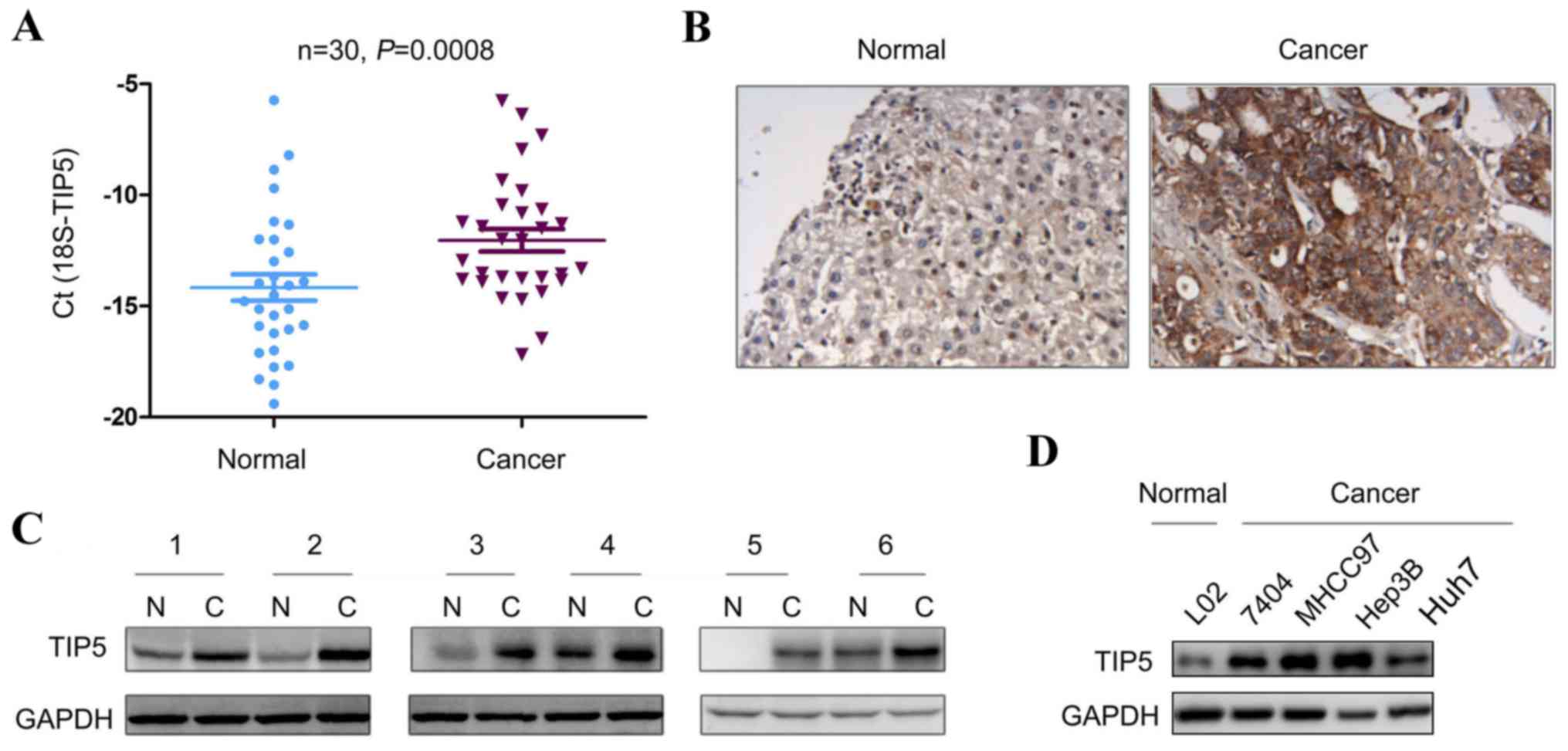
TIP5 is upregulated in HCC
The expression pattern of TIP5 was assessed in HCC.
The mRNA expression of TIP5 in 30 HCC tissues and paired
non-cancerous tissues was examined using RT-qPCR. TIP5 mRNA
expression was significantly elevated in HCC tissues (Fig. 2A). Immunohistochemistry and western
blot analysis demonstrated that the expression of TIP5 protein was
increased in HCC tissues (Fig. 2B and
C). In addition, the protein expression of TIP5 in a panel of
HCC cell lines was examined. The increased expression of TIP5 was
identified in a number of HCC cell lines compared with L02 normal
liver cells (Fig. 2D).
Collectively, these results indicate that TIP5 is upregulated in
HCC.
To study the functions of TIP5 in the progression of
HCC, the expression of TIP5 in 7404 and Huh7 cells was investigated
following the promotion of TIP5 expression (Fig 3A). Furthermore, the role of TIP5 in
the proliferation, migration and colony formation of HCC cells was
evaluated. The results indicated that induced expression of TIP5
significantly promoted the proliferation, migration and colony
formation of 7404 and Huh7 cells (Fig.
3B-D). Notably, upregulating the expression of TIP5 induced a
morphological change in 7404 cells, from an epithelial to shuttle
shape. This suggests that TIP5 may have induced the
epithelial-mesenchymal transition (EMT) in 7404 cells (Fig. 3E). In accordance with these
results, TIP5 knockdown in 7404 and Huh7 cells significantly
inhibited the proliferation, migration and colony formation of HCC
cells (Fig. 4). In summary, these
data suggest that TIP5 promotes HCC progression.
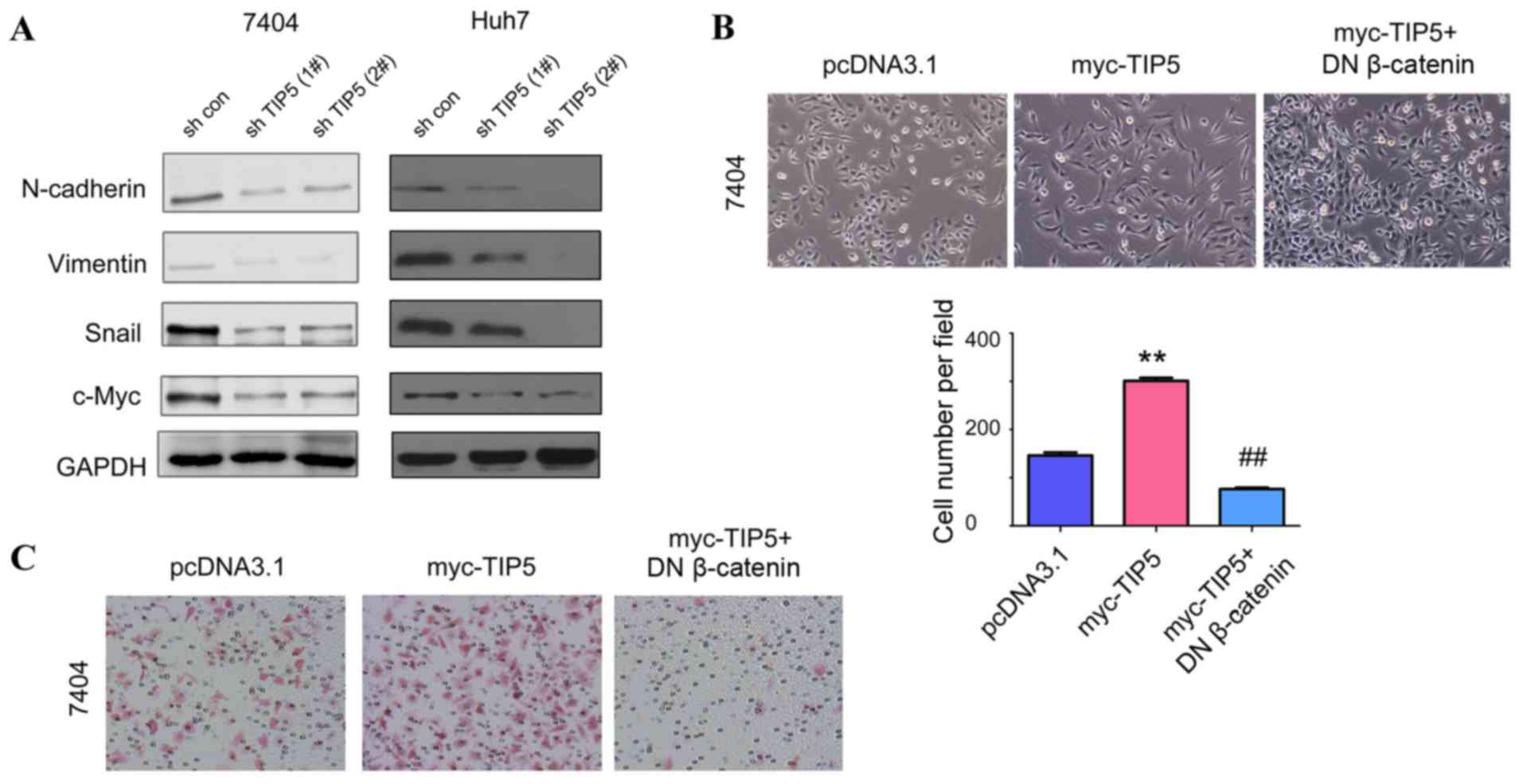
TIP5 promotes the EMT by activating
β-catenin/TCF7L2 signaling
The morphological changes in 7404 cells induced by
TIP5 overexpression indicate that TIP5 may promote the EMT in HCC
cells. As indicated in Fig. 5A,
TIP5 knockdown downregulated the expression of N-cadherin and
vimentin, two markers of mesenchymal cells (12). In addition, TIP5 knockdown
inhibited the expression of c-Myc and Snail, two regulators of the
EMT (13). To examine whether TIP5
promotes the EMT by activating β-catenin/TCF7L2 signaling, a DN
β-catenin, a negative regulator for beta-catenin/TCF7L2 signaling,
was used to rescue the phenotypes of TIP5, as described previously
(6) As indicated in Fig. 5B and C, DN β-catenin significantly
reversed the changes in 7404 cell morphology and migration induced
by TIP5. Taken together, these results demonstrate that TIP5
promotes the EMT by activating β-catenin/TCF7L2 signaling.
Discussion
HCC is one of the most common malignancies in the
world (14). Numerous studies have
identified that β-catenin/TCF7L2 signaling serves an oncogenic role
in the progression of HCC and have indicated that targeting
β-catenin/TCF7L2 signaling may be a promising therapeutic strategy
(15–17). The present study demonstrated that
TIP5 was upregulated in HCC tissues and promoted the proliferation,
colony formation, migration and EMT of HCC cells. Furthermore, TIP5
interacted with TCF7L24 to activate β-catenin/TCF7L2 signaling.
These data suggest that TIP5 promotes the progression of HCC and
may be a potential therapeutic target.
Notably, the present study identified an interaction
between TCF7L24 and TIP5, a chromatin remodeling factor. It was
demonstrated that TIP5 strengthened the interaction between
β-catenin and TCF7L24. Several factors regulate the interaction
between β-catenin and TCF7L24. For example, it has been indicated
that catenin ß interacting protein 1 (ICAT) weakens the interaction
between β-catenin and TCF7L24 (18). As a result, it is possible that
TIP5 and ICAT compete to regulate the interaction between β-catenin
and TCF7L24. Furthermore, TCF7L24 has DNA-binding capabilities and
the interaction that occurs between TCF7L24 and TIP5 suggests that
TCF7L24 may be involved in epigenetic regulation.
The results of the present study suggest that the
overexpression of TIP5 promotes cell migration and the EMT in 7404
cells. The expressionof TIP5 in the non-invasive L02 normal liver
cells was lower compared with in invasive HCC cell lines. These
observations suggest that there is an association between TIP5 and
cell motility. Notably, TIP5 induced a morphological change in 7404
cells and TIP5 knockdown inhibited the expression of the
mesenchymal markers N-cadherin and vimentin, as well as the
expression of the EMT regulator Snail. Furthermore, TIP5 promoted
the migration of HCC cells, which was consistent with the results
of a previous study demonstrating that TIP5 overexpression
predicted prostate cancer recurrence (11). In addition, the promoting effects
of TIP5 on HCC cell migration was blocked by the expression of DN
β-catenin, further supporting the factthat TIP5 promotes the
migration of HCC cells by activating β-catenin/TCF7L2
signaling.
In the present study, TIP5 promoted the growth of
HCC cells in liquid culture and in soft agar, suggesting that TIP5
enhanced the tumorigenicity of HCC cells. Activation of
β-catenin/TCF7L2 signaling by TIP5 may explain why it promotes
effects cell colony formation.
In conclusion, the results of the present study
demonstrated that TIP5 regulates β-catenin/TCF7L2 signaling via
TIP5 during HCC progression and suggests that TIP5 may be a
promising therapeutic target for the treatment of HCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
KX and CL designed this study. CL and WW performed
the cell assays. HD and QL performed the molecular mechanism
experiments.
Ethics approval and consent to
participate
The collection of samples from patients was approved
by the Institutional Ethics Committee of Cangnan People's Hospital
(Wenzhou, China) and written informed consent was received from all
patients in the present study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang W, Pan Q, Fuhler GM, Smits R and
Peppelenbosch MP: Action and function of Wnt/β-catenin signaling in
the progression from chronic hepatitis C to hepatocellular
carcinoma. J Gastroenterol. 52:419–431. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tao J, Zhang R, Singh S, Poddar M, Xu E,
Oertel M, Chen X, Ganesh S, Abrams M and Monga SP: Targeting
β-catenin in hepatocellular cancers induced by coexpression of
mutant β-catenin and K-Ras in mice. Hepatology. 65:1581–1599. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Peng YY, He YH, Chen C, Xu T, Li L, Ni MM,
Meng XM, Huang C and Li J: NLRC5 regulates cell proliferation,
migration and invasion in hepatocellular carcinoma by targeting the
Wnt/β-catenin signaling pathway. Cancer Lett. 376:10–21. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qu C, He, Lu X, Dong L, Zhu Y, Zhao Q,
Jiang X, Chang P, Jiang X, Wang L, et al: Salt-inducible Kinase
(SIK1) regulates HCC progression and WNT/β-catenin activation. J
Hepatol. 64:1076–1089. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Polakis P: More than one way to skin a
catenin. Cell. 105:563–566. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Suda T and Arai F: Wnt signaling in the
niche. Cell. 132:729–730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nardocci G, Simonet NG, Navarro C, Langst
G and Alvarez M: Differential enrichment of TTF-I and Tip5 in the
T-like promoter structures of the rDNA contribute to the epigenetic
response of Cyprinus carpio during environmental adaptation.
Biochem Cell Biol. 94:315–321. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tallant C, Valentini E, Fedorov O,
Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S
and Ciulli A: Molecular basis of histone tail recognition by human
TIP5 PHD finger and bromodomain of the chromatin remodeling complex
NoRC. Structure. 23:80–92. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Anosova I, Melnik S, Tripsianes K, Kateb
F, Grummt I and Sattler M: A novel RNA binding surface of the TAM
domain of TIP5/BAZ2A mediates epigenetic regulation of rRNA genes.
Nucleic Acids Res. 43:5208–5220. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cong R, Das S, Ugrinova I, Kumar S,
Mongelard F, Wong J and Bouvet P: Interaction of nucleolin with
ribosomal RNA genes and its role in RNA polymerase I transcription.
Nucleic Acids Res. 40:9441–9454. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gu L, Frommel SC, Oakes CC, Simon R, Grupp
K, Gerig CY, Bär D, Robinson MD, Baer C, Weiss M, et al: BAZ2A
(TIP5) is involved in epigenetic alterations in prostate cancer and
its overexpression predicts disease recurrence. Nat Genet.
47:22–30. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu L, Liu M, Yu X and Li X: Expressions of
epithelial cell adhesion molecule, vimentin and N-cadherin in
molecular subtypes of breast cancer and the correlation among them.
Zhong Nan Da Xue Xue Bao Yi Xue Ban. 41:1137–1142. 2016.(In
Chinese). PubMed/NCBI
|
|
13
|
Furuya S, Endo K, Takahashi A, Miyazawa K
and Saitoh M: Snail suppresses cellular senescence and promotes
fibroblast-led cancer cell invasion. FEBS Open Bio. 7:1586–1597.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iguchi T, Yamagata M, Sonoda T, Yanagita
K, Fukahori T, Tsujita E, Aishima S, Oda Y and Maehara Y: Malignant
transformation of hepatocellular adenoma with bone marrow
metaplasia arising in glycogen storage disease type I: A case
report. Mol Clin Oncol. 5:599–603. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang JJ, Chen JT, Hua L, Yao KH and Wang
CY: miR-98 inhibits hepatocellular carcinoma cell proliferation via
targeting EZH2 and suppressing Wnt/β-catenin signaling pathway.
Biomed Pharmacother. 85:472–478. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan X, Ma X, Cui L, Dang S, Qu J, Zhang J,
Wang X and Mao Z: CARF activates beta-catenin/TCF signaling in the
hepatocellular carcinoma. Oncotarget. 7:80404–80414. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tago K, Nakamura T, Nishita M, Hyodo J,
Nagai S, Murata Y, Adachi S, Ohwada S, Morishita Y, Shibuya H, et
al: Inhibition of Wnt signaling by ICAT, a novel
β-catenin-interacting protein. Genes Dev. 14:1741–1749.
2000.PubMed/NCBI
|