Introduction
Lung cancer is one of the commonest malignancies all
over the world. Non-small cell lung cancer (NSCLC) is the
predominant form of lung cancer and accounts for the majority of
cancer deaths worldwide, which includes adenocarcinomas and
squamous cell carcinomas (1).
Despite the advanced improvements made in chemotherapy and
radiotherapy over the past few decades, the clinical outcome of
NSCLC is still poor, with only slightly more than 15% of patients
alive 5 years after diagnosis (2).
Therefore, finding new therapeutic markers and better understanding
of the pathway related to cancer progression are warranted to
promote the prognosis of patients with NSCLC, and possibly find a
cure.
With the advent of high-throughput sequencing and
bioinformatic analysis, thousands of circular RNAs (circRNAs) have
been successfully identified in multiple cell lines and across
various species (3,4). circRNAs from back-spliced exons have
been recently identified as a naturally occurring family of
noncoding RNAs (ncRNAs) that is highly prevalent in the eukaryotic
transcriptome; however, they attracted little attention until their
function in post-transcriptional regulation of gene expression was
discovered. circRNAs are conserved and stable, and numerous
circRNAs seem to be specifically expressed in a cell type or
developmental stage (5,6). The progressive stage- and subcellular
type-based expression modern indicate that circRNAs may play
important roles in the mediation of multiple diseases, including
cancer (7,8). Recently, a group of circRNAs have
been found to be significantly deregulated in different cancer
types, including gastric cancer, esophageal squamous cancer, and
NSCLC. Thus, these deregulated circRNAs are suggested to play
important functional role during the cancer development (9). To date, elucidating the deregulated
circRNAs and identifying their functions in NSCLC are still an
ongoing process in cancer investigation.
Forkhead box O class (FOXO) transcription factors
are homeostasis regulators that control cell apoptosis, growth and
chemoresistance. Deregulation of FOXO3 is associated with cancer
development (10), by regulation
of inducing increased AKT activity or PTEN inactivation. FOXO3 is
thus classified as a tumor suppressor (11). Both circular FOXO3 (circRNA-FOXO3)
and linear FOXO3 (FOXO3 mRNA) are encoded by the FOXO3 gene.
Recently, circRNA-FOXO3 was reported lowly expressed in cancer
cells, and could arrest the function of CDK2 and block cell cycle
progression (12). However, the
specific role of circRNA-FOXO3 in NSCLC needs further
investigations.
circRNAs may function as competing endogenous RNAs
(ceRNAs), thus inducing the suppression of genes that targeted by
specific miRNAs (13). In this
study, we hypothesized that circRNA-FOXO3 regulated NSCLC
progression through sponging miR-155 and inducing linear FOXO3. To
verify this hypothesis, we detected the expression level of
circRNA-FOXO3 in NSCLC tissues and cell lines. By performing in
vitro gain and loss-function assays, we further investigated
the functional relevance of circRNA-FOXO3 with NSCLC.
Materials and methods
Clinical samples
Primary tissue samples and adjacent noncancerous
tissues were collected from 45 patients with NSCLC. All the
patients were pathologically confirmed and the clinical tissue
samples were collected before chemotherapy was started. Tissue
samples were classified according to the tumor-node-metastasis
(TNM) classification and WHO grade criteria. They were obtained
during operation and immediately frozen at −80°C until RNA
extraction. Written informed consents obtained from all patients.
The present study was approved by the Ethics Committee of the
Affiliated Zhongshan Hospital of Dalian University (Dalian,
China).
Cell culture
Four NSCLC adenocarcinoma cell lines (A549, SPC-A1,
NCI-H1299, and NCI-H1650), one NSCLC squamous carcinomas cell line
(SK-MES-1), and one normal human bronchial epithelial cell line
(16HBE) were all purchased from the Institute of Biochemistry and
Cell Biology of the Chinese Academy of Sciences (Shanghai, China).
A549, SK-MES-1, NCI-H1299, NCI-H1650 and 16HBE cells were cultured
in RPMI-1640; SPC-A1 cells were cultured in DMEM (Gibco-BRL, Grand
Island, NY, USA) medium supplemented with 10% fetal bovine serum
(FBS), 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C/5%
CO2.
RNA oligoribonucleotides and cell
transfection
The circRNA-FOXO3 overexpression plasmid, miR-155
mimics, and small silencing RNAs (siRNAs) that specifically silence
linear FOXO3 (si-FOXO3) was synthesized by GenePharma (Shanghai,
China). The CRC cells were plated at 5×104 cells/well in
24-well plates ~24 h before transfection. After the cells reached
30–50% confluence, transfection was carried out using Lipofectamine
3000 (Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. Transfection efficiency was evaluated
in every experiment by RT-qPCR 24 h later to ensure that cells were
actually transfected. Functional experiments were then performed
after sufficient transfection for 48 h.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from primary NSCLC cell lines
and tissue samples by using TRIzol reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the instructions of
Invitrogen. RNA was reverse transcribed using the SuperScript
III® (Invitrogen) and then amplified by RT-qPCR based on
the TaqMan method on an BioRad CFX96 Sequence Detection System
(Bio-Rad Laboratories, Inc., Berkeley, CA, USA). The gene
expression levels were normalized by GAPDH/U6 expression. RT-qPCR
results were analysed and expressed relative to CT (threshold
cycle) values, and then converted to fold changes. All the premier
sequences were synthesized by RiboBio Co., Ltd. (Guangzhou, China),
and their sequences are shown as follows: circRNA-FOXO3 forward,
5′-GTGGGGAACTTCACTGGTGCTAAG-3′ and reverse,
5′-GGGTTGATGATCCACCAAGAGCTCTT-3′; linear FOXO3 forward,
5′-GCAAGAGCTCTTGGTGGATCATCAA-3′ and reverse,
5′-TGGGGCTGCCAGGCCACTTGGAGA G-3′; miR-155 forward,
5′-CGGCGGTTTAATGCTAATCGTGAT-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGTAT-3′; U6 forward,
5′-CGGCGGTCGTGAAGCGTTCCAT-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGTAT-3′; GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse,
5′-GAAGATGGTGATGGGATTTC-3′.
Bioinformatics analysis
Predicted targets of miRNAs differentially expressed
in this study were determined using TargetScan (http:www.//targetscan.org) and miRanda (http://www.microrna.com). In addition, we used the
Gene Ontology database (http://www.geneontology.org) to perform Gene Ontology
(GO) analysis on the target genes. Pathway analysis was used to
identify significant pathways for the differentially expressed
genes.
MTT assay
Cell proliferation was evaluated by using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Briefly,
5×103 cells/well were seeded into a 96-well plate. After
transfection and incubation for 12, 24, 36 and 48 h, the cell
growth was measured following addition of 0.5 mg/ml MTT solution
(Sigma-Aldrich; Merck KGaA). Four hours later, the medium was
replaced with 100 µl dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck
KGaA) and vortexed for 10 min. Absorbance was then recorded at 450
nm using a specific microplate reader.
Flow cytometry for apoptosis
assay
Twenty-four hours after transfection, cells were
harvested and stained with Annexin V FITC and propidium iodide (PI)
according to the manufacturer's instructions (BD Biosciences, San
Diego, CA, USA). Then, the relative apoptosis status was evaluated
by flow cytometry on a BD FACSCalibur instrument.
Cell migration and invasion
assays
For wound healing assay, NSCLC cells were seeded in
six well plates and cultured until they reached confluence. Wounds
were scratched on the monolayer of cells using 20 µl pipette tips.
Plates were washed once with fresh medium to remove non-adherent
cells after the cells had been cultured for 48 h, and then
photographed. For transwell invasion assay, 100 µl matrigel (BD
Biosciences) was firstly added onto the bottom of the transwell
chamber (24-well insert; 8-mm pore size; Corning Costar, Corning,
NY, USA), then 1×105 NSCLC cells in reduced serum medium
(Opti-MEM; Gibco) were placed on the coated membrane in the
chamber. RPMI-1640 plus 10% FBS, was placed in the bottom wells as
chemoattractants. After 24 h, cells that did not invade through the
matrigel were removed from the top side of the inserts with a
cotton swab. Cells that migrated through the permeable membrane
were fixed in methanol, stained with crystal violet, and counted
under a microscope at ×20 magnification in random fields in each
well.
CircRNAs immunoprecipitation
(circRIP)
Magna RIP™ RNA-Binding Protein Immunoprecipitation
kit (Millipore, Billerica, MA, USA) were used for RIP. NSCLC cells
were lysed in complete RNA lysis buffer, then cell lysates were
incubated with RIP immunoprecipitation buffer containing magnetic
beads conjugated with human anti-argonaute 2 (AGO2) antibody
(Millipore) or negative control mouse IgG (Millipore). Extracted
RNAs were analyzed by RT-qPCR to identify the presence of
circRNA-FOXO3.
The RIP experiment using FOXO3 antibody (1:1,000;
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) to pull down
miR-155 was also performed. The RIP RNA fraction was digested by
DNase and cDNA was generated using PrimeScript 1st strand cDNA
Synthesis kit (Takara Bio., Inc., Otsu, Shiga, Japan). Final
analysis was investigated by performing RT-qPCR and presented as a
fold enrichment of miR-155.
Western blot analysis and
antibodies
NSLCL cells treated with ESMC for 48 h were lysed
with RIPA lysis buffer containing protease inhibitor cocktail and
phosphates inhibitor cocktail (Sigma-Aldrich; Merck KGaA) on ice
for 30 min, then lysis buffer was collected, and centrifuged at
12,000 g, 4°C for 10 min. The protein lysates were resolved by
SDS-PAGE, and separated proteins were transferred to PVDF membranes
and blocked with 5% skimmed milk for 2 h. The primary antibodies
used for western blotting were rabbit anti-human FOXO3 antibody
(1:1,000; Santa Cruz Biotechnology, Inc.) and rabbit anti-human
β-actin antibody (1:1,000; Santa Cruz Biotechnology, Inc.).
Horseradish peroxidase-conjugated (HRP) anti-rabbit antibodies
(1:5,000; Santa Cruz Biotechnology, Inc.) were used as the
secondary antibodies. The blots were incubated with the respective
antibodies overnight at 4°C under gently shaking. Finally, the
proteins were detected by using horseradish peroxidase labeled
secondary antibodies and an enhanced chemiluminescence detection
system.
Nuclear fractionation
Nuclear fractionation was performed with a PARIS™
Kit (Ambion, Austin, TX, USA). For nuclear fractionation,
1×107 cells were collected and resuspended in the cell
fraction buffer and incubated on ice for 10 min. After
centrifugation, supernatant and nuclear pellet were preserved for
RNA extraction using a cell disruption buffer according to the
manufacturer's instructions.
Fluorescence in situ hybridization
analysis
A549 and SPC-A1 cells were used for RNA FISH
analysis. Nuclear and cytosolic fraction separation was performed
using a PARIS kit (Life Technologies, Foster City, CA, USA), and
RNA FISH probes were designed and synthesized by Bogu according to
the manufacturer's instructions. Briefly, cells were fixed in 4%
formaldehyde for 15 min and then washed with PBS. The fixed cells
were treated with pepsin and dehydrated through ethanol. The
air-dried cells were incubated further with 40 nM of the FISH probe
in hybridization buffer. After hybridization, the slide was washed,
dehydrated and mounted with Prolong Gold Antifade reagent with DAPI
for detection. The slides were visualized for immunofluorescence
with a Olympus microscope.
Statistical analysis
Kolmogorov-Smirnov test was used to determine the
normality of the distribution of data in each group. The
Mann-Whitney U test or Kruskal-Wallis test was used for evaluating
the difference among clinical cohort groups or cell groups.
Spearman test was recruited to investigate the correlation status
between two groups of dates. Receiver operator characteristic (ROC)
curve analysis and area under the curve (AUC) was used to determine
the diagnostic value of circRNA-FOXO3. The results were considered
statistically significant at P<0.05. All statistical analyses
were performed with SPSS 17.0 software (SPSS Inc., Chicago, IL,
USA). Error bars in figures represent standard deviation (SD).
P<0.05, P<0.01, P<0.001 were considered to indicate
statistically significant differnces as indicated in figures.
Results
CircRNA-FOXO3 is downregulated in
NSCLC
To detect the circRNAs expression in NSCLC tissues,
we performed RT-qPCR assays to verify the circular form of FOXO3.
Two sets of FOXO3 primers were designed for this study: A divergent
set that was expected to amplify only the circular form and an
opposite directed set to amplify the linear forms. As expected, the
circular form was amplified by using the divergent primers, and no
amplification was observed when cDNA and genomic DNA were used as
templates (Fig. 1A). GAPDH was
used as a linear control. Thus, we confirmed that circRNA-FOXO3 is
specifically amplified with divergent primers on cDNA.
Then, we detected the expression level of
circRNA-FOXO3 in NSCLC cells. The TaqMan-based RT-qPCR showed that
circRNA-FOXO3 was significantly downregulated in most NSCLC cell
lines when compared to human normal bronchial cell line 16HBE
(Fig. 1B). A similar result was
also observed when circRNA-FOXO3 was determined in NSCLC tissues
and normal tissues (Fig. 1C).
Moreover, the NSCLC tissues in 57.8% (23 of 45) of cases had at
least 2-fold lower expression of circRNA-FOXO3 compared with
non-cancerous tissues (Fig. 1D).
Furthermore, ROC curve indicated that area under the ROC curve
(AUC) of circRNA-FOXO3 was 0.782 (95% CI: 0.682–0.862), and the
sensitivity and specificity of diagnosing NSLCL with circRNA-FOXO3
reached 80.0 and 73.3%, respectively (Fig. 1E).
CircRNA-FOXO3 plays an anti-oncogenic
role in NSCLC cells
The effect of circRNA-FOXO3 on NSCLC progression was
then investigated, and we chose A549 or SPC-A1 cell line for
further experiments. The full-length cDNA of circRNA-FOXO3 from
A549 cells was amplified and cloned into the specific vector
(Fig. 2A). RT-qPCR showed that
circRNA-FOXO3 vector significantly elevated the level of
circRNA-FOXO3 in A549 and SPC-A1 cells (Fig. 2B). Subsequently, the effect of
circRNA-FOXO3 on NSCLC cell proliferation, apoptosis, migration and
invasion were examined. MTT assay showed that overexpression of
circRNA-FOXO3 significantly suppressed cell proliferation when
compared with control vector in both cell lines (Fig. 2C), but promoted the proportion of
apoptotic cells (Fig. 2D). In
addition, circRNA-FOXO3 significantly inhibited the wound healing
ability of NSCLC cells (Fig. 2E).
Matrigel invasion assay showed that overexpression of circRNA-FOXO3
noticeably suppressed invasive ability of both cell lines (Fig. 2F). These indicate that
circRNA-FOXO3 may play an anti-oncogenic role in NSCLC.
CircRNA-FOXO3 positively regulates
FOXO3 gene expression in NSCLC cells
To investigate the underlying regulatory mechanism
of circRNA-FOXO3 in NSCLC, we focus on the potential downstream
target. It is reported that circRNA-FOXO3 is aligned in a sense
orientation to the known protein-coding gene, FOXO3, a member of
FOXO transcription factor family and commonly functions as a tumor
suppressor (14). Hence, we
hypothesized that circRNA-FOXO3 might exert its tumor-suppressive
role through activating its linear isomer, FOXO3. We detected the
expression of FOXO3 mRNA expression, and found that FOXO3 mRNA was
downregulated in the same cohort of NSCLC tissues (Fig. 3A). Spearman correlation analysis
suggested that circRNA-FOXO3 was positively correlated with FOXO3
mRNA expression (Fig. 3B). FOXO3
was also downregulated in NSCLC cell lines at both transcript and
protein level (Fig. 3C). Moreover,
FOXO3 was dramatically upregulated in NSCLC cells after
transfection of circRNA-FOXO3 overexpression vector (Fig. 3D). However, knockdown of FOXO3
showed no significant effect on circRNA-FOXO3 expression (Fig. 3E and F). These suggest that
circRNA-FOXO3 positively regulates FOXO3 expression in a
non-reciprocal way.
CircRNA-FOXO3 regulates FOXO3 gene
expression via a miR-155-dependent manner
It is reported that circRNAs may function as ceRNA
to further regulate the target mRNA of miRNAs. With this
hypothesis, we localized the expression of circRNA-FOXO3 in NSCLC
cells. RT-qPCR analysis of nuclear and cytoplasmic circRNA showed
that circRNA-FOXO3 was mainly enriched in the cytoplasm section of
NSCLC cells (Fig. 4A and B). More
importantly, our in situ RNA FISH analysis also suggested
that circRNA-FOXO3 was expressed predominately in the cytoplasm
part (Fig. 4C). Moreover, the RIP
assay with AGO2 antibody in NSCLC cells showed an enrichment of
both circular form and linear form of FOXO3 (Fig. 4D), indicating that both were
recruited to RNA-induced mediating complexes based on AGO2 and may
sponge with miRNAs. Subsequently, we sought to define the specific
miRNAs. Based on miRanda, we identified that miR-155 targeted the
transcript of FOXO3 (Fig. 4E).
Next, we detected whether circRNA-FOXO3 sponged miR-155, thereby
releasing the linear FOXO3 gene. RIP experiment was performed and
the results showed that the enrichment of FOXO3 and miR-155 was
significantly decreased in A549 cells that were transfected with
circRNA-FOXO3 vector (Fig. 4F).
Taken together, we revealed that circRNA-FOXO3 specifically sponged
miR-155, which induced the release of linear isomer, FOXO3.
circRNA-FOXO3 inhibits cell
proliferation and invasion through sponging miR-155 and activating
FOXO3 gene in NSCLC cells
Based on the above observations, we then sought to
determine the functional mechanism of circRNA-FOXO3 in NSCLC cells.
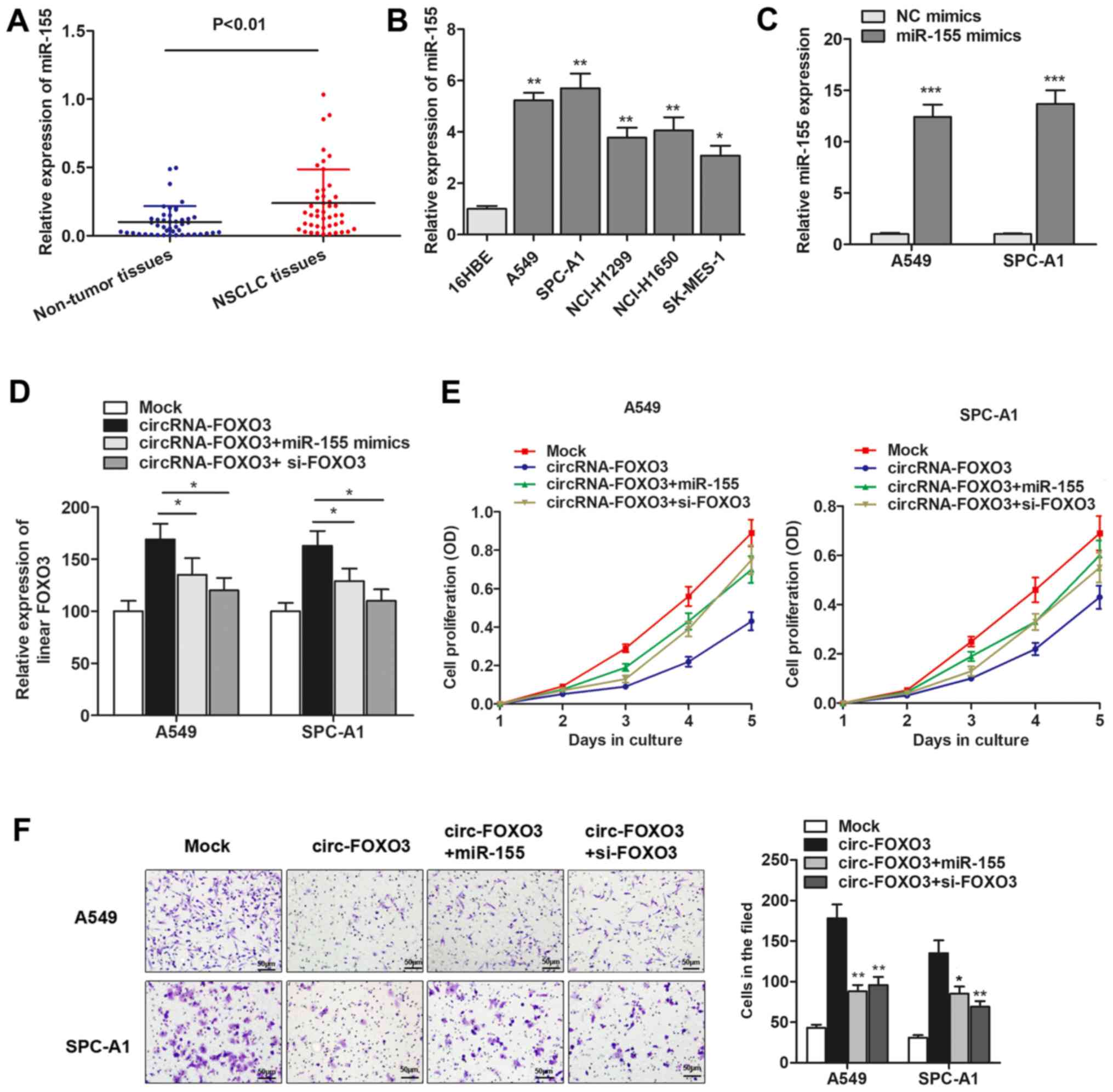
We detected the expression level of miR-155 and found that miR-155
expression was significantly increased in primary NSCLC tissues and
cells (Fig. 5A and B). To
investigate the role of miR-155 and FOXO3 gene during cirRNA-FOXO3
mediated anti-oncogenic effect, we generated miR-155 mimics
(Fig. 5C), and then performed gain
and loss functional assays by cotransfection of miR-155 mimics and
FOXO3 siRNAs. As shown in Fig. 5D,
linear isomer FOXO3 expression was upregulated by circRNA-FOXO3,
however, this upregulation was then partially reversed by
cotransfection of miR-155 mimics or specific FOXO3 siRNAs.
Furthermore, functional experiments showed that the suppression of
cell proliferation induced by circRNA-FOXO3 was dramatically
reversed by co-transfection of miR-155 mimics or si-FOXO3 (Fig. 5E). Similarly, overexpression of
miR-155 or knockdown of FOXO3 abrogated the circRNA-FOXO3-induced
inhibition of cell invasion (Fig.
5F). Collectively, we demonstrated that circRNA-FOXO3 functions
as a tumor-suppressor gene through specifically sponging miR-155
and promoting linear FOXO3 expression in NSCLC.
Discussion
Despite the rapid development of early diagnosis and
treatment in lung cancer, invariably, nearly all patients finally
become metastatic and chemoresistant (15). It is widely accepted that searching
new therapeutic targets and better understanding the pathway
related to cancer initiation and progression are essential for
improving the prognosis of cancer patients. In this study, we
focused on a novel group of gene regulator, circRNAs, and
identified the downregulation of circRNA-FOXO3 in NSCLC patients.
Furthermore, our in vitro investigations suggested that
circRNA-FOXO3 inhibited proliferation, migration and invasion of
NSCLC cells through specifically sponging miR-155 and releasing
FOXO3 gene.
The existence of circular form of RNAs in body fluid
was firstly reported by Sanger et al in 1976. They
demonstrated that this type of single-stranded closed circRNA were
stably expressed from viroids to certain highest species, such as
human beings (16). With the
development of gene investigations, it is recognized that circRNAs
are widely expressed in human cells. CCircRNAs contain highly
conserved sequences and show a potential of stability in cells and
body fluid. (17). These two
properties suggest us that circRNAs can serve as ideal biomarkers
for the diagnosis and prognosis of cancers (18,19).
To date, only a few circRNAs have been explored. In this study, we
identified a novel circRNA termed circRNA-FOXO3 that was
significantly downregulated in NSCLC cells and correlated with
clinical diagnosis. As circRNAs are a class of endogenous RNAs
featuring stable structure making them avoid exonucleolytic
degradation by RNase R, circRNA-FOXO3 may serve a novel biomarker
used for early diagnosis, treatment monitoring and prognosis for
NSCLC patients.
The interaction was further confirmed by an approach
of molecular experiments to explicate the biological functions of
circRNA-FOXO3. We firstly determined the functional role of
circRNA-FOXO3 in NSCLC progression, and found that expression of
circRNA-FOXO3 suppressed cell proliferation, migration and
invasion, and promoted apoptosis. Currently, the biological role of
FOXO3 was largely unknown. Du et al demonstrated that
circ-FOXO3 was highly expressed in noncancer cells and was
associated with cell cycle progression. Ectopic expression of
circFoxo3 repressed cell cycle progression by binding to the cell
cycle proteins CDK2 and p21, resulting in the formation of a
ternary complex (12). They
further revealed that circRNA-FOXO3 increased FOXO3 protein levels,
promoted MDM2-induced p53 ubiquitination and subsequent
degradation, resulting in an overall decrease of p53 (20). Consistent with this conclusion, we
reported that circRNA-FOXO3 positively regulated the expression of
linear FOXO3 gene. Interestingly, we revealed that the linear
isomer of FOXO3 was regulated by circRNA-FOXO3 in a non-reciprocal
way. For this point, we hypothesize that circRNA-FOXO3 may be one
of the upstream regulator of FOXO3 gene, and circRNA-FOXO3 can bind
to miRNAs that targeting FOXO3 but not the other way round.
However, the underlying mechanism of this regulation way should be
comprehensively investigated in future studies.
Take a step further, we then sought to determine how
circRNA-FOXO3 regulates the expression of linear FOXO3 gene. It is
known that circRNAs are novel RNA molecules with different
biological functions and pathological implications. In the nucleus,
circRNAs can act as scaffolds to bind to specific proteins; in the
cytoplasm, lncRNAs may function as competing endogenous RNAs
(ceRNAs). Among these multiple functions, ‘miRNA sponge’ represents
the most conspicuous function. miRNAs, an abundant class of small
noncoding RNAs (~22 nt), posttranscriptionally modulate the
translation of target mRNAs via corresponding miRNA response
elements (MRE) (21).
Computational searches for miRNA target sites in circRNAs
identified a portion of circRNA molecules that contain MREs, which
might act as miRNA sponge, reducing miRNA binding to its target
genes, thereby releasing the expression of the miRNA targets
indirectly (22). Since the first
report of circRNA functioning as a miRNA sponge, the potential of
circRNAs in regulating cancer-related genes through fine-tuning
miRNAs has recently been recognized (23).
To validated the functional interaction between
circRNA-FOXO3 and potential miRNAs, we firstly detected the
sublocation of circRNA-FOXO3 in NSCLC cells and found circRNA-FOXO3
was located at cytoplasm. Furthermore, we identified miR-155 as a
co-target of both circRNA-FOXO3 and linear FOXO3 gene by performing
bioinformatic analysis and a serious of experimental validation.
Previous studies have developed a consensus that miR-155 plays
important functions during cancer initiation, progression and
chemoresistance (24,25). miR-155, as a microRNA, can silence
the downstream target genes, leading to the biological disruptions
(26,27). It is reported that miR-155 is the
most amplified miRNAs in NSCLC, and is critical promoter of NSCLC
progression. In addition, miR-21 and miR-155 share nearly 30% of
their predicted target genes, including SOCS1, SOCS6, and PTEN,
three tumor suppressor genes often silenced in NSCLC. Therefore,
miR-155 is frequently reported as a oncogene in NSCLC. We revealed
that restoration of miR-155 potently reversed the
cancer-suppressive effect induced by circRNA-FOXO3, which are
consistent with previous date. We must admit that there are two
limitations in this study: i) The applied colorectal cancer A549
and SPC-A1 cell lines were inconsistently used in the experiments;
and ii) no in-vivo-experiments were performed to support our
interesting in-vitro-findings. In the future, we will
continue our research and comprehensively validate the observations
at both in-vitro and in-vivo levels.
We finally developed the conclusion that
circRNA-FOXO3 expression was decreased in NSCLC cells and tissue
samples. It can inhibit the development of NSCLC cells as a ceRNA
through sponging miR-155 and releasing FOXO3 level. Therefore, it
can serve as a promising therapeutic target for patients with
NSCLC.
Acknowledgements
Not applicable.
Funding
This study was supported by The National High
Technology Research and Development Program (‘863’ Program) of
China (2015AA020409) and the Project of Liaoning Province Education
Department (L2014358).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ designed and performed the study. HZ and LZ
assisted YZ in the analysis and interpretation of the patient
data.
Ethics approval and consent to
participate
Written informed consents obtained from all patients
were approved according to the guidelines revised by the Ethics
Committee of the Affiliated Zhongshan Hospital of Dalian
University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pastorino U: Lung cancer screening. Br J
Cancer. 102:1681–1686. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen LL and Yang L: Regulation of circRNA
biogenesis. RNA Biol. 12:381–388. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lasda E and Parker R: Circular RNAs:
Diversity of form and function. RNA. 20:1829–1842. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ivanov A, Memczak S, Wyler E, Torti F,
Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M,
Dieterich C and Rajewsky N: Analysis of intron sequences reveals
hallmarks of circular RNA biogenesis in animals. Cell Rep.
10:170–177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xia W, Qiu M, Chen R, Wang S, Leng X, Wang
J, Xu Y, Hu J, Dong G, Xu PL and Yin R: Circular RNA
has_circ_0067934 is upregulated in esophageal squamous cell
carcinoma and promoted proliferation. Sci Rep. 6:355762016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Myatt SS and Lam EW: The emerging roles of
forkhead box (Fox) proteins in cancer. Nat Rev Cancer. 7:847–859.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cho EC, Kuo ML, Liu X, Yang L, Hsieh YC,
Wang J, Cheng Y and Yen Y: Tumor suppressor FOXO3 regulates
ribonucleotide reductase subunit RRM2B and impacts on survival of
cancer patients. Oncotarget. 5:4834–4844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Afonso-Grunz F and Müller S: Principles of
miRNA-mRNA interactions: Beyond sequence complementarity. Cell Mol
Life Sci. 72:3127–3141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Katoh M and Katoh M: Human FOX gene family
(Review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
15
|
Nanavaty P, Alvarez MS and Alberts WM:
Lung cancer screening: Advantages, controversies, and applications.
Cancer Control. 21:9–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Osaki M, Okada F and Ochiya T: miRNA
therapy targeting cancer stem cells: A new paradigm for cancer
treatment and prevention of tumor recurrence. Ther Deliv.
6:323–337. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang YS, Jie N, Zou KJ and Weng Y:
Expression profile of circular RNAs in human gastric cancer
tissues. Mol Med Rep. 16:2469–2476. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang
Z and Yang BB: Induction of tumor apoptosis through a circular RNA
enhancing Foxo3 activity. Cell Death Differ. 24:357–370. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Melo SA, Sugimoto H, O'Connell JT, Kato N,
Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et
al: Cancer exosomes perform cell-independent microRNA biogenesis
and promote tumorigenesis. Cancer Cell. 26:707–721. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu H, Guo S, Li W and Yu P: The circular
RNA Cdr1as, via miR-7 and its targets, regulates insulin
transcription and secretion in islet cells. Sci Rep. 5:124532015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun S, Sun P, Wang C and Sun T:
Downregulation of microRNA-155 accelerates cell growth and invasion
by targeting c-myc in human gastric carcinoma cells. Oncol Rep.
32:951–956. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yu F, Jia X, Du F, Wang J, Wang Y, Ai W
and Fan D: miR-155-deficient bone marrow promotes tumor metastasis.
Mol Cancer Res. 11:923–936. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Li J, Tong L, Zhang J, Zhai A, Xu
K, Wei L and Chu M: The prognostic value of miR-21 and miR-155 in
non-small-cell lung cancer: A meta-analysis. Jpn J Clin Oncol.
43:813–820. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong
W, Liao Y and Du J: High expression of miR-21 and miR-155 predicts
recurrence and unfavourable survival in non-small cell lung cancer.
Eur J Cancer. 49:604–615. 2013. View Article : Google Scholar : PubMed/NCBI
|