Introduction
Hepatitis B virus (HBV) is a widespread human
pathogen associated with liver inflammation, cirrhosis and
hepatocellular carcinoma (HCC). Among HBV proteins, HBx has been
termed the viral oncoprotein and is involved in the initiation and
progression of HCC (1). However,
the underlying mechanism by which HBx contributes to the
development of HCC remains unclear. Various studies have provided
considerable evidence that HBx is a multifunctional protein that
acts on cell cycle regulation, signaling pathways, DNA repair, cell
proliferation, autophagy and apoptosis (2).
The different subcellular localizations of HBx
indicate its different functions. It is primarily localized in the
cytoplasm, with a fraction in the mitochondria and a small amount
in the nucleus (3). A previous
study demonstrated the colocalization of HBx with the inner
mitochondrial membrane protein cytochrome c oxidase III (COXIII)
and, reported an alteration of mitochondrial function and an
upregulation of reactive oxygen species (ROS) generation in HL7702
and HepG2 cells. Subsequently, the key region in HBx for binding
with COXIII was identified to be aa72-117, and ROS from
mitochondria stimulated COX-2 expression (4–7).
COX-2 is an isoform of cyclooxygenase that mediates
the oncogenic actions of HBx. COX-2 activity upregulation results
in induced proliferation, angiogenesis and invasiveness in HCC
(8,9). The Wnt/β-catenin signaling pathway is
involved in cell proliferation, differentiation and oncogenesis.
Previously, studies have documented that HBx serves an important
role in the modulation and induction of the canonical Wnt signaling
pathway. Β-catenin, upregulated by HBx, is associated with the
oncogenic activity of HBx in HBV-associated HCC. Stabilization of
β-catenin in the cytoplasm and its translocation to the nucleus are
the two features of the activation of the canonical Wnt pathway.
The expression of β-catenin targeted genes, including cyclin-D1 and
c-myc proto-oncogene protein (c-myc) are activated following its
translocation to the nucleus (10–12).
A previously study demonstrated that β-catenin is associated with
COX-2 overexpression (13). COX-2
activates the Wnt/β-catenin pathway in gastric cancer (14). However, it remains unclear whether
the COX-2 and β-catenin signaling pathways converge during HCC. A
study on the underlying antitumor mechanism showed that the
Wnt/β-catenin signaling pathway mediated by COX-2 is involved in
HCC (15). Therefore, the aim of
the present study was to clarify the role of the
COX-2/Wnt/β-catenin pathway in HL-7702-HBx cells.
In the present study, the role of HBx in the
oncogenesis of HBV associated with HCC was investigated by stably
expressing HBx in HL-7702 cells. It was concluded that HBx promoted
HL-7702 cell proliferation, and was dependent on the
COX-2/Wnt/β-catenin pathway.
Materials and methods
Antibodies and reagents
Anti-COX-2 antibody was purchased from Abcam
(Cambridge, UK; cat no. ab151571). Anti-β-catenin antibody was
purchased from OriGene Technologies, Inc. (Rockville, MD, USA; cat
no. sc-7963). Anti-c-myc antibody was purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA; cat no. SC-40). Anti-cyclin
D1 antibody was purchased from Santa Cruz Biotechnology, Inc. (cat
no. sc-70899). Anti-β-actin antibody was purchased from OriGene
Technologies, Inc. (cat no. TA-09). Peroxidase-conjugated goat
anti-rabbit immunoglobulin (Ig)G (H+L) was purchased from OriGene
Technologies, Inc. (cat no. ZB-2301). Peroxidase-conjugated goat
anti-mouse IgG (H+L) was purchased from OriGene Technologies, Inc.
(cat no. ZB-2305). XAV939 was purchased from Selleck Chemicals
(Houston, TX, USA). Cells were treated with 20 µmol/l XAV939 (a
β-catenin inhibitor) and DMSO (as a control) for 24 h in cell
proliferation assay. NS-398 was purchased from Beyotime Institute
of Biotechnology (Haimen, China). Cells were treated with 50 µmol/l
NS-398 (a selective COX-2 inhibitor) and DMSO (as a control) for 72
h in western blot analysis and colony formation assay. Primers were
synthesized by Biosune Biotechnology Co., Ltd (Shanghai, China;
www.biosune.com). The enhanced chemiluminescence
kit was purchased from OriGene Technologies, Inc. TRIzol reagent
was purchased from Life Technologies (Thermo Fisher Scientific,
Inc., Waltham, MA, USA). Moloney murine leukemia virus (MMLV)
transcriptase was purchased from New England BioLabs., Inc.
(Ipswich, MA, USA). Dulbecco's Modified Eagle's medium (DMEM) and
fetal bovine serum (FBS) were purchased from HyClone Company; GE
Healthcare (Chicago, IL, USA).
Cell cultures
The human hepatocyte HL-7702 cell line was purchased
from the Cell Bank of the Chinese Academy of Sciences (Shanghai,
China). HL-7702-HBx cells (HL-7702 cells transfected with
lentiviral vector pLOV.flag-HBx to stably express the HBX gene: HBV
subtype ayw) and HL-7702-mock cells (HL-7702 cells transfected with
lentiviral vector pLOV.flag) were constructed previously by the
authors of the present study (5).
HBx, mock and control represent HL-7702-HBx, HL-7702-mock and
HL-7702 cells, respectively. Cells were maintained in DMEM
supplemented with 10% FBS and 1% penicillin-streptomycin at 37°C in
a humidified atmosphere of 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol reagent. The
primer sequences of each gene are listed in Table I. First-strand cDNA was generated
using MMLV transcriptase (2 µg total RNA, 1 µg primer, 5 µl dNTP, 5
µl M-MLV 5X Reaction Buffer and final volume 25 µl), incubated for
60 min at 37°C. The extension temperature is 37°C, and qPCR was
performed using FastStar universal SYBR Green master mix (Roche
Applied Science, Penzberg, Germany) in triplicate using an Applied
Biosystems Step one plus Real time PCR system (Life Technologies;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Thermocycling conditions of the qPCR reaction are as
follows: 95°C for 3 min, then 40 cycles of 95°C for 15 sec followed
by 60°C for 30 sec. Specific primers were used to detect the
relative mRNA expression by the 2−ΔΔCq method. The
expression level was normalized against endogenous β-actin
(16).
 | Table I.Primer sequences for the genes used in
the RT-qPCR. |
Table I.
Primer sequences for the genes used in
the RT-qPCR.
| Gene | Primer sequences
(5′→3′) |
|---|
| COX-2 | F:
TGAAACCCACTCCAAACACA |
|
| R:
GAGAAGGCTTCCCAGCTTTT |
| β-catenin | F:
AAATGGTTGCCTTGCTCAAC |
|
| R:
TCAGCACTCTGCTTGTGGTC |
| c-myc | F:
CCTACCCTCTCAACGACAGC |
|
| R:
CTCTGACCTTTTGCCAGGAG |
| Cyclin D1 | F:
CCCTCGGTGTCCTACTTCAA |
|
| R:
GGGGATGGTCTCCTTCATCT |
| β-actin | F:
CTCCATCCTGGCCTCGCTGT |
|
| R:
GCTGTCACCTTCACCGTTCC |
Western blot analysis
Cells were harvested and lysed by RIPA buffer from
Beyotime Institute of Biotechnology. The protein concentration of
each sample was determined using a bicinchoninic protein assay kit.
A total of 20 ug protein per lane was loaded on a 10% SDS/PAGE gel,
and subsequently transferred to a nitrocellulose membrane. The
membrane was blocked in 5% milk in TBS-Tween 20 [0.1% Tween 20, 20
mM Tris (pH 7.4) and 150 mM NaCl] for 2 h at room temperature,
followed by overnight incubation with the primary antibody at 4°C.
Primary antibodies were used at 1:1,000 (Anti-COX-2 antibody,
Anti-β-catenin antibody, Anti-c-myc antibody and Anti-β-actin) or
1:500 (Anti-cyclin D1 antibody), and secondary antibodies
conjugated with horseradish peroxidase were used at a 1:2,000
dilution in 5% nonfat dry milk at room temperature for 1 h.
Following a final wash, proteins were visualized using the enhanced
chemiluminescence kit. Images of the blots were captured using an
Image Scanner (Epson, Nagano, Japan). Expression levels of protein
were semi-quantitatively analyzed by using Image J Launcher (Broken
Symmetry Software 1.4.3.67), normalized to β-actin density.
Cell proliferation assay
A Cell Counting Kit-8 (CCK-8) assay (Dojindo
Molecular Laboratories, Inc., Kumamoto, Japan) was used to analyze
cell proliferation. Cells were seeded into 96-well plates at
3×103 cells/well in 100 µl complete DMEM and incubated
at 37°C and 5% CO2. The plates were incubated for 24, 48
and 72 h, the medium was removed and 100 µl fresh complete DMEM was
added, containing 10 µl CCK-8, to each well. The plates were
incubated for 3 h at 37°C. The absorbance was measured at a
wavelength of 450 nm using an ELISA plate reader. Proliferation
inhibition rate was calculated according to the following formula:
Proliferation inhibition rate (%)=(1-OD450/OD450
with XAV939) ×100%.
Colony formation assay
Cells were seeded into 6-well plates at 500
cells/well in triplicate and incubated at 37°C and 5%
CO2 for 14 days. Clones were observed in the dish. Each
well was washed with PBS twice, fixed with 1 ml 100% methanol for
15 min, then washed with PBS for three times and stained with
crystal violet staining solution (Beyotime Institute of
Biotechnology) for 10 min at room temperature. Directly counted by
naked eyes based on experience, in case of doubting whether 50 or
more cells formed the suspected colony, counted the colony by
stereomicroscope under lower magnification (×40). Clone formation
rate and Clone formation inhibition rate were calculated according
to the following formula: Clone formation rate=number of formed
colonies/number of seeded cells ×100. Clone formation inhibition
rate=(1-number of formed colonies/number of formed colonies with
NS398) ×100%.
Flow cytometric analysis
Cells were harvested and resuspended in PBS
(1×106 cells/ml). For analysis of the cell cycle, cells
were fixed in 70% ethanol overnight at 4°C. Subsequently, cells
were digested with 50 µl RNase (TakaRa Biotechnology Co. Ltd.
Dalian, China; 10 mg/ml) at 37°C for 30 min and stained with 20 µl
50 µg/ml propidium iodide (Beyotime Institute of Biotechnology) at
37°C for 10 min in the dark. The DNA histograms were determined by
flow cytometry (C6; Becton-Dickinson; BD Biosciences, San Jose,
CA).
Statistical analysis
Experiments were performed in triplicate. All
statistical analyses were performed using the SPSS version 16.0
statistical software package (SPSS, Inc., Chicago, IL, USA).
Continuous variables are expressed as the mean ± standard
deviation. One-way analysis of variance was used for the
statistical analyses, followed by the Fisher's Least Significant
Difference. P≤0.05 was considered to indicate a statistically
significant difference.
Results
HBx promotes the proliferative ability
of HL-7702 cells
Considerable efforts have been made to detect the
role of HBx in cell proliferation (17). A cell viability assay and plate
colony formation assay were performed in the present study to
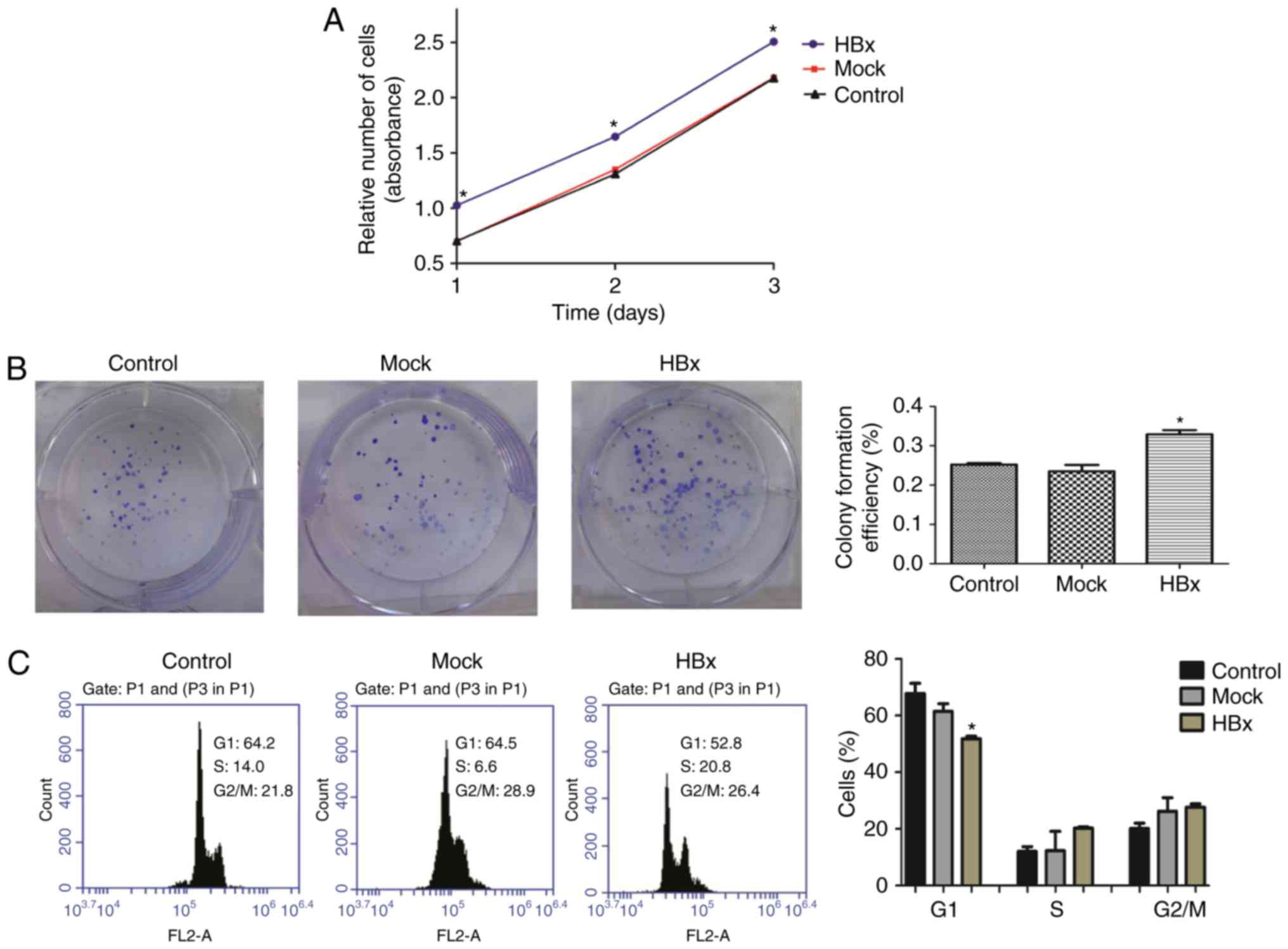
observe the role of HBx in HL-7702 cell proliferation. As
demonstrated in Fig. 1A,
HL-7702-HBx cells grew faster compared with the mock and control
groups on days 1, 2 and 3 (P<0.05). As presented in Fig. 1B, following incubation for 2 weeks,
HL-7702-HBx cells formed more colonies and had higher colony
formation efficiency compared with the mock and control groups. In
addition, HL-7702-HBx cells exhibited a shortened G1 phase of the
cell cycle compared with the HL-7702-mock and HL-7702-control cells
(Fig. 1C). These results suggested
that HBx promoted the proliferation of HL-7702 cells.
Upregulation of COX-2 promotes the
proliferation ability of HL-7702 cells
Previous studies have demonstrated that HBx promoted
the levels of mitochondrial ROS in HL-7702 cells (4,5). ROS
from mitochondria lead to COX-2 induction (18). As demonstrated in Fig. 2A and B, HBx enhanced the mRNA and
protein expression levels of COX-2 in HL-7702 cells. COX-2
increases proliferation in various types of tumor and its
expression appears to be associated with early HCC events (19). The present study investigated
whether upregulation of COX-2 promoted the proliferative ability of
HL-7702 cells and treated cells with NS-398. COX-2 protein
expression levels and colony formation efficiency were analyzed. As
presented in Fig. 2C and D, NS-398
suppressed the colony formation efficiency in the three groups
investigated, the colony formation inhibition rate of HL-7702-HBx
cells was significantly higher than that of other two groups after
downregulation of COX-2 at the protein level (Fig. 2E). Therefore, HBx may promote cell
proliferation through upregulation of COX-2.
 | Figure 2.Upregulation of COX-2 promotes the
proliferative ability of HL-7702 cells. (A) The mRNA expression
levels of COX-2 in cells were examined using the reverse
transcription-quantitative polymerase chain reaction. (B) The
expression levels of the COX-2 protein in cells were detected by
western blotting. (C) Western blot analysis of COX-2 protein
expression levels. A total of 50 µmol/l COX-2 inhibitor, NS-398,
was added to cells in the HBx, mock and control groups, or DMSO was
added instead, for 72 h. (D) A total of 50 µmol/l NS-398 was added
to cells, or DMSO as a control, for 72 h. A plate clone formation
assay was performed to detect the colony formation efficiency. (E)
Clone formation inhibition rate was calculated. Clone formation
inhibition rate=(1-number of formed colonies/number of formed
colonies with NS398) ×100% *P<0.05 vs. HL-7702-mock cells and
#P<0.05 vs. HL-7702 cells. $P<0.05 vs.
HL-7702-HBx cells with NS398. COX-2, cyclooxygenase-2; DMSO,
dimethyl sulfoxide; HBx, hepatitis B virus X protein. |
HBx activates the Wnt/β-catenin
signaling pathway in HL-7702 cells
The Wnt/β-catenin signaling pathway is involved in
cell proliferation, differentiation and oncogenesis (10,12).
The present study questioned whether aberrant activation of the
Wnt/β-catenin signaling pathway occurs in HBx-transfected cells.
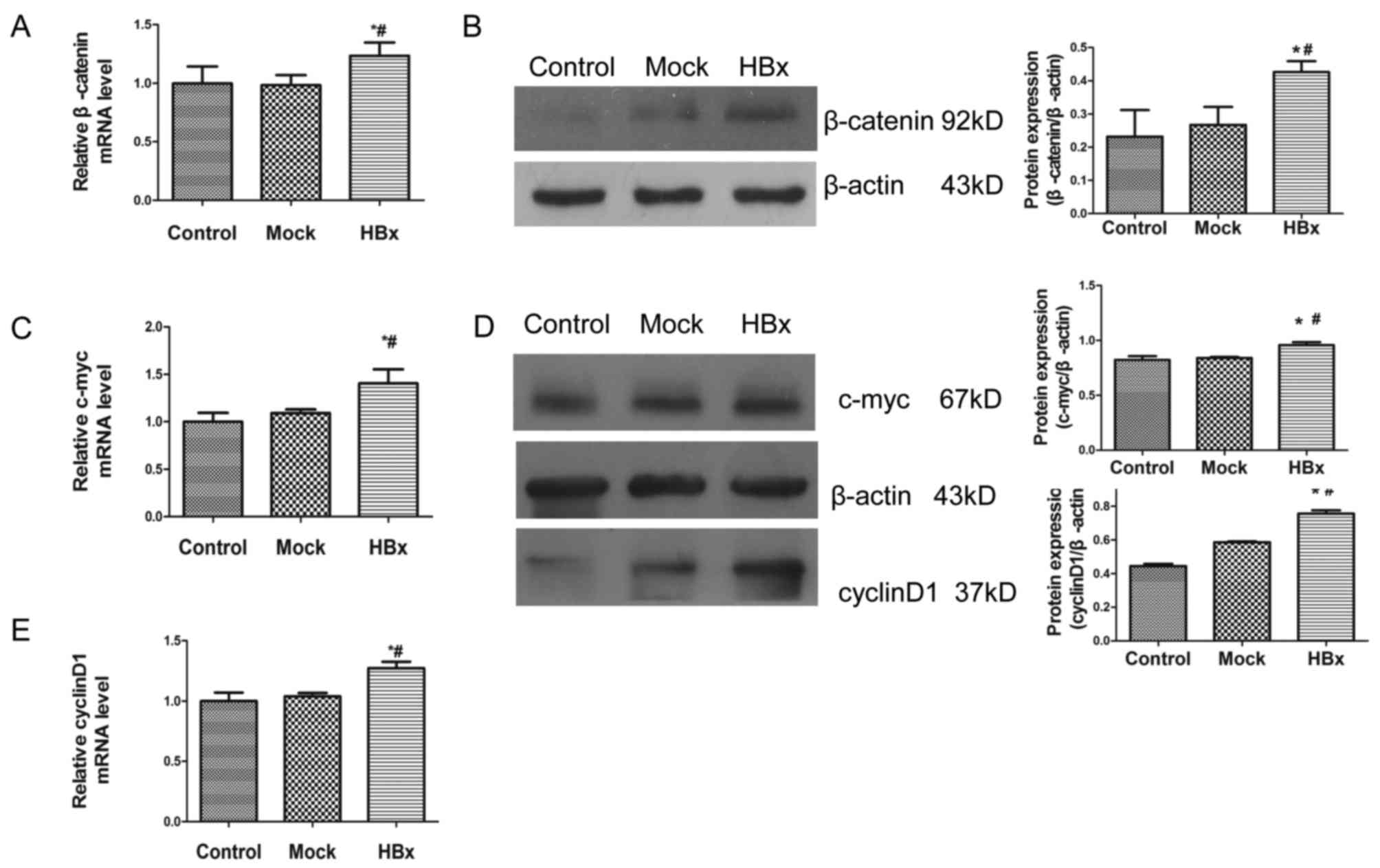
The mRNA expression level of β-catenin was increased in HL-7702-HBx
cells compared with the control and mock groups (Fig. 3A). In support of this upregulation
at the mRNA level, the protein expression of β-catenin was
increased in HL-7702-HBx cells compared with the other two cell
groups (Fig. 3B). The results
demonstrated that the mRNA and protein expression levels of
β-catenin responsive genes, c-myc and cyclin-D1, were activated
compared with the mock and control groups (Fig. 3C-E). These data suggested that HBx
may activate Wnt/β-catenin signaling in HL7702 cells.
HBx promotes the proliferative ability
of HL-7702 cells via the COX-2/Wnt/β-catenin signaling pathway
To further examine the association between β-catenin
and COX-2 in HL-7702-HBx cell proliferation, cells were treated
with NS-398 (50 µmol/l) for 72 h, and as presented in Fig 3B and Fig. 4A, NS398 attenuated the effects of
HBx on β-catenin expression in HL7702-HBx cells. The present study
considered whether the upregulation of β-catenin resulted in
increased proliferation of HL-7702-HBx cells. To support this
hypothesis, cell viability assays were performed following
treatment of the cells with a β-catenin inhibitor, XAV939 (20
µmol/l), for 24 h. As indicated in Fig. 4B, treatment with XAV939 suppressed
cell proliferation in the three groups. HL7702-HBx cells treated
with XAV939 demonstrated higher proliferative inhibition rate than
the mock and control groups (Fig.
4C). Therefore, it was concluded that upregulation of COX-2 may
promote the proliferation of HL-7702-HBx cells via activation of
the Wnt/β-catenin signaling pathway.
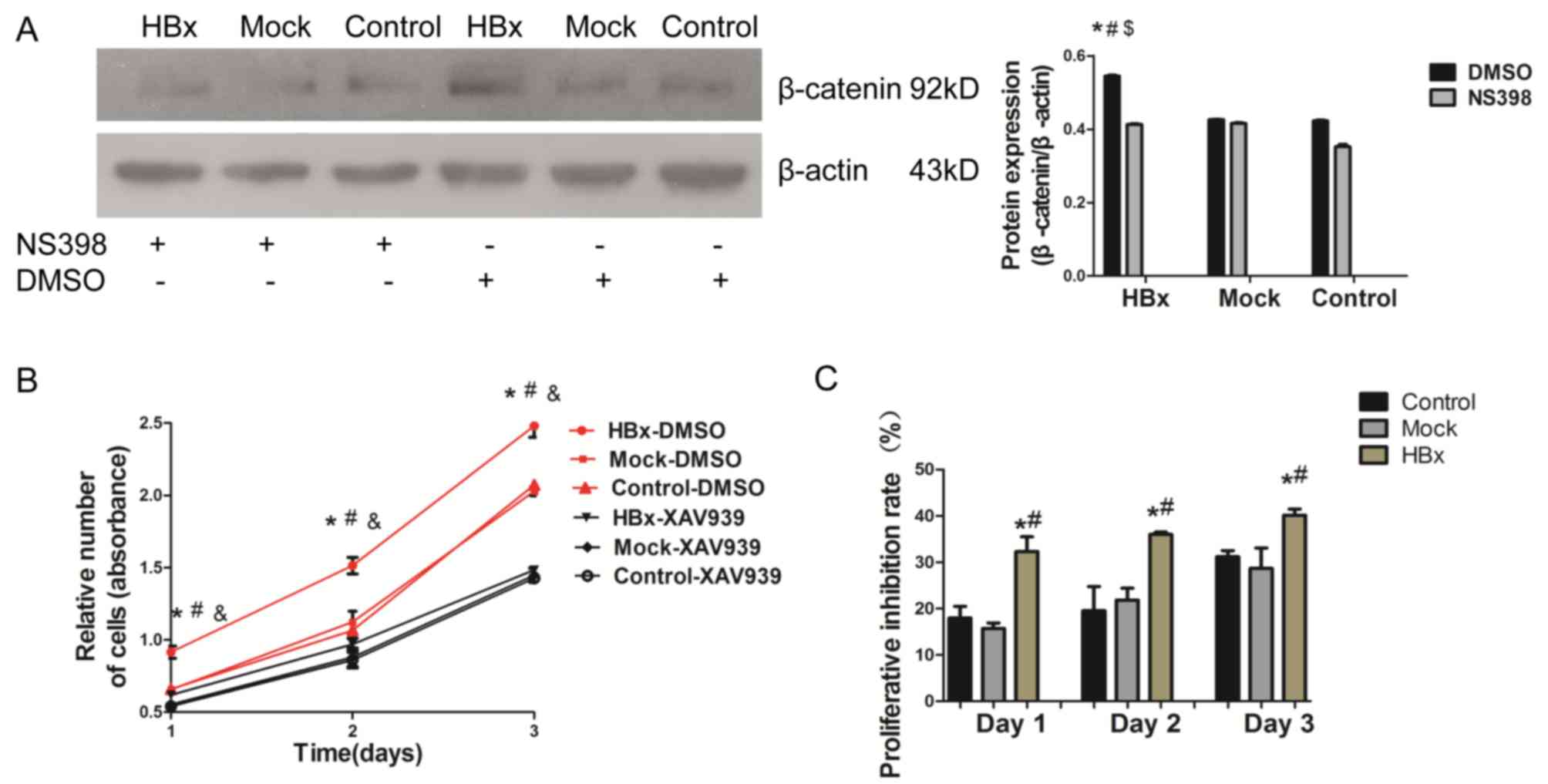 | Figure 4.HBx promotes the proliferative ability
of HL-7702 cells via the COX-2/Wnt/β-catenin pathway. (A) Following
the addition of 50 µmol/l NS-398 and, DMSO as a control, to cells
for 72 h, the expression of the β-catenin protein was examined in
the lysates of HL-7702, HL-7702-mock and HL-7702-HBx cells by
western blotting. (B) Cells were treated with a β-catenin
inhibitor, XAV939 (20 µmol/l), or DMSO as a control. Following
incubation for 24,48 and 72 h, cell proliferation was determined by
a Cell Counting Kit-8 assay. (C) Proliferation inhibition rate was
calculated. Proliferation inhibition rate (%)=(1-
OD450/OD450 with XAV939)×100%. *P<0.05 vs.
HL-7702-mock cells and #P<0.05 vs. HL-7702 cells.
$P<0.05 vs. HL-7702-HBx cells with NS398.
&P<0.05 vs. HL-7702-HBx cells with XAV939. DMSO,
dimethyl sulfoxide; HBx, hepatitis B virus X protein. |
Discussion
HBx, a key regulatory HBV protein that is important
for HBV replication, is involved in the initiation and progression
of HCC (20). Research has
demonstrated that HBx leads to cell proliferation and is involved
in the initiation of HCC (20,21).
A previous study confirmed the colocalization of HBx with COXIII
and the upregulation of ROS generation in HL7702 and HepG2 cells,
and ROS from mitochondria induced COX-2 expression that promoted
HepG2 cell growth (5,6). However, the role of HBx in HL-7702
cells, which are human normal hepatocytes and serve as a
biologically relevant system for examining the role of HBx, remains
unclear. The results of the present study demonstrated that HBx
increased the proliferation rate, led to the formation of more
colonies and shortened the G1 phase in HL-7702 cells. Therefore, it
was concluded that HBx may promote HL-7702 cell proliferation.
Increased COX-2 activity induces proliferation and
mediates the oncogenic action of HBx (8). The present study demonstrated that
COX-2 mRNA and protein expression levels were increased by HBx, and
when cells were treated with NS-398, the colony formation
efficiency in HL-7702-HBx cells was significantly suppressed
following reduced expression of the COX-2 protein. Therefore, it
was concluded that the upregulation of COX-2 may promote
HL-7702-HBx cell proliferation. However, the underlying molecular
mechanism remains unknown. COX-2 was observed to promote cell
proliferation by mediating the activation of downstream oncogenic
pathways (22). Thus, it is
important to investigate the downstream pathways of COX-2 and the
role of HBx in HCC.
The Wnt/β-catenin signaling pathway has long been
considered to be an important developmental pathway in human HCC
(23). It has been reported that
HBx upregulates β-catenin expression during the oncogenesis of
HBV-associated HCC (24). The
present study examined the mRNA and protein expression levels of
β-catenin, which were increased in HL-7702-HBx cells. When
activated, β-catenin is translocated to the nucleus leading to
increased expression of β-catenin targeted genes, including
cyclin-D1 and c-myc (10), which
serve important roles in oncogenesis (12). The present study detected that the
mRNA and protein expression levels of cyclin-D1 and c-myc were
increased. Experimental data has demonstrated that β-catenin is
associated with COX-2 overexpression (13). The Wnt/β-catenin pathway is
activated by COX-2 in gastric cancer (14). The COX-2-mediated Wnt/β-catenin
signaling pathway is involved in HCC (15). These previous results question the
effect of COX-2 in the regulation of β-catenin in HL-7702-HBx
cells. The present study treated cells with NS-398, and the
expression of β-catenin protein was decreased with the
downregulated expression of COX-2. Therefore, the results of the
present study may provide evidence that the upregulation of
β-catenin is activated by COX-2. In addition, the present study
demonstrated that the upregulation of β-catenin resulted in
increased proliferation of HL-7702-HBx cells by adding the
β-catenin inhibitor, XAV939, to the cells. Treatment with XAV939
significantly suppressed cell proliferation. Thus, the results of
the present study demonstrated that that activation of COX-2 may
lead to the upregulation of β-catenin, resulting in HL-7702 cell
proliferation.
In conclusion, the present study demonstrated that
HBx activated the expression of COX-2. In addition, it upregulated
the expression levels of β-catenin and ultimately promoted
HL-7702-HBx cell proliferation. The authors of the present study
hypothesize that therapies aimed at simultaneous disruption of the
COX-2/Wnt/β-catenin pathways may produce effective chemopreventive
and antitumorigenic effects. The present study may provide a novel
insight into the underlying molecular mechanism of HBV-associated
HCC. In future studies, small interfering RNA knockdown may be used
to specifically silence COX-2 and β-catenin, in order to further
examine the role of the COX-2/Wnt/β-catenin pathways in the
oncogenesis of HBV-associated HCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural
Science Foundation of Fujian Province, China (grant no.
2016J05189), the Youth Project of Fujian Provincial Health and
Family Planning Commission (grant no. 2016-1-42) and the Key
Clinical Specialty Discipline Construction Program of Fujian, China
(Min Wei Ke Jiao 2012, No. 49).
Availability of data and materials
The materials described in the manuscript, including
all relevant raw data, will be freely available to any scientist
wishing to use them for non-commercial purposes, without breaching
participant confidentiality.
Author contributions
BYZ and XZW conceived and designed the experiments.
BYZ and WYG performed the experiments; XYH, LYL, XFF and ZXC
analyzed the data; BYZ wrote the paper.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Buendia MA and Neuveut C: Hepatocellular
carcinoma. Cold Spring Harb Perspect Med. 5:a0214442015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lamontagne RJ, Bagga S and Bouchard MJ:
Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res.
2:163–186. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma J, Sun T, Park S, Shen G and Liu J: The
role of hepatitis B virus X protein is related to its differential
intracellular localization. Acta Biochim Biophys Sin (Shanghai).
43:583–588. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gao WY, Li D, Cai DE, Huang XY, Zheng BY,
Huang YH, Chen ZX and Wang XZ: Hepatitis B virus X protein
sensitizes HL-7702 cells to oxidative stress-induced apoptosis
through modulation of the mitochondrial permeability transition
pore. Oncol Rep. 37:48–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zou LY, Zheng BY, Fang XF, Li D, Huang YH,
Chen ZX, Zhou LY and Wang XZ: HBx co-localizes with COXIII in
HL-7702 cells to upregulate mitochondrial function and ROS
generation. Oncol Rep. 33:2461–2467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng BY, Fang XF, Zou LY, Huang YH, Chen
ZX, Li D, Zhou LY, Chen H and Wang XZ: The co-localization of HBx
and COXIII upregulates COX-2 promoting HepG2 cell growth. Int J
Oncol. 45:1143–1150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li D, Ding J, Chen Z, Chen Y, Lin N, Chen
F and Wang X: Accurately mapping the location of the binding site
for the interaction between hepatitis B virus X protein and
cytochrome c oxidase III. Int J Mol Med. 35:319–324. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cheng AS, Yu J, Lai PB, Chan HL and Sung
JJ: COX-2 mediates hepatitis B virus X protein abrogation of
p53-induced apoptosis. Biochem Biophys Res Commun. 374:175–180.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen H, Cai W, Chu ESH, Tang J, Wong CC,
Wong SH, Sun W, Liang Q, Fang J, Sun Z and Yu J: Hepatic
cyclooxygenase-2 overexpression induced spontaneous hepatocellular
carcinoma formation in mice. Oncogene. 36:4415–4426. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vilchez V, Turcios L, Marti F and Gedaly
R: Targeting Wnt/β-catenin pathway in hepatocellular carcinoma
treatment. World J Gastroenterol. 22:823–832. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xie Q, Chen L, Shan X, Shan X, Tang J,
Zhou F, Chen Q, Quan H, Nie D, Zhang W, et al: Epigenetic silencing
of SFRP1 and SFRP5 by hepatitis B virus X protein enhances hepatoma
cell tumorigenicity through Wnt signaling pathway. Int J Cancer.
135:635–646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Daud M, Rana MA, Husnain T and Ijaz B:
Modulation of Wnt signaling pathway by hepatitis B virus. Arch
Virol. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lim K, Han C, Xu L, Isse K, Demetris AJ
and Wu T: Cyclooxygenase-2-derived prostaglandin E2 activates
beta-catenin in human cholangiocarcinoma cells: Evidence for
inhibition of these signaling pathways by omega 3 polyunsaturated
fatty acids. Cancer Res. 68:553–560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu N, Zhou N, Chai N, Liu X, Jiang H, Wu
Q and Li Q: Helicobacter pylori promotes angiogenesis depending on
Wnt/beta-catenin-mediated vascular endothelial growth factor via
the cyclooxygenase-2 pathway in gastric cancer. BMC Cancer.
16:3212016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qinglin L, Xin W, Zhong L, Fang L, Cao G
and Huang P: A study on the anti-tumor mechanism of total
flavonoids from Radix Tetrastigmae against additional cell line
based on COX-2-mediated Wnt/β-catenin signaling pathway.
Oncotarget. 8:54304–54319. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang R, Kong F, Hu L, You H, Zhang P, Du W
and Zheng K: Role of hepatitis B virus X protein in regulating LIM
and SH3 protein 1 (LASP-1) expression to mediate proliferation and
migration of hepatoma cells. Virol J. 9:1632012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lim W, Kwon SH and Cho H, Kim S, Lee S,
Ryu WS and Cho H: HBx targeting to mitochondria and ROS generation
are necessary but insufficient for HBV-induced cyclooxygenase-2
expression. J Mol Med (Berl). 88:359–369. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang HJ, Jiang JH, Yang YT, Yang XD, Guo
Z, Qi YP, Zeng FH, Zhang KL, Chen NZ, Xiang BD and Li LQ:
Cyclooxygenase-2 expression is associated with initiation of
hepatocellular carcinoma, while prostaglandin receptor-1 expression
predicts survival. World J Gastroenterol. 22:8798–8805. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tian Y, Xiao X, Gong X, Peng F, Xu Y,
Jiang Y and Gong G: HBx promotes cell proliferation by disturbing
the cross-talk between miR-181a and PTEN. Sci Rep. 7:400892017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shen L, Zhang X, Hu D, Feng T, Li H, Lu Y
and Huang J: Hepatitis B virus X (HBx) play an anti-apoptosis role
in hepatic progenitor cells by activating Wnt/β-catenin pathway.
Mol Cell Biochem. 383:213–222. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sobolewski C, Cerella C, Dicato M,
Ghibelli L and Diederich M: The role of cyclooxygenase-2 in cell
proliferation and cell death in human malignancies. Int J Cell
Biol. 2010:2151582010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang X, Wang Y, Fan Z, Ji G, Wang M, Lin
J, Huang S and Meltzer SJ: Klotho: A tumor suppressor and modulator
of the Wnt/β-catenin pathway in human hepatocellular carcinoma. Lab
Invest. 96:197–205. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun Q, Wang R, Luo J, Wang P, Xiong S, Liu
M and Cheng B: Notch1 promotes hepatitis B virus X protein-induced
hepatocarcinogenesis via Wnt/β-catenin pathway. Int J Oncol.
45:1638–1648. 2014. View Article : Google Scholar : PubMed/NCBI
|