Introduction
Nasopharyngeal carcinoma (NPC) is the most common
cancer occurring in certain regions of East Asia and Africa
(1). Over 84,000 new NPC cases are
diagnosed annually, with ~80% occurred in Asia (2,3). All
of viral, dietary, genetic and environmental factors contribute to
the risk of NPC development (3,4). The
main strategies against primary NPC are radiotherapy and
comprehensive chemotherapy (5,6).
However, the 5-year survival rate was improved to 70%, it is
estimated that 15–58% of patients with NPC experience recurrence of
the disease and have to undergo re-treatment (7,8).
Local relapse and distant metastasis continue to be a global
problem in the treatment of NPC due to the failure to eradicate all
tumor cells with conventional treatment. Drug-resistant tumor cells
survive after treatment and develop into a new tumor mass or become
metastatic (9). Therefore, it is
important to develop potential treatments as an efficient and novel
therapy against NPC.
The tripartite motif protein, tripartite
motif-containing 24 (TRIM24) was identified as a co-regulator of
retinoid signaling. It was previously determined to be involved in
the progression of several human cancers (10). The aberrant overexpression of
TRIM24 may promote tumor development by multiple mechanisms. TRIM24
was reported as a target of chromosomal translocations to form
oncogenic fusion proteins in acute promyelocytic leukaemia,
papillary thyroid carcinoma and myeloproliferative syndrome
(11,12). Additionally, elevated expression of
TRIM24 may promote progression of prostate cancer (13) and may be negatively correlated with
survival of patients with breast cancer (14). A previous study determined that
TRIM24 binds to chromatin and oestrogen receptor to activate
oestrogen-dependent genes-associated to cellular proliferation and
tumor development (15).
Additionally, the expression level of p53 may be negatively
regulated by TRIM24 resulting in the inhibited tumor suppression
ability (16,17). However, previous studies have
determined the important role of TRIM24 in several human cancers
(13–15), and the association between TRIM24
and NPC was not fully reported, which suggested that further
investigation in the role of TRIM24 in NPC is required in order to
determine the possible action mechanisms in tumor development of
NPC. The present study detected the expression level of TRIM24 in
NPC tissues and evaluated the inhibition ability of TRIM24 small
interfering RNA (siRNA) in cell proliferation of NPC cells using
NPC cell lines HONE1 and CNE1 transfected with TRIM24 siRNA.
Biochemical tests, western blotting and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) were
used to determine the expression level of apoptosis, metastasis and
signaling pathway-associated proteins in order to determine the
role of TRIM24 in tumor development of NPC. The present study aimed
to determine the association between TRIM24 with tumor development
in NPC and provide the theoretical basis of potential therapies for
clinical treatment NPC.
Materials and methods
Cell culture and transfection
A total of 5 NPC cell lines HONE1 (Shanghai Jining
Shiye, Shanghai, China), C666-1 (Shanghai Gaining Biotechnology
Co., Ltd., Shanghai, China), CNE2, CNE1 [both from Sai Bai Sheng
(Shanghai) Biotechnology Co., Ltd., Shanghai, China] and SUNE-1
[Kelton Biotech (Shanghai) Co., Ltd., Shanghai, China] were
cultured in Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA) containing 10% fetal calf
serum (Gibco; Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) and 1% 100 × mycillin (Beijing Solarbio Science &
Technology Co., Ltd., Beijing, China) with 5% CO2 at
37°C. NPC cell lines with high expression level of TRIM24 were
determined by using RT-qPCR analysis. Cell viability of selected
cell lines (HONE1 and CNE1) was detected at 95% using Trypan blue
staining for 5 min at room temperature. Then the cells were
digested and seeded into 6-well plate (5×105 cells/well)
prior to transfection. Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to transfect 5 µl TRIM24 siRNA
(Shanghai GenePharma Co., Ltd., Shanghai, China) or negative
control siRNA (Shanghai GenePharma Co., Ltd.) into HONE1 and CNE1
cell lines. After incubation for 48 h, the transfected cells were
collected and processed for cell viability, apoptosis, biochemical
tests, RT-qPCR and western blot assay.
Western blot assay
Western blot assay was used to determine the
expression levels of TRIM24 and associated proteins in NPC cell
lines. Cultured or transfected cells were washed with 1xPBS twice,
followed by the radioimmunoprecipitation assay lysis buffer
(Beijing Solarbio Science & Technology Co., Ltd.) at 4°C.
Following lysis, samples were centrifuged at 12,000 × g for 15 min
at 4°C and the supernatant was collected. Proteins were quantified
by using the bicinchoninic acid assay. Protein (15 µl) with loading
buffer were heated in a boiling water bath for 10 min and then
centrifuged at 12,000 × g for 10 min at room temperature. The
supernatant (30 µg protein) was collected and loaded on 10%
SDS-polyacrylamide gels. Following electrophoresis, proteins on the
gel were transferred onto a nitrocellulose blotting membrane (EMD
Millipore, Billerica, MA, USA) and incubated with 5% non-fat milk
at 4°C overnight. The membrane was washed and incubated with TRIM24
(Abcam, Cambridge, UK; cat. no. Ab174287; 1:1,000), vascular
endothelial growth factor (VEGF; Abcam; cat. no. Ab46154; 1:1,000),
VEGF receptor 2 (VEGFR2; Affinity, Cincinnati, OH, USA; cat. no.
AF6281; 1:1,000), caspase-3 (Abcam; cat. no. Ab44976; 1:500),
caspase-9 (Abcam; cat. no. Ab2013; 1:1,000), janus kinase 2 (JAK2;
Cell Signaling Technology, Inc., Danvers, MA, USA; cat. no. 3230;
1:1,000), phosphorylated (p)-JAK2 (Cell Signaling Technology, Inc.;
cat. no. 8082; 1:1,000), signal transducer and activator of
transcription 3 (STAT3; Abcam; cat. no. Ab76315; 1:2,000), p-STAT3
(cat. no. 9139; 1:1,500) and GAPDH (both Cell Signaling Technology,
Inc.; cat. no. 5174; 1:2,000) antibodies for 2 h at room
temperature. Then the membrane was washed with TBS-0.05% Tween-20
and incubated with anti-horseradish peroxidase secondary antibody
(Beyotime Institute of Biotechnology, Shanghai, China; cat. nos.
A0208 and A0216; 1:1,000 dilution) for 1 h. Immunoreactive bands
were detected using an enhanced chemiluminescence detection kit and
an LAS-4000 mini system (Fujifilm Corporation, Kumamoto, Japan) was
used for visualization. Western blotting was repeated three times.
The films of western blotting were then scanned by using a Bio-Rad
imaging densitometer (ModelGS-700; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) and the densities of the bands were
semi-quantified using Quantity One software Version 4.62 (Bio-Rad
Laboratories, Inc.).
RT-qPCR analysis
PCR analysis was preformed to determine the
expression level of TRIM24 in NPC tissue samples and associated
proteins in NPC cell lines. The gene expression of TRIM24 in 35
tissue samples with para-carcinoma tissues were determined using
RT-qPCR. Paired human NPC tissues and para-carcinoma tissues were
collected from 35 patients (including 23 males and 12 females, age
range: 31 to 74 years) that underwent standard surgical procedures
between January 2014 and December 2014 in Department of
Otolaryngology, Zhongshan Hospital (Shanghai, China) after the
written informed consent of the patients was obtained. The
experimental protocols were approved by the Institutional Review
Committees of Zhongshan Hospital (Shanghai, China). Tissue samples
were collected from patients with complete clinical and
pathological follow-up data.
Total RNA was extracted from tissue samples by using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
Reverse transcription kit (Fermentas; Thermo Fisher Scientific,
Inc.) was used to syntheses cDNA through a total volume of 25 µl
(12 µl RNA-primer mix, 5 µl 5XRT reaction buffer, 1 µl 25 mM dNTPs,
1 µl 25 U/µl RNase inhibitor, 1 µl 200 U/µl M-MLV reverse
transcriptase, 1 µl Oligo(dt)18 and 4 µl DNase-free
ddH2O) and the thermocycling conditions were: 37°C for
60 min, 85°C for 5 min and 4°C for 5 min for the RT step. Then cDNA
was amplified by using SYBR-Green PCR kit (Thermo Fisher
Scientific, Inc.) using a total PCR system of 25 µl (12.5 µl
SYBR-Green Mix, 0.5 µl forward primer, 0.5 µl reverse primer, 9.5
µl ddH2O and 2 µl cDNA template) with the PCR
thermocycling conditions were as follows: 10 min at 95°C, and then
15 sec at 95°C and 45 sec at 60°C for 40 cycles. Amplification
kinetic curves were obtained through 15 sec at 95°C, 1 min at 60°C,
15 sec at 95°C and 15 sec at 60°C. Data was collected by using ABI
Prism 7300 SDS software (Applied Biosystem, Thermo Fisher
Scientific, Inc.) and quantified as previously described (18). Primers used for the amplification
were listed in Table I.
 | Table I.Primers for quantitative polymerase
chain reaction analysis. |
Table I.
Primers for quantitative polymerase
chain reaction analysis.
| Gene | Forward (5′-3′) | Reverse (5′-3′) | Amplicon size
(bp) |
|---|
| TRIM24 |
CCAGCCAAGACCACCCTCAAAC |
CAGAGCTTCCTCGGCTTCCAAC | 149 |
| VEGF |
ATTTCTGGGATTCCTGTAG |
CAGTGAAGACACCAATAAC | 157 |
| VEGFR2 |
CTCAGCAGGATGGCAAAG |
ACTGTCCGTCTGGTTGTC | 278 |
| Caspase-3 |
AACTGGACTGTGGCATTGAG |
ACAAAGCGACTGGATGAACC | 161 |
| Caspase-9 |
CCTCACCCTGCCTTATCTTG |
TCCCTCTTCCTCCACTGTTC | 189 |
| GAPDH |
AATCCCATCACCATCTTC |
AGGCTGTTGTCATACTTC | 218 |
Cell Counting kit-8 (CCK-8) assay
Cultured HONE1 and CNE1 cells were trypsinized and
diluted to 2×104 cells/ml. Then cells were plated into
96-well plates (2×103 cells/well) and incubated at 37°C
for 12 h, followed by the transfection with TRIM24 siRNA or
negative control siRNA, respectively. Transfected cells in each
well were mixed with 100 µl DMEM containing 10% CCK-8 (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) at 0, 24, 48 and 72
h, and then incubated in 5% CO2 at 37°C for 1 h. The
optical density (OD)450 nm of cell suspension was
measured using a spectrophotometer in order to evaluate the cell
viability of HONE1 and CNE1 cells.
Flow cytometry
Transfected cells were formed into a single cell
suspension by 0.25% trypsin and washed by 10% PBS, followed by the
centrifugation at 1,000 × g for 5 min at 4°C. Supernatant was
discarded and then the cells were incubated with Annexin-V
fluorescein isothiocyanate (FITC) apoptosis detection kit (BD
Biosciences, Franklin Lakes, NJ, USA) for 10 min at room
temperature without light. Cell apoptosis rate was measured by
using flow cytometry (FACS Calibur, BD Biosciences) and FlowJo
Software version 7.6 (FlowJo LLC, Ashland, OR, USA).
Biochemical test
Caspase-3 and caspase-9 concentrations were assessed
using Caspase-3 and caspase-9 colorimetric assay kits (KeyGen
Biotech Co., Ltd., Shanghai, China). Cultured and transfected cells
were collected and washed by using PBS for twice, followed with
lysis with 100 µl pre-cooled lysis buffer containing 1 µl
dithiothreitol (DTT). Lysed cells were centrifuged at 12,000 × g
for 1 min. Supernatant was collected and protein concentrations
were determined using the Bradford method. 50 µl supernatant was
mixed with 50 µl 2X Reaction Buffer containing 0.5 µl DTT and 5 µl
Caspase-3 or Caspase-9 substrate, respectively. After 4 h of
incubation at 37°C, the concentrations of Caspase-3 and Caspase-9
were measured at OD 400 nm.
Statistical analysis
All the experiments were repeated three times and
the values are expressed as the mean ± standard deviation.
Statistical comparisons were performed using one-way analysis of
variance followed by the Tukey post hoc test with GraphPad Prism
software (version 6.0; Graphpad Software, Inc., San Diego, CA,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Screening and identification of
nasopharyngeal cancer cell lines
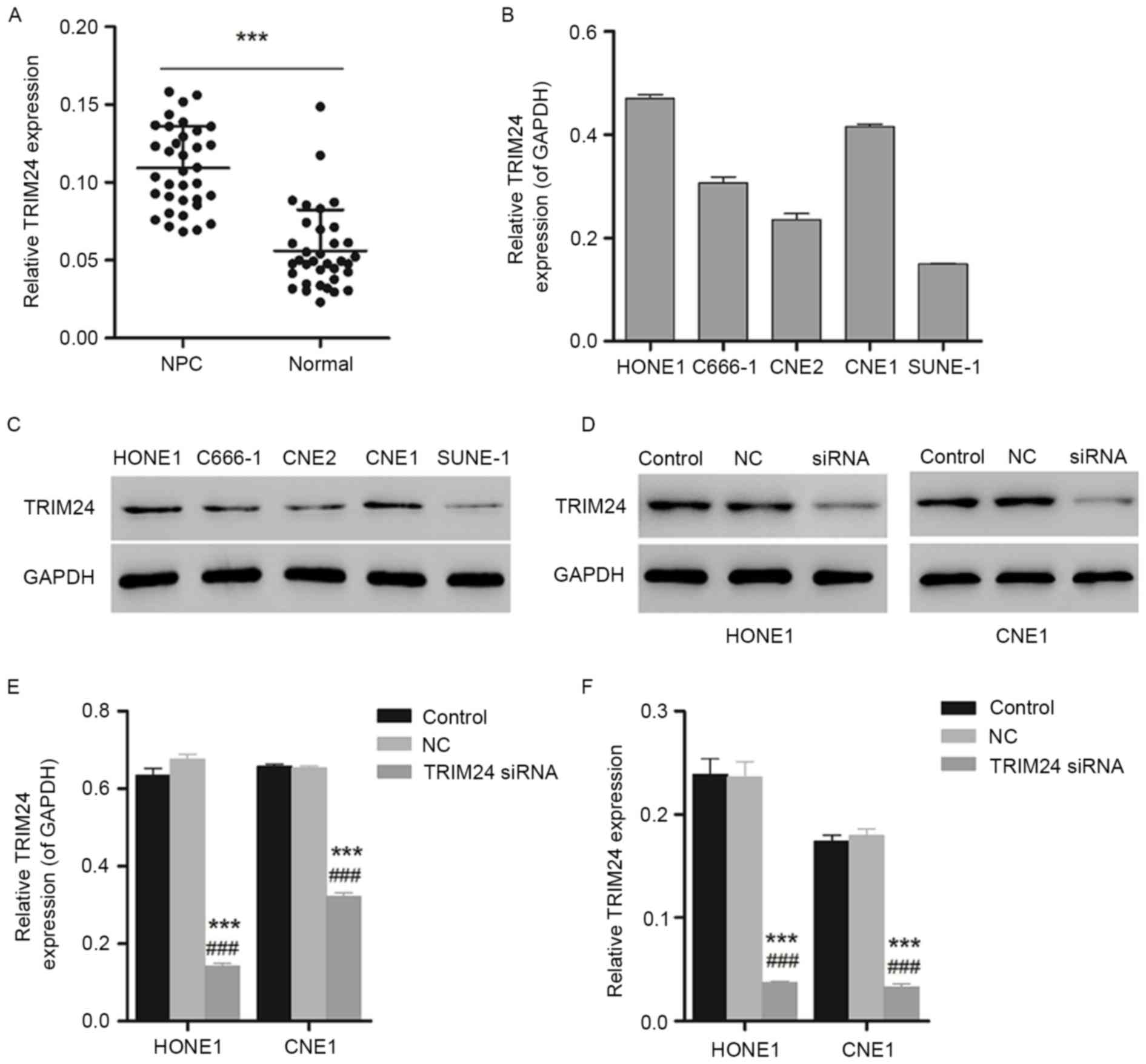
As presented in Fig.
1A, the increased expression level of TRIM24 measured by RT-PCR
in NPC tissues demonstrated a significant difference compared with
the paracancerous tissues, indicating the overexpression of TRIM24
in NPC tissues and suggesting an association between TRIM24 and
NPC. To further investigate the association between TIRM24 and NPC,
NPC cell lines HONE1 and CNE1 with higher expression levels of
TRIM24, as determined using western blotting, were selected for
further experiments (Fig. 1B and
C).
Furthermore, western blotting and qPCR were used to
identify the interference effect of TRIM24 siRNA on the expression
of TRIM24 in HONE1 and CNE1 cells. As presented in Fig. 1D and E, the expression level of
TRIM24 declined significantly in HONE1 and CNE1 cells (P<0.05 or
P<0.001). The gene expression of TRIM24 was additionally
decreased, with a significant difference compared with the negative
control group (Fig. 1F;
P<0.001), indicating the efficient gene silencing of TRIM24
siRNA.
TRIM24 siRNA inhibits cell
proliferation
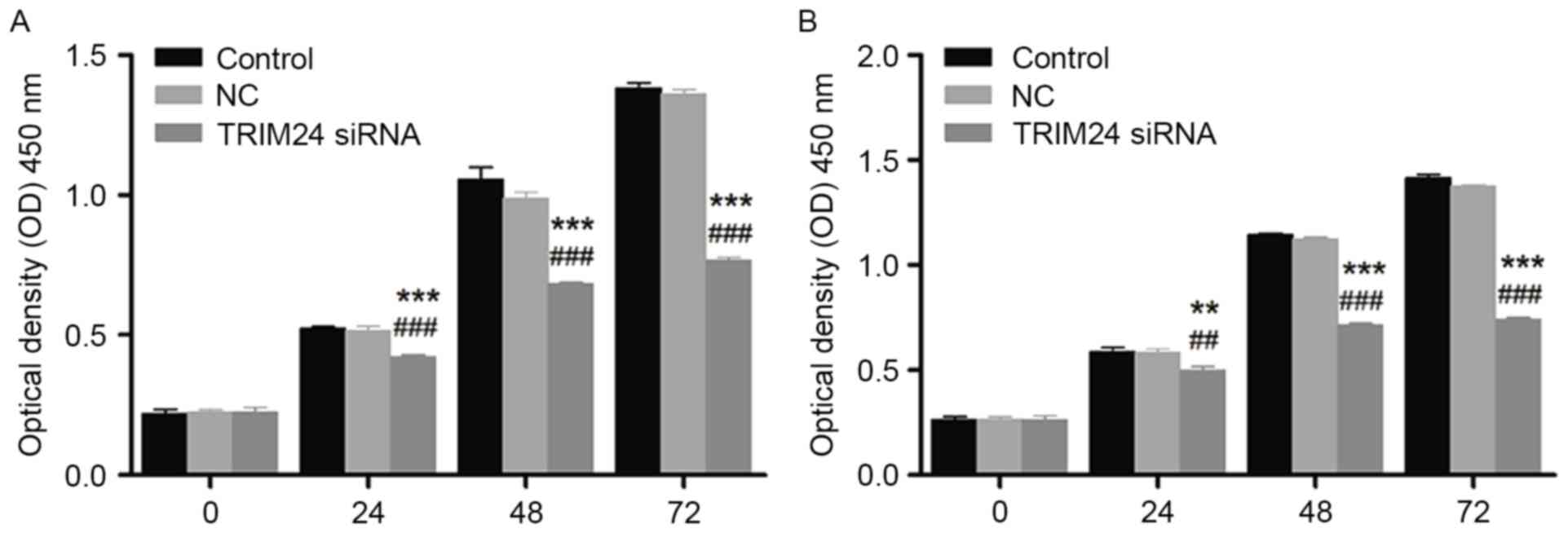
As presented in Fig.
2, the cell viability of HONE1 and CNE1 cells detected via CCK8
assay was decreased following treatment with TRIM24 siRNA, in a
time-dependent manner. The decreased cell viability was significant
compared with the control and negative control groups, indicating
the inhibitory effect of TRIM24 siRNA on cell proliferation in
HONE1 and CNE1 cells.
TRIM24 siRNA induces cellular
apoptosis
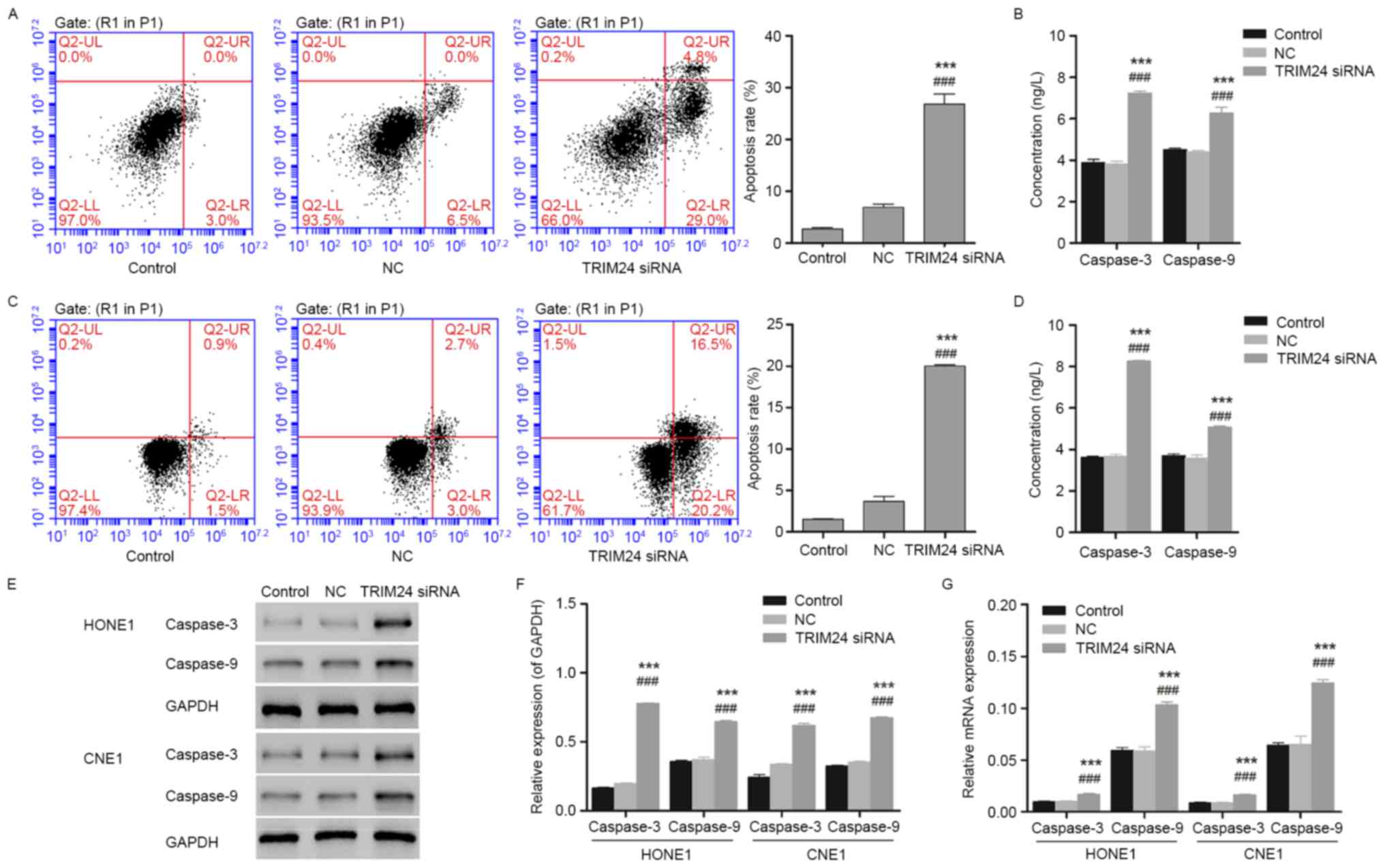
The results of the flow cytometry demonstrated
cellular apoptosis in HONE1 and CNE1 cells induced by TRIM24 siRNA
(Fig. 3). The percentages of early
apoptotic cells presented in the lower right quadrant of the
histograms indicated the induced cellular apoptosis in the TRIM24
siRNA group, suggesting an apoptosis-promoting effect of TRIM24
siRNA on HONE1 and CNE1 cells. Furthermore, the expression of the
pro-apoptotic proteins caspase-3 and caspase-9, measured using
biochemical analysis, exhibited an upregulation following treatment
with TRIM24 siRNA, which corresponded to the results of the flow
cytometry (Fig. 3B and D).
Additionally, the upregulated caspase-3 and caspase-9 measured by
western blotting and qPCR indicated the pro-apoptotic ability of
TRIM24 siRNA on HONE1 and CNE1 cells, suggesting an important role
for TRIM24 in NPC (Fig. 3E-G).
TRIM24 siRNA blocks the JAK2/STAT3
signaling pathway
To further investigate the possible mechanisms
involved in inhibited cellular apoptosis and cell proliferation due
to TRIM24 siRNA, the expression levels of the cellular apoptosis-
and tumorigenesis-associated proteins caspase-3, caspase-9, VEGF
and VEGFR2 in HONE1 and CNE1 cells was measured by western blotting
and qPCR. Compared with the control group, the expression levels of
VEGF and VEGFR2 exhibited a downregulation with a significant
difference in HONE1 and CNE1 cells transfected with TRIM24 siRNA
(Fig. 4A and B; n=3; P<0.001).
Similar results were observed with the PCR analysis (Fig. 4C). Besides, the expression levels
of the pro-apoptotic proteins caspase-3 and caspase-9 exhibited an
upregulation, corresponding to the results of the flow cytometry.
In addition, the expression of the JAK2/STAT3 signaling
pathway-associated proteins JAK2 and STAT3 was measured using
western blotting and qPCR. As presented in Fig. 4A, D and E, decreased expression of
JAK2 and STAT3 indicated the inhibition of the JAK2/STAT3 signaling
pathway. However, further evidence is required to demonstrate the
direct action of siTRIM24 on the inactivation of JAK2/STAT3
signaling pathway.
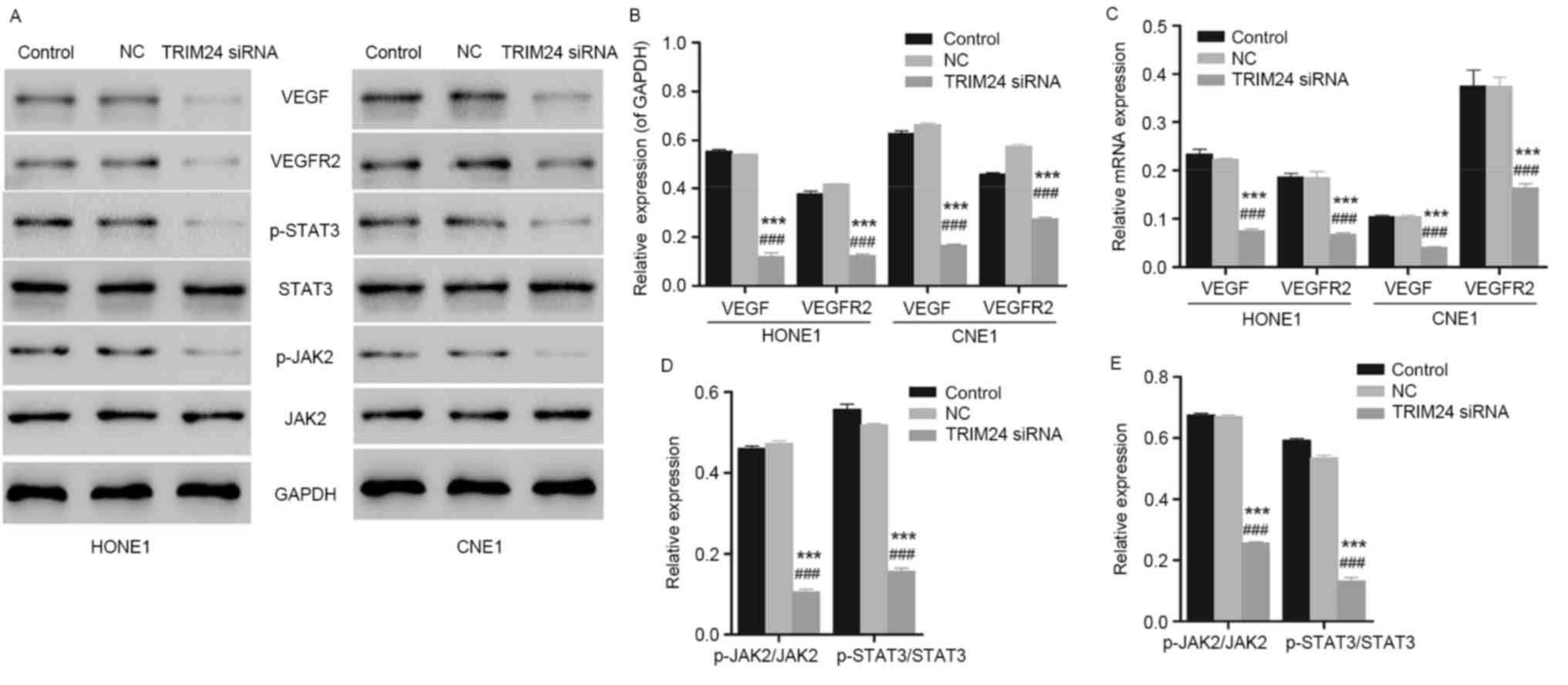 | Figure 4.Expression level of VEGF, VEGFR2, JAK2
and STAT3 in HONE1 and CNE1 cells, determined using western
blotting and qPCR. (A) Detection and visualization of
immunoreactive bands via western blot analysis. (B) HONE1 and CNE1
cells with TRIM24 silenced had downregulated protein expression of
VEGF and VEGFR2, indicating the inhibited angiogenesis.
***P<0.001 vs. control group. ###P<0.001 vs. NC
group (n=3). (C) HONE1 and CNE1 cells transfected with TRIM24 siRNA
exhibited reduced mRNA expression of VEGF and VEGFR2, corresponding
to the results obtained in the western blot analysis. ***P<0.001
vs. control group. ###P<0.001 vs. NC group (n=3). (D)
HONE1 cells with TRIM24 silenced presented downregulated JAK2 and
STAT3, indicating the blocked JAK2/STAT3 signaling pathway.
***P<0.001 vs. control group. ###P<0.001 vs. NC
group (n=3). (E) Similar results were obtained in CNE1 cells.
***P<0.001 vs. control group. ###P<0.001 vs. NC
group (n=3). NC, negative control; TRIM24, tripartite motif
containing 24; siRNA, small interfering RNA; VEGF, vascular
endothelial growth factor; VEGFR2, VEGF receptor 2; STAT3, signal
transducer and activator of transcription 3; p-STAT3,
phosphorylated-STAT3; JAK2, janus kinase 2; p-JAK2,
phosphorylated-JAK2. |
Discussion
TRIM24 belongs to the tripartite motif superfamily
and was initially identified as a fusion partner of the B-raf
protein in the oncoprotein T18, observed in mouse hepatocellular
carcinoma (19). TRIM24 has been
reported to be overexpressed in a number of types of cancer.
Upregulated TRIM24 was observed in acute promyelocytic leukaemia,
myeloproliferative syndrome, papillary thyroid carcinoma, breast
cancer and non-small cell lung cancer (11,12,19,20),
which indicated that TRIM24 may serve an oncogenic role in
carcinogenesis. In addition, overexpression of TRIM24 is associated
with survival in breast cancer patients (14), and with pTNM stage and
differentiation in non-small cell lung cancer (20). These previous studies suggested
that the TRIM24 gene may serve an important role in tumorigenesis.
In the present study, it was demonstrated that the expression of
TRIM24 in NPC tissues was increased compared with noncancerous
tissues. These findings suggested that TRIM24 may serve an
important role in the progression of human NPC.
In order to confirm the potential role of TRIM24 in
human NPC development, the present study employed siRNA to knock
down TRIM24 in HONE1 and CNE1 cell lines for further study. A CCK-8
assay was used to investigate the role of TRIM24 in cell
proliferation. The cell proliferation of HONE1 and CNE1 cells
following TRIM24 silencing was downregulated compared with the
control group, which suggested that TRIM24 may promote cell
proliferation in HONE1 and CNE1 cells. Subsequently, cell apoptosis
of HONE1 and CNE1 cells with TRIM24 knockdown, and the
apoptosis-associated proteins caspase-3 and caspase-9, were
investigated using an Annexin/V kit and biochemical assays.
Caspase-3 activated by apoptotic signals acts on peptide chains and
splits substrates, resulting in the apoptosis of cells (21,22).
Caspase-3 serves a direct role in cellular apoptosis as one of the
effector caspases. It was previously reported that caspase-3
induced the activation of caspase-activated deoxyribonuclease (CAD)
in the degradation of DNA. When apoptosis occurs, CAD with nuclease
activity is released by its inhibitor ICAD, activated by
caspase-3 (23). The nuclear
lamina, which is responsible for the stability of chromatin, may be
cut by caspase-3 at a single site, resulting in the degradation of
chromatin (23). The results of
the flow cytometry analysis performed in the present study
demonstrated a significant increase in the population of apoptotic
HONE1 cells following treatment with TRIM24 siRNA compared with the
negative control, in addition to CNE1 cells. Biochemical and
western blot analyses demonstrated that the depletion of TRIM24
upregulated caspase-3 and caspase-9 expression. These results
suggested that TRIM24 may suppress cellular apoptosis and
differentiation via caspase-3 and caspase-9. The above results may
explain the mechanism of TRIM24 in human NPC cell
differentiation.
One of the most intractable characteristics of NPC
is metastasis (24). Tumor
angiogenesis factors involved in migration serve an important role
in the generation of tumor vessels, resulting in the uncontrolled
growth of the tumor (25). The
process of angiogenesis, including the proliferation and migration
of endothelial cells, the degradation of the basement membrane and
the formation of the cavity, is a complex process regulated by a
number of positive and negative regulatory factors (25). VEGF is a type of specific
heparin-binding growth factor in vascular endothelial cells. VEGF,
VEGFA and VEGFB have been demonstrated to induce angiogenesis
through the recruitment of tumor-associated macrophages, which
serve an important role in the angiogenesis and growth of tumor
cells in gastric cancer (26). The
association between the high expression of VEGF and metastasis,
angiogenesis and survival in rectal cancer has additionally been
reported (27). According to the
results obtained in the western blot and PCR analyses in the
present study, downregulated expression levels of VEGF and VEGFR2,
in HONE1 and CNE1 cells following treatment with TRIM24 siRNA,
suggested that silenced TRIM24 decreased angiogenesis and the
metastatic ability of NPC cells significantly compared with the
negative control. Furthermore, these results indicated an
association between TRIM24, and a higher incidence of metastasis
and recurrence in NPC.
The JAK2/STAS3 signaling pathway, with a range of
functions including proliferation, differentiation, apoptosis, cell
cycle and immunoregulation, is a signal transduction pathway
stimulated by cytokines (28,29).
JAK2 is a type of non-transmembrane tyrosine kinase. STAT3 serves
an important role in signal transduction and transcriptional
activation. Constitutive activation of JAK2 has been reported in
childhood T cell acute lymphoblastic leukemia (30). STAT3 has been observed to be
constitutively activated in breast carcinoma and non-small cell
lung cancer, correlated with induced cell proliferation and
inhibited cellular apoptosis (31). It has been reported that
suppression of the JAK2/STAT3 signaling pathway inhibited cell
growth and induced cellular apoptosis in various cancer cells
(32). Previous studies revealed
suppressed cell growth, induced cellular apoptosis and arrest of
the cell cycle in colorectal cancer cells due to blocked JAK2/STAT3
signaling (33). In the present
study, decreased JAK2 and STAT3 expression levels indicated the
inhibition of the JAK2/STAS3 signaling pathway in HONE1 and CNE1
cells, resulting in the inhibited cell proliferation and induced
cellular apoptosis. However, there is no direct evidence indicating
the association between the regulation of siTRIM24 and the
inhibition of the JAK2/STAT3 signaling pathway at present.
Overexpression of TRIM24 was identified in tumor growth due to the
activation of the upstream extracellular signal-regulated kinase
signaling pathway (34), while the
interaction between TRIM24 and JAK2/STAT3 remains unclear. It was
hypothesized that the inhibition of the JAK2/STAT3 signaling
pathway may be involved in the regulation of TRIM24. Further
studies are required to investigate the possible mechanisms,
including the functional channels involved.
In conclusion, the present demonstrated the
overexpression of TRIM24 in human NPC and the important role of
TRIM24 in the growth of NPC cells. Furthermore, silencing of TRIM24
may cause inhibited cell proliferation and induced cellular
apoptosis in NPC cells. The limitation of the present study was
that HONE 1, CNE1 and CNE2 cells may have been contaminated with
HeLa cells or cells of unknown origin. Further experiments with
validated NPC cells may be needed.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the study
are available from the corresponding author on reasonable
request.
Authors' contributions
PW conceived and designed the study. PW, NS, DL, XN,
DW and XH performed the experiments. PW and NS wrote the
manuscript. All authors read and approved the manuscript.
Ethics approval and consent to
participate
The experimental protocols were approved by the
Institutional Review Committees of ZhongShan Hospital (Shanghai,
China). Written informed consent of the patients was obtained.
Consent for publication
Written informed consent of the patients was
obtained.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Lee AW, Ng W, Chan Y, Sze H, Chan C and
Lam T: The battle against nasopharyngeal cancer. Radiother Oncol.
104:272–278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qu S, Liang ZG and Zhu XD: Advances and
challenges in intensity-modulated radiotherapy for nasopharyngeal
carcinoma. Asian Pac J Cancer Prev. 16:1687–1692. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hashim Nor NA, Ramzi NH, Velapasamy S,
Alex L, Chahil JK, Lye SH, Munretnam K, Haron MR and Ler LW:
Identification of genetic and non-genetic risk factors for
nasopharyngeal carcinoma in a southeast asian population. Asian Pac
J Cancer Prev. 13:6005–6010. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li JX, Lu TX, Huang Y and Han F: Clinical
characteristics of recurrent nasopharyngeal carcinoma in
high-incidence area. ScientificWorldJournal. 2012:7197542012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chau RM, Teo PM, Kam MK, Leung S, Cheung K
and Chan AT: Dosimetric comparison between 2-dimensional radiation
therapy and intensity modulated radiation therapy in treatment of
advanced T-stage nasopharyngeal carcinoma: To treat less or more in
the planning organ-at-risk volume of the brainstem and spinal cord.
Med Dosim. 32:263–270. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen AM, Yang C, Marsano J, Liu T and
Purdy J: Intensity-modulated radiotherapy for nasopharyngeal
carcinoma: Improvement of the therapeutic ratio with helical
tomotherapy vs segmental multileaf collimator-based techniques. Br
J Radiol. 85:e537–e543. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Du L, Zhang XX, Ma L, Feng LC, Li F, Zhou
GX, Qu BL, Xu SP, Xie CB and Yang J: Clinical study of
nasopharyngeal carcinoma treated by helical tomotherapy in China:
5-year outcomes. Biomed Res Int. 2014:9807672014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fang FM, Chien CY, Tsai WL, Chen HC, Hsu
HC, Lui CC, Huang TL and Huang HY: Quality of life and survival
outcome for patients with nasopharyngeal carcinoma receiving
three-dimensional conformal radiotherapy vs. intensity-modulated
radiotherapy-a longitudinal study. Int J Radiat Oncol Biol Phys.
72:356–364. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He X, Ye M, Guo X, Pan Z, Zhang Z, He S
and Liu T: Treatment outcome of patients with stages I–II
nasopharyngeal carcinoma after late course accelerated
hyperfractionation radiotherapy alone. Oral Oncol. 48:1058–1063.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hatakeyama S: TRIM proteins and cancer.
Nat Rev Cancer. 11:792–804. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Belloni E, Trubia M, Gasparini P, Micucci
C, Tapinassi C, Confalonieri S, Nuciforo P, Martino B, Lo-Coco F,
Pelicci PG and Di Fiore PP: 8p11 myeloproliferative syndrome with a
novel t(7; 8) translocation leading to fusion of the FGFR1 and TIF1
genes. Genes Chromosomes Cancer. 42:320–325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhong S, Delva L, Rachez C, Cenciarelli C,
Gandini D, Zhang H, Kalantry S, Freedman LP and Pandolfi PP: A
RA-dependent, tumour-growth suppressive transcription complex is
the target of the PML-RARα and T18 oncoproteins. Nat Genet.
23:287–295. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Groner AC, Cato L, de Tribolet-Hardy J,
Bernasocchi T, Janouskova H, Melchers D, Houtman R, Cato ACB,
Tschopp P, Gu L, et al: TRIM24 is an oncogenic transcriptional
activator in prostate cancer. Cancer Cell. 29:846–858. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chambon M, Orsetti B, Berthe ML,
Bascoul-Mollevi C, Rodriguez C, Duong V, Gleizes M, Thénot S,
Bibeau F, Theillet C and Cavaillès V: Prognostic significance of
TRIM24/TIF-1α gene expression in breast cancer. Am J Pathol.
178:1461–1469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia
W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al: TRIM24
links a non-canonical histone signature to breast cancer. Nature.
468:927–932. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Allton K, Jain AK, Herz HM, Tsai WW, Jung
SY, Qin J, Bergmann A, Johnson RL and Barton MC: Trim24 targets
endogenous p53 for degradation. Proc Natl Acad Sci USA.
106:11612–11616. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jain AK and Barton MC: Regulation of p53:
TRIM24 enters the RING. Cell Cycle. 8:3668–3674. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang S, Minter LC, Stratton SA, Yang P,
Abbas HA, Akdemir ZC, Pant V, Post S, Gagea M, Lee RG, et al:
TRIM24 suppresses development of spontaneous hepatic lipid
accumulation and hepatocellular carcinoma in mice. J Hepatol.
62:371–379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li H, Sun L, Tang Z, Fu L, Xu Y, Li Z, Luo
W, Qiu X and Wang E: Overexpression of TRIM24 correlates with tumor
progression in non-small cell lung cancer. PLoS One. 7:e376572012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Debatin KM: Apoptosis pathways in cancer
and cancer therapy. Cancer Immunol Immunother. 53:153–159. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stennicke HR and Salvesen GS: Biochemical
characteristics of caspases-3,-6,-7, and-8. J Biol Chem.
272:25719–25723. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lavrik IN, Golks A and Krammer PH:
Caspases: Pharmacological manipulation of cell death. J Clin
Invest. 115:2665–2672. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hong B, Lui VW, Hashiguchi M, Hui EP and
Chan AT: Targeting tumor hypoxia in nasopharyngeal carcinoma. Head
Neck. 35:133–145. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nyberg P, Salo T and Kalluri R: Tumor
microenvironment and angiogenesis. Front Biosci. 13:6537–6553.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bai Y, Zhu X, Chao J, Zhang Y, Qian C, Li
P, Liu D, Han B, Zhao L, Zhang J, et al: Pericytes contribute to
the disruption of the cerebral endothelial barrier via increasing
VEGF expression: Implications for stroke. PloS one.
10:e01243622015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Des Guetz G, Uzzan B, Nicolas P, Cucherat
M, Morere JF, Benamouzig R, Breau JL and Perret GY: Microvessel
density and VEGF expression are prognostic factors in colorectal
cancer. Meta-analysis of the literature. Br J Cancer. 94:1823–1832.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Alvarez JV, Greulich H, Sellers WR,
Meyerson M and Frank DA: Signal transducer and activator of
transcription 3 is required for the oncogenic effects of
non-small-cell lung cancer-associated mutations of the epidermal
growth factor receptor. Cancer Res. 66:3162–3168. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Buettner R, Mora LB and Jove R: Activated
STAT signaling in human tumors provides novel molecular targets for
therapeutic intervention. Clin Cancer Res. 8:945–954.
2002.PubMed/NCBI
|
|
30
|
Mullighan CG, Zhang J, Harvey RC,
Collins-Underwood JR, Schulman BA, Phillips LA, Tasian SK, Loh ML,
Su X, Liu W, et al: JAK mutations in high-risk childhood acute
lymphoblastic leukemia. Proc Natl Acad Sci USA. 106:9414–9418.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grandis JR, Drenning SD, Zeng Q, Watkins
SC, Melhem MF, Endo S, Johnson DE, Huang L, He Y and Kim JD:
Constitutive activation of Stat3 signaling abrogates apoptosis in
squamous cell carcinogenesis in vivo. Proc Natl Acad Sci.
97:4227–4232. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xiong H, Zhang ZG, Tian XQ, Sun DF, Liang
QC, Zhang YJ, Lu R, Chen YX and Fang JY: Inhibition of JAK1,
2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces
tumor cell invasion in colorectal cancer cells. Neoplasia.
10:287–297. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Du W, Hong J, Wang YC, Zhang YJ, Wang P,
Su WY, Lin YW, Lu R, Zou WP, Xiong H and Fang JY: Inhibition of
JAK2/STAT3 signalling induces colorectal cancer cell apoptosis via
mitochondrial pathway. J Cell Mol Med. 16:1878–1888. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lv D, Li Y, Zhang W, Alvarez AA, Song L,
Tang J, Gao WQ, Hu B, Cheng SY and Feng H: TRIM24 is an oncogenic
transcriptional co-activator of STAT3 in glioblastoma. Nat Commun.
8:14542017. View Article : Google Scholar : PubMed/NCBI
|