Introduction
Ovarian cancer (OC) is a common gynecological
malignancy and the most common cause of gynecological
cancer-associated death in women worldwide (1). Despite advances in surgical
techniques and chemotherapeutic agents, the prognosis for OC
remains poor, with a 5-year survival rate of <30% (2). The poor prognosis of OC is associated
with early metastasis and recurrence (3); it is therefore important to explore
the underlying molecular mechanisms associated with ovarian
carcinogenesis and progression in order to develop more effective
treatment strategies and personalized therapies for OC.
Long non-coding RNAs (lncRNAs), a heterogeneous
group of genomic transcripts with length >200 nucleotides, have
little or no protein-coding ability (4). The functional roles of lncRNAs have
been underestimated, as they were initially thought to be
transcriptional noise in the genome (5). However, accumulating evidence has
demonstrated that they serve crucial roles in physiological and
pathological processes (6),
including cancer progression and metastasis (7–10).
P73 antisense RNA 1T (non-protein coding), also called TP73-AS1, is
a long non-coding RNA that regulates apoptosis via the modulation
of p53-dependent anti-apoptotic gene and is abnormally expressed in
cancer (11,12). However, the biological role of
TP73-AS1 in patients with OC remains to be elucidated.
In the present study, the expression of TP73-AS1 in
OC tissues and cell lines was assessed and it was investigated
whether TP73-AS1 expression was associated with disease prognosis.
Functional assays were also performed to determine the impact of
TP73-AS1 on cell proliferation, cell cycle and apoptosis.
Materials and methods
Clinical samples
The present study was approved by the Ethics
Committee of the Department of Gynecology and Obstetrics, Binzhou
Central Hospital (Binzhou, China). A total of 62 pairs of OC
tissues and their adjacent noncancerous tissues were collected from
62 patients (age 36–87 years; all female) who had undergone
ovariectomy at the Department of Gynecology and Obstetrics, Binzhou
Central Hospital, between March 2006 and April 2011. All cases were
diagnosed with OC by two independent pathologists. Signed informed
consent was provided by all patients enrolled in this study.
Patients with other known tumors and those receiving medical
treatment and/or previous cancer treatment were excluded from the
present study. Patients' clinicopathological features are
summarized in Table I.
 | Table I.Association between lncRNA-TP73-AS1
expression and clinical features (n=62). |
Table I.
Association between lncRNA-TP73-AS1
expression and clinical features (n=62).
|
| LncRNA-TP73-AS1
expression |
|---|
|
|
|
|---|
| Variable | Low | High | P-value |
|---|
| Age (years) |
|
|
|
|
<50 | 19 | 21 | 0.288 |
| ≥50 | 7 | 15 |
|
| Residual tumor
diameter (cm) |
|
|
|
|
<1 | 17 | 19 | 0.435 |
| ≥1 | 9 | 17 |
|
| Histological
grade |
|
|
|
|
G1-G2 | 21 | 13 | 0.001 |
| G3 | 5 | 23 |
|
| Lymph node
metastasis |
|
|
|
|
Negative | 21 | 7 | <0.001 |
|
Positive | 5 | 29 |
|
| CA125 level
(U/ml) |
|
|
|
|
<600 | 17 | 23 | 1.000 |
|
≥600 | 9 | 13 |
|
| International
Federation of Gynecology and Obstetrics stage |
|
|
|
|
I–II | 22 | 8 | <0.001 |
|
III–IV | 4 | 28 |
|
Cell culture and cell
transfection
The human OC cell lines SKOV3, A2780, HO8910 and
CAOV3, as well as one normal human ovarian surface epithelial cell
line, HOSEPiCs, were purchased from the American Type Culture
Collection (Manassas, VA, USA). Cells were cultured for 3 days in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc. Waltham,
MA, USA) supplemented with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) and 100 U/ml
streptomycin/penicillin in a humidified atmosphere containing 5% of
CO2 at 37°C. Full-length human TP73-AS1 transcript cDNA
(NCBI Reference Sequence: NM_001204189.1) was amplified by
polymerase chain reaction (PCR) and subcloned into a pcDNA3.1
vector, according to the manufacturer's protocol (Thermo Fisher
Scientific, Inc.). The pcDNA3.1 empty vector was used as the
control. phi29 DNA polymerase (Pure-one Bio Technology, Co., Ltd.,
Shanghai, China; 10 U/µl) was used for PCR reaction. The forward
and reverse primer sequences for LnRNATP73-AS1 were
5′-CCGGTTTTCCAGTTCTTGCAC-3′ and 5′-GCCTCACAGGGAAACTTCATGC3-3′,
respectively. The following primers were used for amplification of
BDH2: Forward, 5′-TTCCAGCGTCAAAGGAGTTGT-3′ and reverse,
5′-TTCCTGGGCACACACAGTTG-3′. The following thermocycling conditions
were used for the PCR: 94°C for 60 sec, 37°C for 60 sec, 72°C for
120 sec; 25 cycles. Anza restriction enzyme cloning system (Thermo
Fisher Scientific, Inc.) was used to cut DNA. Small interfering
(si)RNA for TP73-AS1 (si-TP73-AS1) and negative control siRNA (neg
siRNA) were purchased from Shanghai GenePharma Co., Ltd. (Shanghai,
China). Cells (5×104 cells/well) were seeded into 6-well
plates and transfected with 20 nM siRNA using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. At 48 h following transfection, the transfected cells
were used in in vitro assays. The target siRNA sequence for
TP73-AS1 was as follows: 5′-TAAGGTTATCCGAATAACGGTATCGTT-3′. The
sequence of neg siRNA was as follows: 5′-TTCTCCGAACGTGTCACGT-3′.
All experiments were performed three times.
Western blotting
Total protein lysates were obtained using RIPA lysis
buffer (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Bicinchoninic acid method was used for protein determination.
Proteins (10–15 µg/lane) were separated by 10% SDS-PAGE, and
electrophoretically transferred to polyvinylidene difluoride
membranes (Roche Applied Science, Penzberg, Germany). Membranes
were probed with primary antibodies against anti-caspase-8
(1:2,000; cat. no. ab25901), anti-caspase-9 (1:2,000; cat. no.
ab32539), anti-B-cell lymphoma 2 (Bcl-2; 1:2,000; cat. no.
ab32124), anti-Bcl-2-associated X protein (Bax; 1:2,000; cat. no.
ab32503) and GAPDH (1:3,000; cat. no. ab8245; all Abcam, Cambridge,
UK) at 4°C overnight. Membranes were blocked with 5% of non-fat
(v/v) milk for 1 h at room temperature. Followed primary antibody
incubation, membranes were probed with the corresponding goat anti
rabbit immunoglobulin G Alexa Fluor® 488-conjugated
secondary antibody (1:3,000; cat. no. ab150077; Abcam) at 4°C for 1
h. Blots were washed with PBS three times for 15 min and visualized
using an enhanced chemiluminescence detection kit (Thermo Fisher
Scientific, Inc.), followed by the exposure with a Tanon 5200
instrument (Tanon Science and Technology Co., Ltd., Shanghai,
China). Image Lab (version 4.0; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was used for quantitative analysis. All
experiments were repeated at least three times.
RNA extraction and RT-qPCR
Total RNA was extracted from tissues using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. cDNA synthesis was
carried out using M-MLV reverse transcriptase (Promega Corporation,
Madison, WI, USA). Reverse transcription was conducted at 70°C for
10 min and at 4°C for 5 min. TP73-AS1 expression was measured using
SYBR-Green fluorescent dye and STEP ONE RT-PCR apparatus (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
conditions were as follows: 20–30 cycles of initial denaturation at
94°C for 30 sec, annealing at 55°C for 30 sec and final extension
at 70–72°C for 30–60 sec. The relative expression levels of genes
were quantified using 2−ΔΔCq method (13). GAPDH was used as an internal
reference. The primers used were as follows: TP73-AS1 forward,
5′-CCGGTTTTCCAGTTCTTGCAC-3′ and reverse,
5′-GCCTCACAGGGAAACTTCATGC-3′; GAPDH forward,
5′-GTCAACGGATTTGGTCTGTATT-3′ and reverse,
5′-AGTCTTCTGGGTGGCAGTGAT-3′. All assays were repeated at least
three times.
Relative cell viability
Negative siRNA and si-TP73-A1 were transfected into
SKOV3 and A2780 cells, while pcDNA-TP73-AS1 and control pcDNA
vector were transfected into HOSEPiC cell by
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). Ovarian cancer cells
(3×103 to 6×103 cells/µl) were inoculated in
a 96-well plate (200 µl per well, 6 repeated wells) at 37°C in an
atmosphere containing 5% CO2 for 24–72 h and 20 µl MTT
solution (5 mg/ml; Sigma-Aldrich; Merck KGaA) was added into each
well. Following 4 h of incubation at 37°C, the incubation was
terminated and the culture medium was discarded. 10% dimethyl
sulfoxide (150 µl; Sigma-Aldrich; Merck KGaA) was added to each
well and plates were gently shaken for 10 min dissolve formazan
crystals. Absorbance values were determined with a plate detector
at 0, 24, 48, 72 and 96 h at a wavelength of 450 nm. The DMSO was
used as the control. The experiment was performed in
triplicate.
Colony formation assay
Negative siRNA and si-TP73-A1 were transfected into
SKOV3 and A2780 cells, while pcDNA-TP73-AS1 and control pcDNA
vector were transfected into HOSEPiCs using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). Transfections were terminated at a
final concentration of 50 nM, as described above. Cells (500
cells/well) were plated in 6-well plates and incubated in RPMI-1640
with 10% FBS at 37°C for two weeks and the medium was changed every
3 days. Then, the cells were fixed with 4% paraformaldehyde for 15
min at room temperature and stained with 0.1% crystal violet for 30
min at a room temperature. The number of visible colonies was
counted manually through observing the violet dots. Cell clone
formation was observed under a light microscope, the cell number
was observed at a magnification of ×10 in one field of view. All
independent assays were repeated at least three times.
Flow cytometric analysis of
apoptosis
Apoptosis was determined in transfected cells using
flow cytometric analysis with an Annexin V: Fluorescein
isothiocyanate Apoptosis Detection kit (BD Biosciences, Franklin
Lakes, NJ, USA), according to the manufacturer's protocol. The
apoptotic cells were analyzed by using FlowJo 10 software and
Attune® NxT cytometer (both Thermo Fisher Scientific,
Inc.). All samples were assayed in triplicate.
Flow cytometric analysis of cell cycle
distribution
Cells were collected directly or 48 h following
transfection, washed with ice-cold PBS and fixed with 70% ethanol
overnight at −20°C. Fixed cells were rehydrated in PBS for 10 min
and incubated in 1 mg/ml of RNase A (Thermo Fisher Scientific,
Inc.) for 30 min at 37°C. Subsequently, the cells were subjected to
propidium iodide/RNase staining at 4°C for 30 min followed by flow
cytometric analysis with a FACScan instrument (BD Biosciences) and
Cell Quest software version 3.0 (BD Biosciences) as previously
described (14). All samples were
assayed in triplicate.
Statistical analysis
SPSS version 19.0 (IBM Corp., Armonk, NY, USA) was
used for statistical analysis. Data were presented as the mean ±
standard deviation. The Pearson χ2 test was used to
assess the association between TP73-AS1 expression and
clinicopathological factors. Data between two groups were analyzed
using the Student's t-test. Multiple comparisons were made by
one-way analysis of variance with the LSD post hoc test. Survival
analysis was performed using the Kaplan-Meier method and the
log-rank test was used to compare differences among patient groups.
A cox proportional hazards regression model was generated to
identify factors associated with overall survival through a
multivariate survival analysis of OC. P<0.05 was considered to
indicate a statistically significant difference.
Results
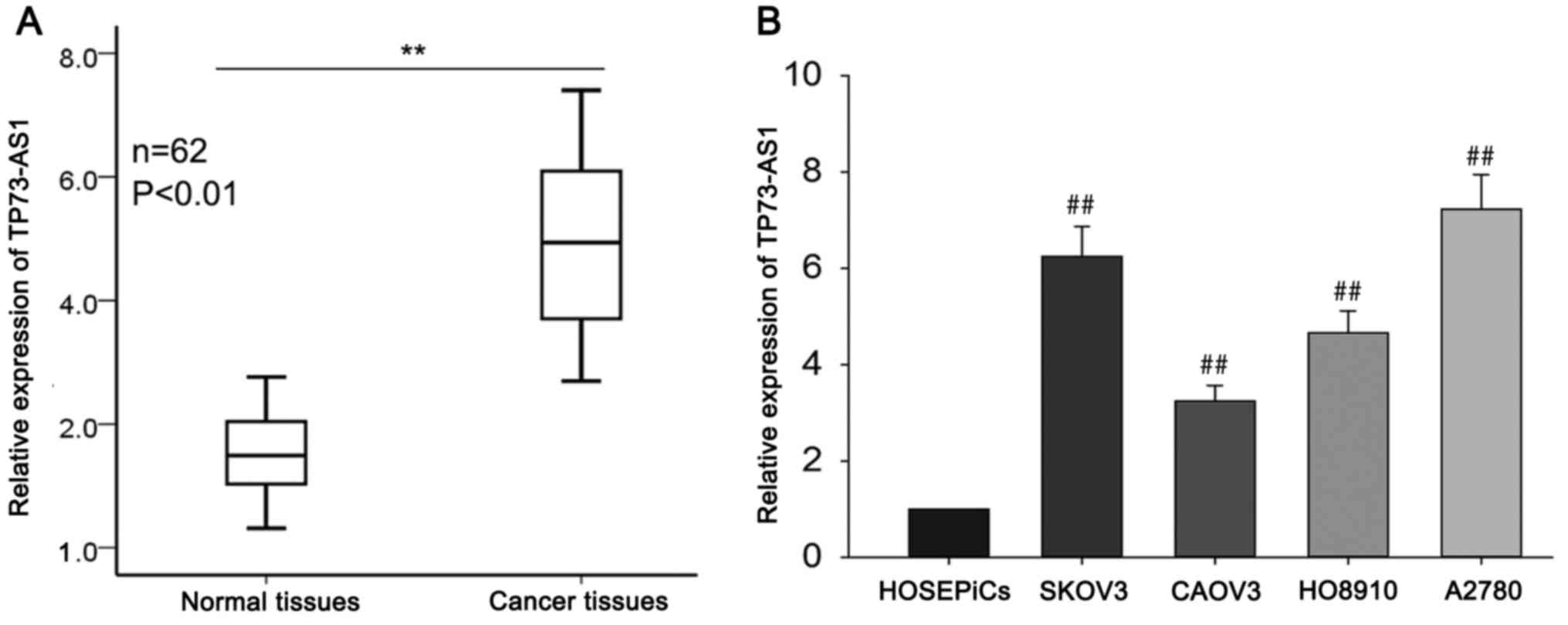
TP73-AS1 is upregulated in OC tissues
and cell lines
To determine the biological function of TP73-AS1,
RT-qPCR was performed to measure the relative expression of
TP73-AS1 in OC tissues (n=62) compared with corresponding normal
tissues (n=62). TP73-AS1 was significantly overexpressed in OC
tissues compared with the corresponding adjacent non-tumorous
tissues (Fig. 1A). TP73-AS1
expression in four OC cell lines (SKOV3, CAOV3, HO8910 and A2780)
and one normal human ovarian surface epithelial cell line
(HOSEPiCs) was assessed using RT-qPCR. The results revealed a
significant increase in TP73-AS1 expression in OC cell lines
compared with HOSEPiCs (Fig. 1B).
Of the four OC cell lines, TP73-AS1 expression was highest in the
SKOV3 and A2780 cells, and so SKOV3 and A2780 cells were chosen for
the following experiments. These findings suggest that TP73-AS1 may
act as an oncogene and contribute to the progression of OC.
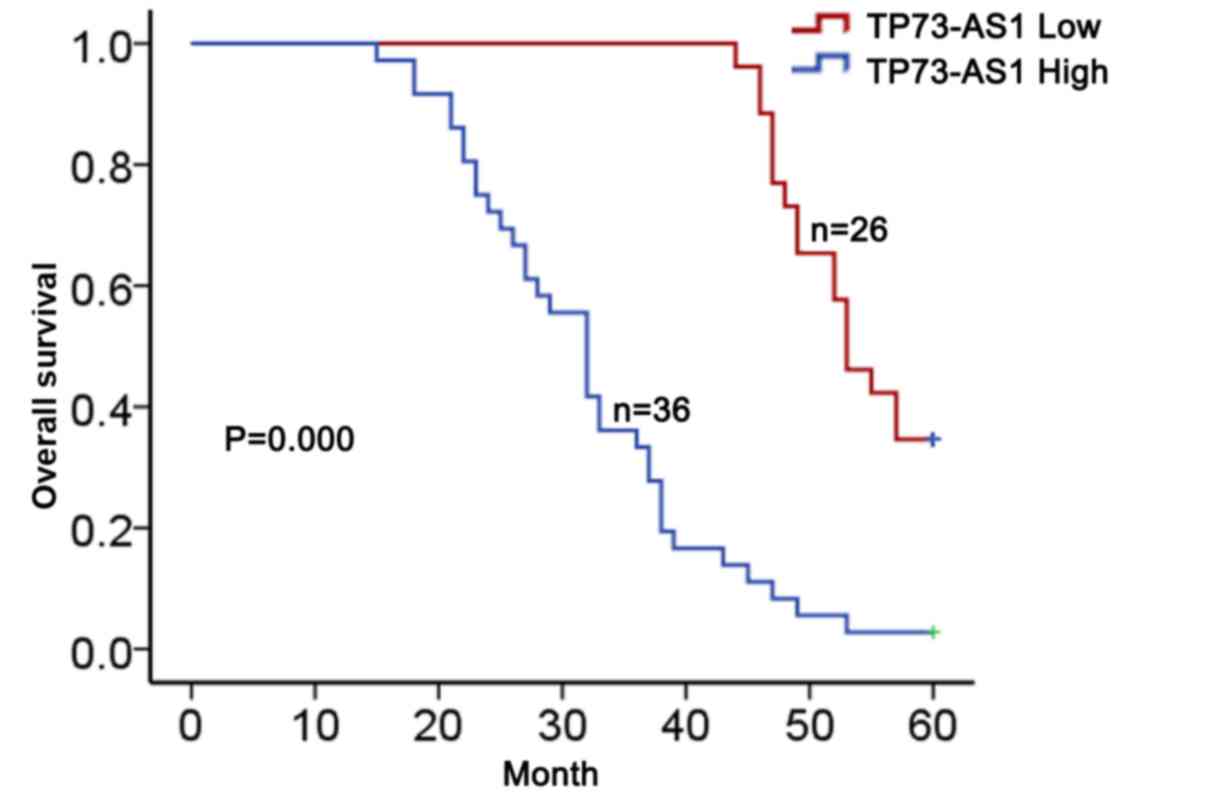
High expression of TP73-AS1 predicts
poor prognosis in OC patients
To determine the clinical relevance of TP73-AS1
expression in OC, the association between the levels of TP73-AS1
and clinicopathological parameters was analyzed. According to the
mean value of TP73-AS1 expression (4.99) in OC tissues, TP73-AS1
expression was divided into two groups (high group, n=36; and low
group, n=26). TP73-AS1 expression was significantly associated with
histological grade, lymph node metastasis and International
Federation of Gynecology and Obstetrics (FIGO) stage (Table I); however, no significant
associations with age, residual tumor diameter or CA125 levels were
observed (Table I). A Kaplan-Meier
method analysis (log-rank test) was performed and revealed that
higher expression of TP73-AS1 was associated with a lower overall
survival in patients with OC (Fig.
2). Proportional hazards method analysis revealed that high
levels of TP73-AS1 may be a prognostic factor in addition to the
independent prognostic impact of histological grade and FIGO stage
(Table II). These results suggest
that TP73-AS1 serves a role in OC and may be considered as a
specific biomarker of poor prognosis.
 | Table II.Multivariate analysis of prognostic
parameters in patients with ovarian cancer by Cox regression
analysis. |
Table II.
Multivariate analysis of prognostic
parameters in patients with ovarian cancer by Cox regression
analysis.
| Variable | Category | P-value |
|---|
| Age (years) | <50 | 0.264 |
|
| ≥50 |
|
| Residual tumor
diameter (cm) | <1 | 0.211 |
|
| ≥1 |
|
| Histological
grade | G1-G2 | 0.026 |
|
| G3 |
|
| Lymph node
metastasis | Negative | 0.852 |
|
| Positive |
|
| CA125 level
(U/ml) | <600 | 0.430 |
|
| ≥600 |
|
| International
Federation of | I–II | <0.001 |
| Gynecology and
Obstetrics stage | III–IV |
|
| TP73-AS1
expression | Low | 0.003 |
|
| High |
|
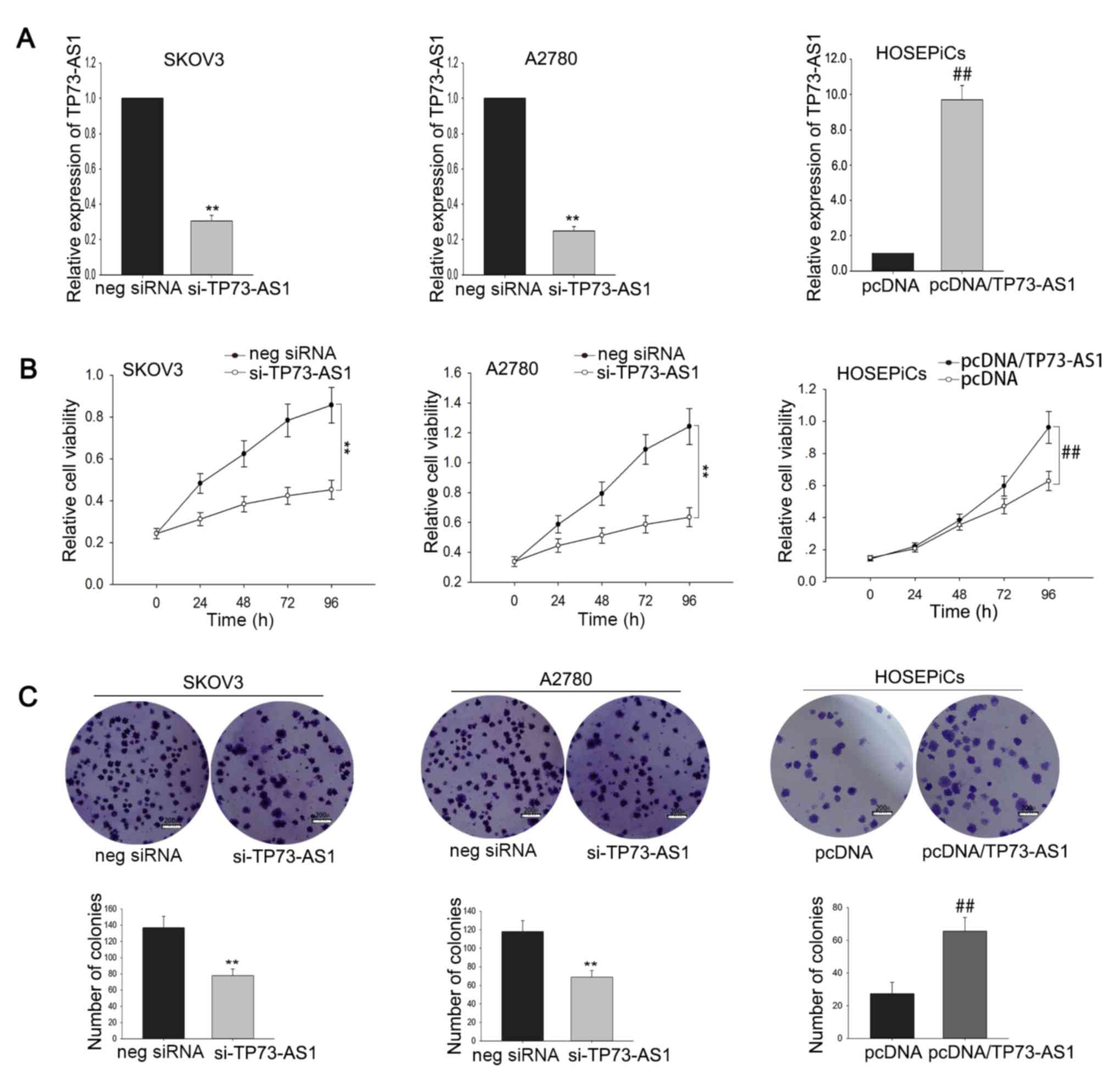
Dysregulation of TP73-AS1 affects OC
cell proliferation
To explore the biological role of TP73-AS1 in OC
cell proliferation, SKOV3 and A2780 cells were transfected with
TP73-AS1-specific siRNA and HOSEPiCs cells were transfected with a
TP73-AS1 expression vector (pcDNA/TP73-AS1). Transfection
efficiency was determined after 48 h (Fig. 3A). MTT assays were performed to
determine the relative viability of SKOV3 and A2780 cells
transfected with si-TP73-AS1, and HOSEPiCs cells transfected with
pcDNA/TP73-AS1. TP73-AS1 silencing reduced the viability of OC
cells compared with cells transfected with negative siRNA, whereas
TP73-AS1 overexpression enhanced the viability of HOSEPiCs cells
(Fig. 3B). Consistent with the MTT
assays, the results of colony formation assays also demonstrated a
growth-inhibition effect of TP73-AS1 knockdown and a
pro-proliferation effect of TP73-AS1 overexpression (Fig. 3C). Collectively, these findings
suggest that TP73-AS1 may function as an oncogene via regulating
cell proliferation.
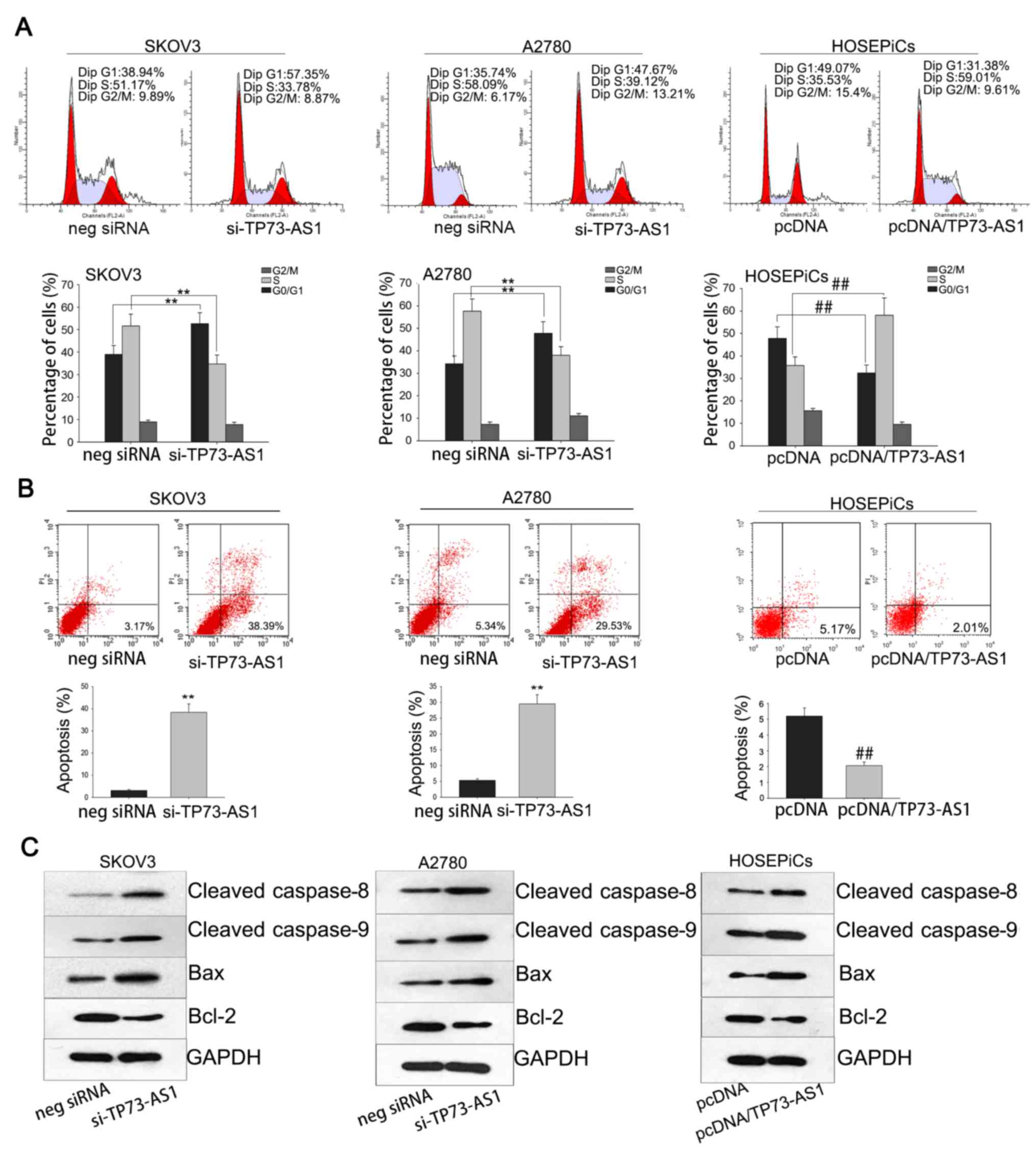
Growth-modulation mediated by
dysregulation of TP73-AS1 is attributed to its effect on cell cycle
and apoptosis
As cell cycle dysregulation serves a role in tumor
proliferation (15), it was
hypothesized that growth modulation mediated by TP73-AS1
dysregulation may occur by affecting the cell cycle and apoptosis.
To verify this hypothesis, flow cytometric analysis was performed.
SKOV3 and A2780 cells transfected with si-TP73-AS1 and exhibited a
high percentage of G1 cells compared with the negative siRNA group,
whereas HOSEPiCs cell transfected with pcDNA/TP72-AS1 had a higher
percentage of S phase cells compared with the pcDNA group (Fig. 4A). In addition, flow cytometric
analysis of apoptosis revealed that TP73-AS1 knockdown increased
the percentage of apoptotic cells in SKOV3 and A2780 assays. By
comparison, forced overexpression of TP73-AS1 significantly reduced
the number of apoptotic cells in the HOSEPiCs experiment (Fig. 4B). Results of western blotting
revealed that the expression of apoptosis-associated proteins
(caspase-8, caspase-9, Bcl-2 and Bax) was affected by TP73-AS1
(Fig. 4C). The protein levels of
caspase-8, caspase-9 and Bax were upregulated by knockdown of
TP3-AS1 in SKOV3 and A2780 cells, and they were also upregulated by
silencing of TP3-AS1 in HOSEPiCs cell. By contrast, the protein
level of Bcl-2 was downregulated by TP73-AS1 knockdown in SKOV3 and
A2780 cells, and downregulated by TP73-AS1 silencing in HOSEPiCs
cells. These data suggest that the pro-growth effect of TP73-AS1 in
OC cells may be attributed to its influence on the cell cycle and
apoptosis.
Discussion
OC is the most common cause of cancer-associated
death in women worldwide (16). As
there is no effective method for early diagnosis, the majority of
patients with OC are diagnosed at an advanced stage, and so the
prognosis of OC is poor (17,18).
The molecular mechanism underlying the initiation and progression
of OC is generally complex, involving dysregulated oncogenes and
tumor suppressor genes (19–21).
Previous reports have identified that the
dysregulation of lncRNAs contributes to a range of biological
processes and is associated with a variety of cancers (22–30),
including OC. For example, in 2016, Kuang et al (31) reported that lncRNA TUG1 regulated
OC proliferation and metastasis via affecting
epithelial-mesenchymal transition. Richards et al (32) demonstrated that a functional
variant of HOXA11-antisense, a novel long non-coding RNA, inhibited
the oncogenic phenotype of epithelial OC. Furthermore, Liu et
al (33) reported that
overexpression of long non-coding RNA PVT1 in OC cells promoted
cisplatin resistance by regulating apoptotic pathways. However, the
exact molecular mechanisms underlying the tumorigenesis of OC
remain unclear. TP73-AS1, transcribed from chromosome 1p36, has
been reported to be associated with cell proliferation and tumor
progression in glioma and esophageal squamous cell carcinoma
(11,12). However, its biological roles in OC
remain unknown.
The results of the present study demonstrate that
TP73-AS1 was overexpressed in OC tissues and cell lines compared
with controls and increased TP73-AS1 expression was associated with
histological grade, lymph node metastasis, FIGO stage and poor
prognosis in patients with OC. Furthermore, proportional hazards
method analysis revealed that high TP73-AS1 expression may be a
specific biomarker of poor prognosis for OC. Loss-of-function
assays revealed that TP73-AS1 knockdown inhibited the proliferation
ability of OC cells. Additionally, flow cytometric analysis
revealed that the proliferation-inhibition mediated by TP73-AS1
knockdown may be due to changes in the cell cycle and apoptosis.
These results suggest that TP73-AS1 expression may be used as a
biomarker for predicting OC prognosis. TP73-AS1 may therefore be
used as a potential prognostic and therapeutic target of OC. Future
research should focus on analyzing other pathways and signaling
molecules associated with TP73-AS1.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and XW analyzed and interpreted the patient data.
LM, SZ and HW analyzed and interpreted the in vitro data
obtained using cell lines. All authors contributed to experiments.
XL was the major contributor in writing the manuscript. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Signed informed consent to participate in the study
was obtained from all participants. This study has approved by the
ethics committee of Binzhou Central Hospital (Binzhou, China).
Consent for publication
All patients have provided written informed consent
for publication.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA A
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
2
|
Menon U: Ovarian cancer: challenges of
early detection. Nat Clin Pract Oncol. 4:498–499. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi C, Qin L, Gao H, Gu L, Yang C, Liu H
and Liu T: NUCKS nuclear elevated expression indicates progression
and prognosis of ovarian cancer. Tumour Biol.
39:10104283177146312017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen G, Wang Z, Wang D, Qiu C, Liu M, Chen
X, Zhang Q, Yan G and Cui Q: LncRNADisease: A database for
long-non-coding RNA-associated diseases. Nucleic Acids Res.
41:D983–D986. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu H, Lv Z and Guo E: Knockdown of long
noncoding RNA SPRY4-IT1 suppresses glioma cell proliferation,
metastasis and epithelial-mesenchymal transition. Int J Clin Exp
Pathol. 8:9140–9146. 2015.PubMed/NCBI
|
|
8
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang Y, Li H, Hou S, Hu B, Liu J and Wang
J: The noncoding RNA expression profile and the effect of lncRNA
AK126698 on cisplatin resistance in non-small-cell lung cancer
cell. PLoS One. 8:e653092013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 LncRNA
antagonizes Let-7 MicroRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang R, Jin H and Lou F: The long
non-coding RNA TP73-AS1 interacted with miR-142 to modulate brain
glioma growth through HMGB1/RAGE pathway. J Cell Biochem.
119:3007–3017. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zang W, Wang T, Wang Y, Chen X, Du Y, Sun
Q, Li M, Dong Z and Zhao G: Knockdown of long non-coding RNA
TP73-AS1 inhibits cell proliferation and induces apoptosis in
esophageal squamous cell carcinoma. Oncotarget. 7:19960–19974.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang SZ, Pan FY, Xu JF, Yuan J, Guo SY,
Dai G, Xue B, Shen WG, Wen CJ, Zhao DH and Li CJ: Knockdown of
c-Met by adenovirus-delivered small interfering RNA inhibits
hepatocellular carcinoma growth in vitro and in vivo. Mol Cancer
Ther. 4:1577–1584. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang Z, Zhang L, Liao Q, Wang Y, Yu F,
Feng M, Xiang X and Xiong J: Regulation of TRIM24 by miR-511
modulates cell proliferation in gastric cancer. J Exp Clin Cancer
Res. 36:172017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA A Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar
|
|
17
|
Puvanenthiran S, Essapen S, Seddon AM and
Modjtahedi H: Impact of the putative cancer stem cell markers and
growth factor receptor expression on the sensitivity of ovarian
cancer cells to treatment with various forms of small molecule
tyrosine kinase inhibitors and cytotoxic drugs. Int J Oncol.
49:1825–1838. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dillman RO, McClay EF, Barth NM, Amatruda
TT, Schwartzberg LS, Mahdavi K, de Leon C, Ellis RE and DePriest C:
Dendritic versus tumor cell presentation of autologous tumor
antigens for active specific immunotherapy in metastatic melanoma:
Impact on long-term survival by extent of disease at the time of
treatment. Cancer Biother Radiopharm. 30:187–194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zou Z, Zou R, Zong D, Shi Y, Chen J, Huang
J, Zhu J, Chen L, Bao X, Liu Y, et al: miR-495 sensitizes MDR
cancer cells to the combination of doxorubicin and taxol by
inhibiting MDR1 expression. J Cell Mol Med. 21:1929–1943. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang YW, Cheng HL, Ding YR, Chou LH and
Chow NH: EMP1, EMP 2 and EMP3 as novel therapeutic targets in human
cancer. Biochim Biophys Acta. 1868:199–211. 2017.PubMed/NCBI
|
|
21
|
Eccleston A, Bentley A, Dyer M, Strydom A,
Vereecken W, George A and Rahman N: A cost-effectiveness evaluation
of germline BRCA1 and BRCA2 testing in UK women with ovarian
cancer. Value Health. 20:567–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yan H, Xia JY and Feng FZ: Long non-coding
RNA ENST00000457645 reverses cisplatin resistance in CP70 ovarian
cancer cells. Genet Mol Res. 16:2017.doi: 10.4238/gmr16019411.
View Article : Google Scholar
|
|
23
|
Liu RT, Cao JL, Yan CQ, Wang Y, An CJ and
Lv HT: Effects of LncRNA-HOST2 on cell proliferation, migration,
invasion and apoptosis of human hepatocellular carcinoma cell line
SMMC-7721. Biosci Rep. 37:BSR201605322017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu R, Zeng Y, Zhou CF, Wang Y, Li X, Liu
ZQ, Chen XP, Zhang W and Zhou HH: Long noncoding RNA expression
signature to predict platinum-based chemotherapeutic sensitivity of
ovarian cancer patients. Sci Rep. 7:182017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Liu C, Lu Z, Chen L, Wang J, Li Y
and Ma H: Upregulation of the long non-coding RNA SPRY4-IT1
indicates a poor prognosis and promotes tumorigenesis in ovarian
cancer. Biomed Pharmacother. 88:529–534. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zuo ZK, Gong Y, Chen XH, Ye F, Yin ZM,
Gong QN and Huang JS: TGFβ1-induced LncRNA UCA1 upregulation
promotes gastric cancer invasion and migration. DNA Cell Biol.
36:159–167. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou C, Huang C, Wang J, Huang H, Li J,
Xie Q, Liu Y, Zhu J, Li Y, Zhang D, et al: LncRNA MEG3
downregulation mediated by DNMT3b contributes to nickel malignant
transformation of human bronchial epithelial cells via modulating
PHLPP1 transcription and HIF-1α translation. Oncogene.
36:3878–3889. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang CL, Zhu KP and Ma XL: Antisense
lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by
increasing the expression of FOXC2. Cancer Lett. 396:66–75. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yu H, Xue Y, Wang P, Liu X, Ma J, Zheng J,
Li Z, Li Z, Cai H and Liu Y: Knockdown of long non-coding RNA XIST
increases blood-tumor barrier permeability and inhibits glioma
angiogenesis by targeting miR-137. Oncogenesis. 6:e3032017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiong H, Ni Z, He J, Jiang S, Li X, He J,
Gong W, Zheng L, Chen S, Li B, et al: LncRNA HULC triggers
autophagy via stabilizing Sirt1 and attenuates the chemosensitivity
of HCC cells. Oncogene. 36:3528–3540. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kuang D, Zhang X, Hua S, Dong W and Li Z:
Long non-coding RNA TUG1 regulates ovarian cancer proliferation and
metastasis via affecting epithelial-mesenchymal transition. Exp Mol
Pathol. 101:267–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Richards EJ, Permuth-Wey J, Li Y, Chen YA,
Coppola D, Reid BM, Lin HY, Teer JK, Berchuck A, Birrer MJ, et al:
A functional variant in HOXA11-AS, a novel long non-coding RNA,
inhibits the oncogenic phenotype of epithelial ovarian cancer.
Oncotarget. 6:34745–34757. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu E, Liu Z, Zhou Y, Mi R and Wang D:
Overexpression of long non-coding RNA PVT1 in ovarian cancer cells
promotes cisplatin resistance by regulating apoptotic pathways. Int
J Clin Exp Med. 8:20565–20572. 2015.PubMed/NCBI
|