Introduction
The coiled-coil-helix-coiled-coil-helix domain
(CHCHD)-containing proteins are small mitochondrial proteins with
important functions. Mutations of CHCHD genes have been identified
to be associated with various human neurodegenerative diseases
(1). CHCHD10, which is a CHCHD
protein, was identified to be associated with amyotrophic lateral
sclerosis (ALS), frontotemporal dementia (FTD) and Alzheimer's
disease (AD) in Chinese population (2,3).
Recently, the CHCHD2 gene was identified as a possible
causative gene for Parkinson's disease (PD). A missense mutation
(Thr61Ile) in this gene was first detected in a multigenerational
Japanese family with autosomal dominant PD (ADPD) (4). Several subsequent efforts have been
made to confirm the association between the CHCHD2 gene and
PD in other ethnicities, including European and Chinese
populations. Jansen et al (5) reported three novel putative
pathogenic variants (Ala32Thr, Pro34Leu, and Ile80Val) in patients
with PD from a western European population; another study
identified a heterozygous variant (182C>T; Thr61Ile) in an ADPD
pedigree in a Chinese population (6). The CHCHD2 gene is located on
chromosome 7p11.2 and contains four exons encoding 151 amino acids
with a predicted N-terminal mitochondrial targeting sequence
(7). The CHCHD2 protein is a small
mitochondrial protein that serves as one of the negative regulators
of mitochondria-mediated apoptosis. The knockdown of CHCHD2
promoted a significant increase in nuclear fragmentation and in
phosphatidylserine exposure, both of which are hallmarks of
apoptosis (8).
Neurodegenerative disorders are conditions that have
yet to be fully elucidated. Nevertheless, different types of
neurodegenerative disorders are closely related, including the four
major types: PD, AD, ALS and FTD. Although they are different
diseases with distinct features, they manifest overlapping clinical
phenotypes, pathologic features and genetic backgrounds. For
instance, the PD-related genetic variant rs76904798 of leucine-rich
repeat kinase 2 (LRRK2) is found to be a common genetic risk
variant for late-onset AD (LOAD) susceptibility in a northern Han
Chinese people (9). Expansions in
the C9orf72 gene are most frequently associated with ALS-FTD
and can be combined with symmetrical Parkinsonism; FTD in patients
with mutations in the gene that encodes microtubule-associated
protein tau (MAPT) can also manifest as symmetrical
Parkinsonism (10).
In addition, mitochondrial dysfunction has been
described in neurodegenerative disorders. In addition to CHCHD2,
the proteins that are associated with familial PD-PTEN-induced
putative kinase 1 (PINK1), DJ-1, alpha-synuclein and LRRK2-are
either mitochondrial proteins or associated with mitochondria, and
all interface with the pathways of oxidative stress and free
radical damage (11). In AD,
aggregation of β-amyloid (Aβ) is central to initiating AD
pathogenesis. In the brain and in isolated mitochondria, exposure
to Aβ inhibits key mitochondrial enzymes (12). As mentioned above, mutations of
CHCHD10 are associated with ALS and/or FTD, AD.
In this study, to confirm the potential role of
CHCHD2 in these three diseases, we assessed the prevalence
of CHCHD2 mutations in AD, ALS and FTD patients.
Patients and methods
Patients
This study recruited a total of 511 AD patients (436
sporadic AD and 75 probands from FAD families, mean age at onset
was 66.2±5.5 years, male: 44.0%), 181 ALS patients (166 sporadic
ALS and 15 familial ALS, mean age at onset was 48.1±13.4 years,
male: 67.9%) and 88 FTD patients (77 sporadic FTD and 11 familial
FTD, mean age at onset was 53.3±9.7 years, male: 42.0%) from
mainland China in 2014 to 2016. The diagnoses of probable or
possible AD according to the NINCDS-ADRDA criteria were made by 2
or more experienced neurologists in Xiangya Hospital. The diagnoses
of ALS were made according to the El Escorial revised criteria. The
diagnoses of FTD met the Lund-Manchester criteria (13–15).
Neuroradiological examinations for example MRI, was assessed with
patients diagnosed as probable or possible AD and FTD in this
study. We have excluded AD, FTD and ALS patients carrying
disease-causing gene like PSEN1, PSEN2, APP, MAPT, GRN, C9orf72,
TREM2, CHCHD10, SOD1, TARDBP, FUS (3,16–20).
Additionally, the APOE genotype was available for all AD
patients. A total of 500 healthy unrelated age-matched Chinese
individuals without a history of neurodegenerative disease were
recruited from the Xiangya Wellness Center as a control group.
Written informed consent for participation in the study was
obtained from all subjects. For patients who can understand our
study, we asked for the consent of the patients, and for severe
patients, we sought the consent of the patient's guardian or
immediate family members. This study was conducted in accordance
with the Declaration of Helsinki and was approved by the Expert
Committee of Xiangya Hospital of Central South University in
China.
DNA isolation and genotyping
methods
Genomic DNA was extracted from peripheral blood
leukocytes from all patients and controls. The quality and quantity
of DNA were assessed with a fluorometer. All DNA samples were
diluted to 50 ng/ml. Polymerase chain reaction (PCR) was performed
on the exonic regions of CHCHD2. The CHCHD2
sequencing (exons 1–4) was amplified using primers designed
according to the GenBank entries (data not shown). Each PCR product
was sequenced using forward and reverse primers identical to the
ones used in PCR with BigDye terminator v3.1 sequencing chemistry
on an ABI 3730xl DNA analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The DNA sequences were
analyzed using Sequencher software.
Statistical analysis
When a mutation was detected, we first confirmed
whether it was a novel or rare (MAF<1%) mutation by comparison
with sequences in the ExAC (exac.broadinstitute.org/) and 1000 Genomes Project
databases (www.1000genomes.org/) and with direct sequencing of
healthy controls. We then use bioinformatics prediction tools like
Mutation Taster (mutationtaster.org), REVEL (sites.google.com/site/revelgenomics/downloads), CADD
(cadd.gs.washington.edu/download) to predict the
pathogenicity of the mutation. Statistical analysis of clinical
data was performed using IBM SPSS 19.0 (IBM Corp., Armonk, NY,
USA). To compare the differences among the patients and controls,
the Fisher's exact test was used. The threshold of statistical
significance was set at P<0.05. Descriptive statistics were
expressed as the mean ± the standard deviation.
Results
The demographic features of 511 AD, 181 ALS and 88
FTD cases and 500 controls are shown in Table I. The Clinical features and
APOE genotype of the carriers is presented in Table II.
 | Table I.Demographic information of patients
and control groups. |
Table I.
Demographic information of patients
and control groups.
| Variable | AD | ALS | FTD | Control |
|---|
| Cases, n | 511 | 181 | 88 | 500 |
| No. of male cases, n
(%) | 225 (44.0) | 123 (68.0) | 37 (42.0) | 253 (50.6) |
| Age at onset,
years | 66.2±5.5 | 48.1±13.4 | 53.3±9.7 | – |
| Age at examination,
years | 70.0±5.7 | 50.8±12.3 | 58.5±11.5 | 69.3±6.1 |
| MMSE score | 18.13±7.78 | – | 16.31±9.45 | 28.7±1.4 |
 | Table II.Clinical features of Alzheimer's
disease patients carrying variants of the CHCHD2 gene. |
Table II.
Clinical features of Alzheimer's
disease patients carrying variants of the CHCHD2 gene.
| Characteristic | M6937 | M14200 | M14851 | M24736 | M31801 |
|---|
| Sex | Female | Female | Female | Female | Male |
| Family history | Y | N | N | N | Y |
| Age at onset
(years) | 67 | 60 | 82 | 53 | 67 |
| Age at examination
(years) | 79 | 64 | 91 | 58 | 70 |
| MMSE | 5 | 1 | 4 | 22 | 5 |
| MoCA | 4 | 1 | 6 | 9 | 0 |
| Variant |
5C>T(Pro2Leu) |
5C>T(Pro2Leu) |
5C>T(Pro2Leu) |
5C>T(Pro2Leu) |
238A>G(Ile80Val) |
| APOE allele | ε4/ε2 | ε3/ε3 | ε3/ε3 | ε3/ε3 | ε3/ε3 |
Two rare heterozygous variants of
CHCHD2, 5C>T (Pro2Leu) and 238A>G (Ile80Val), were found in
five of 511 AD patients
The variant Pro2Leu was identified in four patients
with typical symptoms of cognitive impairment; three of them had
sporadic AD, and one had a family history of AD. The patient with a
family history developed progressive memory impairment at the age
of 67. At the onset, she easily forgot what she had just done.
Then, she began to forget her relatives' names and could not find
her way home. Finally, multiple cognitive domains were impaired.
Her Mini-Mental State Examination (MMSE) score was 5 of 30 points;
her Montreal cognitive assessment scale (MoCA) score was 4 of 30
points. Her mother also had symptoms of memory loss, according to
the recollection of the patient's family. One of the proband's
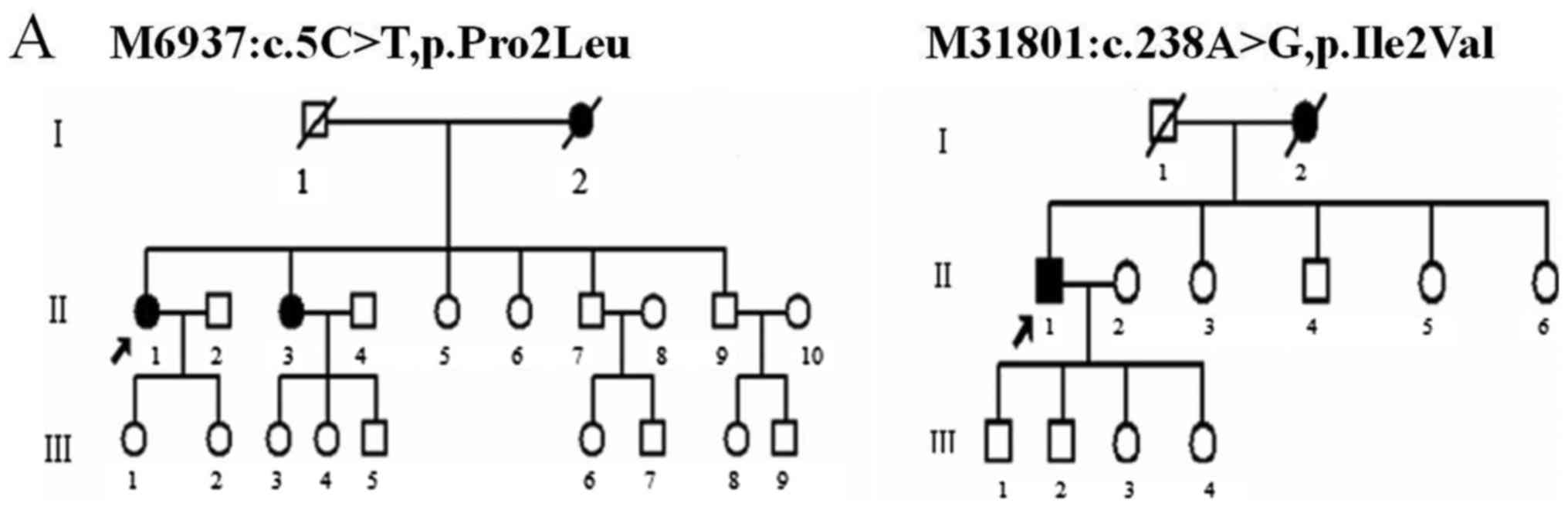
sisters (Fig. 1A, M6937: II3) who
had similar symptoms also carried the same variant. Unfortunately,
DNA samples from other family members were unavailable for genetic
analysis. This variant showed a frequency of 0.007475 in the ExAC
database. We also detected this variant with equal frequency in our
control group (Table III), which
indicated that there was no significant association between Pro2Leu
and the risk for AD in our cohort.
 | Table III.Frequencies and in silico
analyses of CHCHD2 variants. |
Table III.
Frequencies and in silico
analyses of CHCHD2 variants.
|
| Minor allele
frequency |
|
| CADD |
|---|
|
|
|
|
|
|
|---|
| Variants | AD | Control | P-value | Mutation
taster | REVEL
scoreb | Raw score | PHREDc |
|---|
|
5C>T(Pro2Leu) | 0.0078 | 0.006 | 1a | Disease
causing | 0.275 | 4.554172 | 24.4 |
|
238A>G(Ile80Val) | 0.0020 | – | – | Disease
causing | 0.009 | 1.187786 | 11.68 |
The variant Ile80Val was identified in a male
patient who developed progressive memory loss at the age of 67. A
year later, he had a change of personality. Specifically, he became
less talkative and more aggressive than he used to be. The patient
went to see a doctor at the age of 70; his MMSE score was 5 of 30
points, and his MoCA score was 0 of 30 points. His mother began
suffering from memory loss at the age of 81; two years later, she
lost the ability to live independently and died because of
pulmonary infection. One of the proband's younger sister (Fig. 1A, M31801: II-3) and his two sons
(III-1 and III-2) agreed to participate in our study. We performed
cognitive evaluation and genetic analysis of these three members,
it turned out that one of his son (III-2) carried this variant
while others did not (all of them are cognitively normal). We will
continue to monitor whether he develops any symptoms of AD. This
variant showed a frequency of 0.000025 in ExAC and was not detected
in data from the 1000 Genomes Project and was also absent in our
500 healthy control individuals. The variant sequences in patients
and references are presented in Fig.
1B.
Both of these two variants were highly conserved
among the primary species (Fig.
1C) and were predicted to be detrimental based on the Mutation
Taster. The variant 5C>T (Pro2Leu) was predicted to be the 1%
most deleterious and the variant 238A>G (Ile80Val) to be the 10%
most deleterious according to CADD. However, these two variants are
less likely to cause disease in REVEL prediction (Table III).
No CHCHD2 variant was detected in
patients with ALS or FTD
In our 181 ALS and 88 FTD samples, Sanger sequencing
ruled out coding mutations in CHCHD2, suggesting that
CHCHD2 might not be a risk gene in these two diseases.
Discussion
CHCHD2 co-expresses with other genes of the
oxidative phosphorylation pathway, and the CHCHD2 protein serves as
a transcription factor to activate the oxygen response element
(ORE) in the COX4I2 gene (21). Funayama et al (4) first identified a missense mutation
(Thr61Ile) of CHCHD2 in a multigenerational Japanese family
with ADPD. Subsequently, they found two more mutations, one
missense mutation (434G>A, Arg145Gln) and one splice-site
mutation (300+5G>A), in two other families. And none of these
three mutations was noted in the 559 unaffected Japanese controls
(4). Of these mutations, only the
Thr61Ile mutation was confirmed to cosegregate in two independent
families with ADPD (22). In
addition to PD, recent studies have reported that CHCHD2
expression was increased in neural stem cell lines derived from a
patient with Huntington's disease, and gene variants of
CHCHD2 were also detected in patients with Lewy body disease
(LBD) (23,24) Based on these observations, the
CHCHD2 gene might be involved in various neurodegenerative
diseases.
In this study, we detected two single nucleotide
variants of CHCHD2 (Pro2 Leu and Ile80 Val) in five AD
patients. All of these five patients had typical symptoms of AD.
Pro2Leu was confirmed to have different frequencies in patients
with sporadic PD and controls by Funayama et al (4) and Shi et al (6). Another similar research that may be
carried out at the same time with this study found four variants of
CHCHD2 gene in AD and FTD patients and one of them is
Pro2Leu (25). However, in terms
of our results, this variant might not be significantly associated
with AD. Ile80Val was first identified in a PD patient in a western
European population (5). According
to the ExAC database and the 1000 Genomes Project, this variant has
not been previously detected in an East Asian population. Ile80Val
is located in a transmembrane domain and was predicted to be
detrimental based on the Mutation Taster and CADD (8). Nonetheless, the REVEL predicted a
score of 0.009 for this mutation, which might not cause disease.
Considering the reasons given above, CHCHD2 is not likely to
be a causative gene of AD in the Chinese population but might be
associated with AD and AD might share a common pathway with PD in
mitochondrial dysfunction. However, the pathogenicity of this
variant remains uncertain. Further functional experiments are
needed to investigate how CHCHD2 plays a role in AD and
PD.
No mutation of CHCHD2 was observed in either
the 181 ALS or the 88 FTD patients. There are several possible
explanations for these negative results. The first possibility is
that CHCHD2 might not be associated with these two diseases
in the Chinese population, although many pathogenic gene mutations
can be detected in ALS, FTD and PD. Another explanation for these
results is that the CHCHD2 gene might have genetic
heterogeneity in different ethnic groups and since some
CHCHD2 gene mutations are very rare, more samples should be
included to provide more solid evidence.
Rare coding variants play major roles in disease
causation and might contribute to the missing heritability from
genome-wide association studies. In this study, to find out whether
these mutations play a role in the disease, we use bioinformatics
prediction tools to predict the pathogenicity of the mutation and
compared its frequency with healthy control. However, the present
study has several limitations. Firstly, association study requires
a larger sample size to detect rare variants with modest effect
sizes with high statistical power. Secondly, bioinformatics
prediction tools rely on pathogenicity assertions from existing
databases, which might be inaccurate and incomplete.
In general, this study identified a novel mutation
of the CHCHD2 gene in Chinese AD patients, while no mutation
of CHCHD2 was observed in either ALS or FTD patients,
suggesting that the CHCHD2 gene might be associated with AD
in the Chinese Han population. Further screening should be
conducted in a large number of samples and in different ethnicities
and further functional experiments are needed.
Acknowledgements
Not applicable.
Funding
The present study was supported through the National
Natural Science Foundation of China (grant nos. 81671075 and
81701134).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL, BJ, LS and BT designed the study. XL, WZ, TX,
LH, CP and BT conducted the experiments, and analyzed and
interpreted the data. XL and LS wrote the manuscript. BT and LS
supervised the study. BJ and LS provided financial support.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki and was approved by the Expert
Committee of Xiangya Hospital of Central South University in China
(Hunan, China; ref. no. 201603198). Written informed consent for
participation in the study was obtained from all subjects; consent
was obtained from the patient's guardian or immediate family member
for those without the capacity to consent.
Consent for publication
Written informed consent was obtained from all
subjects; consent was obtained from the patient's guardian or
immediate family member for those without the capacity to
consent.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhou ZD, Saw WT and Tan EK: Mitochondrial
CHCHD-containing proteins: Physiologic functions and link with
neurodegenerative diseases. Mol Neurobiol. 54:5534–5546. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bannwarth S, Ait-El-Mkadem S, Chaussenot
A, Genin EC, Lacas-Gervais S, Fragaki K, Berg-Alonso L, Kageyama Y,
Serre V, Moore DG, et al: A mitochondrial origin for frontotemporal
dementia and amyotrophic lateral sclerosis through CHCHD10
involvement. Brain. 137:2329–2345. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xiao T, Jiao B, Zhang W, Pan C, Wei J, Liu
X, Zhou Y, Zhou L, Tang B and Shen L: Identification of CHCHD10
mutation in chinese patients with Alzheimer disease. Mol Neurobiol.
54:5243–5247. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Funayama M, Ohe K, Amo T, Furuya N,
Yamaguchi J, Saiki S, Li Y, Ogaki K, Ando M, Yoshino H, et al:
CHCHD2 mutations in autosomal dominant late-onset Parkinson's
disease: A genome-wide linkage and sequencing study. Lancet Neurol.
14:274–282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jansen IE, Bras JM, Lesage S, Schulte C,
Gibbs JR, Nalls MA, Brice A, Wood NW, Morris H, Hardy JA, et al:
CHCHD2 and Parkinson's disease. Lancet Neurol. 14:678–679. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shi CH, Mao CY, Zhang SY, Yang J, Song B,
Wu P, Zuo CT, Liu YT, Ji Y, Yang ZH, et al: CHCHD2 gene mutations
in familial and sporadic Parkinson's disease. Neurobiol Aging.
38:217.e9–e217.e13. 2016. View Article : Google Scholar
|
|
7
|
Puschmann A, Dickson DW, Englund E,
Wszolek ZK and Ross OA: CHCHD2 and Parkinson's disease. Lancet
Neurol. 14:6792015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Clegg HV, Leslie PL, Di J, Tollini
LA, He Y, Kim TH, Jin A, Graves LM, Zheng J and Zhang Y: CHCHD2
inhibits apoptosis by interacting with Bcl-x L to regulate Bax
activation. Cell Death Differ. 22:1035–1046. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu XC, Cao L, Tan MS, Jiang T, Wang HF,
Lu H, Tan CC, Zhang W, Tan L and Yu JT: Association of Parkinson's
Disease GWAS-linked loci with Alzheimer's disease in Han Chinese.
Mol Neurobiol. 54:308–318. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baizabal-Carvallo JF and Jankovic J:
Parkinsonism, movement disorders and genetics in frontotemporal
dementia. Nat Rev Neurol. 12:175–185. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schapira AH: Mitochondria in the aetiology
and pathogenesis of Parkinson's disease. Lancet Neurol. 7:97–109.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Querfurth HW and LaFerla FM: Alzheimer's
disease. N Engl J Med. 362:329–344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
McKhann G, Drachman D, Folstein M, Katzman
R, Price D and Stadlan EM: Clinical diagnosis of Alzheimer's
disease: Report of the NINCDS-ADRDA Work Group under the auspices
of Department of Health and Human Services Task Force on
Alzheimer's Disease. Neurology. 34:939–944. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brooks BR: El Escorial World Federation of
Neurology criteria for the diagnosis of amyotrophic lateral
sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic
Lateral Sclerosis of the World Federation of Neurology Research
Group on Neuromuscular Diseases and the El Escorial ‘Clinical
limits of amyotrophic lateral sclerosis’ workshop contributors. J
Neurol Sci. 124 Suppl:S96–S107. 1994. View Article : Google Scholar
|
|
15
|
Clinical and neuropathological criteria
for frontotemporal dementia. The Lund and Manchester Groups. J
Neurol Neurosurg Psychiatry. 57:416–418. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang M, Gu X, Wei J, Jiao B, Zhou L, Zhou
Y, Weng L, Yan X, Tang B, Xu J and Shen L: Analyses MAPT, GRN, and
C9orf72 mutations in Chinese patients with frontotemporal dementia.
Neurobiol Aging. 46(235): e11–e15. 2016.
|
|
17
|
Jiao B, Xiao T, Hou L, Gu X, Zhou Y, Zhou
L, Tang B, Xu J and Shen L: High prevalence of CHCHD10 mutation in
patients with frontotemporal dementia from China. Brain.
139:e212016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiao B, Liu X, Tang B, Hou L, Zhou L,
Zhang F, Zhou Y, Guo J, Yan X and Shen L: Investigation of TREM2,
PLD3, and UNC5C variants in patients with Alzheimer's disease from
mainland China. Neurobiol Aging. 35:2422.e9–e2422.e11. 2014.
View Article : Google Scholar
|
|
19
|
Jiao B, Tang B, Liu X, Xu J, Wang Y, Zhou
L, Zhang F, Yan X, Zhou Y and Shen L: Mutational analysis in
early-onset familial Alzheimer's disease in Mainland China.
Neurobiol Aging. 35(1957): e1–e6. 2014.PubMed/NCBI
|
|
20
|
Hou L, Jiao B, Xiao T, Zhou L, Zhou Z, Du
J, Yan X, Wang J, Tang B and Shen L: Screening of SOD1, FUS and
TARDBP genes in patients with amyotrophic lateral sclerosis in
central-southern China. Sci Rep. 6:324782016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aras S, Pak O, Sommer N, Finley R Jr,
Hüttemann M, Weissmann N and Grossman LI: Oxygen-dependent
expression of cytochrome c oxidase subunit 4-2 gene expression is
mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic
Acids Res. 41:2255–2266. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Funayama M and Hattori N: CHCHD2 and
Parkinson's disease-authors' reply. Lancet Neurol. 14:682–683.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ogaki K, Koga S, Heckman MG, Fiesel FC,
Ando M, Labbé C, Lorenzo-Betancor O, Moussaud-Lamodière EL,
Soto-Ortolaza AI, Walton RL, et al: Mitochondrial targeting
sequence variants of the CHCHD2 gene are a risk for Lewy body
disorders. Neurology. 85:2016–2025. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Feyeux M, Bourgois-Rocha F, Redfern A,
Giles P, Lefort N, Aubert S, Bonnefond C, Bugi A, Ruiz M, Deglon N,
et al: Early transcriptional changes linked to naturally occurring
Huntington's disease mutations in neural derivatives of human
embryonic stem cells. Hum Mol Genet. 21:3883–3895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Che XQ, Zhao QH, Huang Y, Li X, Ren RJ,
Chen SD, Guo QH and Wang G: Mutation screening of the CHCHD2 gene
for Alzheimer's Disease and Frontotemporal Dementia in Chinese
Mainland Population. J Alzheimers Dis. 61:1283–1288. 2018.
View Article : Google Scholar : PubMed/NCBI
|