Introduction
LIN28 was first identified in Caenorhabditis
elegans as a key control element of embryonic-stage development
(1). Mammals have two homologs of
LIN28, LIN28A and LIN28B; LIN28B is expressed in the nucleus and
regulates the development of embryos by controlling the maturation
of LET-7 (2). LIN28A is primarily
expressed in embryonic stem cells (ESCs). LIN28A is also able to
re-program human fibroblasts into induced pluripotent stem cells,
along with octamer-binding protein 4 (OCT4), SOX2 and NANOG, which
may indicate a stemness function (3,4).
LIN28A may function primarily by inhibiting the maturation of LET-7
microRNA (miRNA) precursors (2).
A number of previous studies have reported that
LIN28A is not only expressed in ESCs, but also serves a key role in
tumor development (5–9). LIN28A was demonstrated to promote
malignant cell transformation and tumor progression, and is also
associated with the advanced stages of many tumors, including
ovarian carcinoma, hepatocarcinoma, germ cell tumors and lung
carcinoma (6–10); however, the role in bladder cancer
remains poorly understood.
A previous study (11) have suggested that in addition to
targeting LET-7 precursors, LIN28A also targets several other
miRNAs and is an inhibitor of endoplasmic reticulum (ER)-related
protein translation in ESCs LAMP1 is a membrane glycoprotein and an
ER-related protein that functions to maintain the integrity of the
lysosomal membrane and serves a role in protein transport, membrane
fusion and the transport of intracellular degradation of protein
(12,13). However, whether LIN28A affects the
translation of LAMP1 proteins in ESCs or in cancer cells is
unclear. The process of ESC differentiation is accompanied by a
loss of immortality, and the formation of tumor cells indicates
that the cells acquire immortality and multi-differentiation
(14), which further suggested
that a similar functional status may exist between ESCs and tumor
cells.
The present study aimed to determine the effects of
LIN28A on LAMP1 expression in mouse (m)ESCs and in human bladder
cancer cells, and the results demonstrated that LIN28A inhibited
LAMP1 protein synthesis in mESCs and in human cancer cells. In
addition, it was also demonstrated that LIN28A is able to maintain
human bladder cancer cell proliferation migration and invasion.
Materials and methods
Cell culture
mESCs (National Key Laboratory of Stem Cell and
Reproductive Biology, Chinese academy of sciences; Beijing, China)
were cultured in mESC media: Dulbecco's modified Eagle medium
(DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing
15% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.), 100
mM Minimum Essential Medium/Non-essential Amino Acids medium
(Thermo Fisher Scientific, Inc.), 1,000 U/ml mLIF (Millipore,
Darmstadt, Germany) and 50 mM 2-mercaptoethanol (Thermo Fisher
Scientific, Inc.).
Human bladder cancer cell lines 5637, SW780, T24,
J82 and UM-UC3 and human cervical cancer cell line HeLa (Type
Culture Collection of the Chinese Academy of Sciences, Shanghai,
China) were cultured in DMEM with 1% L-glutamine (Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Thermo Fisher
Scientific, Inc.). The normal human urothelial cell line SV-HUC-1
(Type Culture Collection of the Chinese Academy of Sciences) was
cultured in F12K medium (Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS and 1% L-glutamine. Cells were cultured
in a standard humidified incubator at 37°C in a 5% CO2
atmosphere.
Transfection of siRNA
One day prior to transfection, 3×104
cells were plated in 200 µl DMEM with 1% L-glutamine and 10% FBS in
48-well plates. After 24 h, 0.6 µl LAMP1 siRNA (stock solution 20
pmol/µl; Shanghai GenePharma Co., Ltd., Shanghai, China) was added
to 40 µl Opti-MEM I Reduced Serum medium (Invitrogen; Thermo Fisher
Scientific, Inc.) to a final concentration of 50 pM. siRNA was
gently mixed with Lipofectamine 3000 (Invitrogen, CA, USA) and
incubated for 20 min at room temperature. LIN28A-targeted siRNA
(mouse) was used to knock down LIN28A in mESCs, and LIN28A-targeted
siRNA (human) and LAMP1-targeted (human) siRNA were used to knock
down LIN28A and LAMP1 in human bladder cancer cells. siRNA was
designed with the following primers: LIN28A-targeted siRNA (human)
(5′-GCAUCUGUAAGUGGUUCAATT-3′), LIN28A-targeted siRNA (mouse)
(5′-GCAGTGGAGTTCACCTTTAAG-3′), LAMP1-targeted (human) siRNA
(5′-GCCACAGUCGGCAAUUCCUACAAGU-3′) and negative control (NC)-siRNA
(5′-UUCUCCGAACGUGUCACGUTT-3′). Cells were harvested 24 h
post-transfection. The cells in the wild type (WT) group did not
undergo any treatment.
Total RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cells when the
confluence reached 80%, using TRIzol reagent (Takara Bio, Inc.,
Otsu, Japan), according to the manufacturer's protocol. Total RNA
(10 µg) was reverse transcribed to cDNA using the PrimeScript RT
Reagent kit with gDNA Eraser (Takara Bio, Inc.), according to the
manufacturer's protocol. qPCR was performed using SYBR Premix Ex
Taq (TaKaRa Bio, Inc.). In detail, the cDNAs acquired following RT
were diluted 10 times, then 2 µl was mixed with 5 µl AceQ qPCR SYBR
Green Master Mix and 50 µM primers (0.15 µl each), and distilled
deionized water was added to make a total volume of 10 µl. The
thermocycling conditions used were as follows: Initial denaturation
at 95°C for 10 min; followed by 35 cycles of amplification at 95°C
for 10 sec and 58°C for 30 sec. The samples were amplified using
the LightCycler® Nano system (Roche Diagnostics, Basel,
Switzerland). The primers listed in Table I; experiments were performed in
triplicate. mRNA expression levels were normalized to GAPDH and
calculated using the 2−ΔΔCq method (15).
 | Table I.Sequences of primers used for PCR,
position of primers on the gene-specific mRNA and expected
amplification length of PCR products. |
Table I.
Sequences of primers used for PCR,
position of primers on the gene-specific mRNA and expected
amplification length of PCR products.
| Gene | Primer (5′→3′) | Position (nt) | Size (bp) |
|---|
| LAMP1 | F:
CAGATGTGTTAGTGGCACCCA | 459–479 | 84 |
|
| R:
TTGGAAAGGTACGCCTGGATG | 542–522 |
|
| LIN28A | F:
TGCGGGCATCTGTAAGTGG | 120–138 | 134 |
|
| R:
GGAACCCTTCCATGTGCAG | 253–235 |
|
| GAPDH | F:
TGTGGGCATCAATGGATTTGG | 231–251 | 116 |
|
| R:
ACACCATGTATTCCGGGTCAAT | 346–325 |
|
Gel electrophoresis
A total of 1 µg of cDNA obtained from cells was
mixed with 2 µl of primers (Table
I), 10 µl of PCR Master Mix (Thermo Fisher Scientific, Inc.)
and 8 µl of RNase-free water. A Bio-Rad T100 (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) thermocycler was used for amplification.
Amplification was performed under the following conditions: Initial
denaturation at 98°C for 3 min; followed by 40 cycles of
denaturation at 98°C for 30 sec, annealing at 60°C for 30 sec and
extension at 72°C for 30 sec; and a final extension at 72°C for 5
min. Electrophoresis (85 V) was performed using 1.2% agarose gels
containing ethidium bromide at room temperature for 120 min.
Following completion, the gel was placed in an Ulttima 16si
detector (Hoefer, Inc., Holliston, MA, USA) in order to visualize
the DNA bands by irradiation using ultraviolet light (254 nm).
Densitometric analysis was performed using E-Editor 2.0 software
(Thermo Fisher Scientific, Inc.).
Fluorescence immunocytochemistry
For immunofluorescence staining, 3×104
cells were placed onto coverslips and fixed with 4%
paraformaldehyde at room temperature for 30 min, permeabilized in
0.1% Triton for 15 min, blocked by 5% bovine serum albumin (Thermo
Fisher Scientific, Inc.) at 37°C for 30 min, incubated with
anti-LAMP1 (1:100; cat. no. 21997-1-AP; ProteinTech Group, Inc.,
Chicago, IL, USA) or anti-LIN28A primary antibodies (1:100; cat.
no. ab46020; Abcam, Cambridge, UK) at 37°C for 2 h, followed by
incubation with an Alexa Fluor 594-conjugated goat anti-rabbit
secondary antibody (1:500; A-11012; Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 1 h. Nuclei were stained with Hoechst
33342 (1:2,000) at 37°C for 5 min. Images were captured with a
Zeiss LSM710 fluorescence microscope (Oberkochen, Germany). mESCs
were used as a positive control for LIN28A and LAMP1 expression,
SV-HUC-1 cells were used as negative control for LAMP1 expression
and HeLa cells were used as a negative control for LIN28A
expression (16,17).
Western blot analysis
Proteins were extracted from cells when the
confluence reached 80% using radioimmunoprecipitation assay buffer
(Beyotime Institute of Biotechnology, Shanghai, China) supplemented
with protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany). Protein concentrations were determined using
the Pierce BCA Protein assay kit (cat. no. 23227; Thermo Fisher
Scientific, Inc.). Protein extracts (50 µg) were separated by 12%
SDS-PAGE and transferred to polyvinylidene difluoride membranes
(EMD Millipore, Billerica, MA, USA). The proteins were then
transferred onto PVDF membranes (EMD Millipore). Following blocking
with 5% non-fat milk (blocking buffer) at room temperature for 1 h,
the membranes were incubated overnight at 4°C with appropriate
primary antibodies diluted in blocking buffer as follows: Rabbit
monoclonal lysosome-associated membrane glycoprotein 1 (LAMP1;
1:500; cat. no. 9091S; Cell Signaling Technology, Inc., Danvers,
MA, USA), mouse monoclonal LIN28A (1:500; cat. no. 11724-1-AP;
ProteinTech Group, Inc., Chicago, IL, USA) and mouse monoclonal
GAPDH (1:5,000; cat. no. 51332S; Cell Signaling Technology, Inc.).
Following washing with PBS with 0.1% Tween-20 three times, the
membranes were incubated at room temperature for 1 h with
anti-mouse (1:2,000; cat. no. sc-2005; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) or anti-rabbit (1:2,000; cat. no. sc-2004;
Santa Cruz Biotechnology, Inc.) horseradish peroxidase-conjugated
secondary antibodies. Subsequently, the bands of target proteins
were visualized using Chemistar ECL Western Blotting Substrate and
Tanon-5200 automatic chemiluminescence image analysis system (both
Tanon Science & Technology, Ltd., Shanghai, China). The
relative intensity of the target bands were quantified by
densitometric scanning using Image J 1.32j software (National
Institutes of Health, Bethesda, MD, USA).
Co-immunoprecipitation (Co-IP)
Proteins were extracted from cells when the
confluence reached 70–80% using 100 µl RIPA buffer (Beyotime
Institute of Biotechnology) supplemented with 1% protease inhibitor
cocktail (Sigma-Aldrich; Merck KGaA). The concentration of protein
was determined using a BCA kit according to the aforementioned
protocol. Protein extracted from cells was mixed with Protein A
agarose (Santa Cruz Biotechnology, Inc.) and the respective
antibodies for 8 h at 4°C. The antibodies used were: Rabbit
monoclonal LAMP1, mouse monoclonal LIN28A and mouse monoclonal as
aforementioned. Subsequently, the combined product was incubated
with Dynabeads Protein G (Santa Cruz Biotechnology, Inc.) for 1 h
at 4°C. The bound proteins were analyzed using western blot, as
aforementioned.
Transwell invasion assay
A total of 40 µl Matrigel (0.5 mg/ml; Beckman
Coulter, Inc., Brea, CA, USA) was spread onto the upper Transwell
chamber and incubated for 4 h at 37°C. A total of 1×105
cells, harvested 24 h post-siRNA transfection, were seeded in the
upper compartment of a Transwell chamber (Corning Inc., Corning,
NY, USA) containing OptiMEM I Reduced Serum Medium (Thermo Fisher
Scientific, Inc.) and incubated at 37°C for 24 h. The cells in the
upper membrane were removed via rinsing with PBS three times. The
migrated cells underneath the membrane were stained with 0.1%
crystal violet at room temperature for 30 min, and cells were
counted from three random fields per membrane under a light
microscope (magnification, ×20); the average number of cells was
calculated from three independent experiments.
Wound-healing assay
A total of 3×105 siRNA-transfected cells
were cultured in 6-well plates to form a monolayer at 37°C, 5%
CO2 for 24 h. A scratch was made in the cell monolayer
and the migration distances were measured at 0 and 24 h; the
average migration distance was calculated from three independent
experiments.
Cell viability assay
The viability of bladder cancer cells was
quantitatively assessed using the Cell Counting kit-8 (CCK-8) assay
according to the manufacturer's protocol at 0, 24, 48 and 72 h
post-transfection with siRNA. The cells were incubated in 10 µl of
CCK-8 solution (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) at 37°C for 1 h. The optical density of each well was
determined using a microplate reader (Bio-Rad Laboratories, Inc.);
the absorbance was measured at 450 nm.
Statistical analysis
To compare data between different groups, one-way
analysis of variance was performed followed by Fisher's least
significant difference (for equal variances) or Games-Howell (for
unequal variances) post hoc test using SPSS 17.0 software (SPSS
Inc., IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Lamp1 and lin28A are expressed in
mESCs and human bladder cancer cells
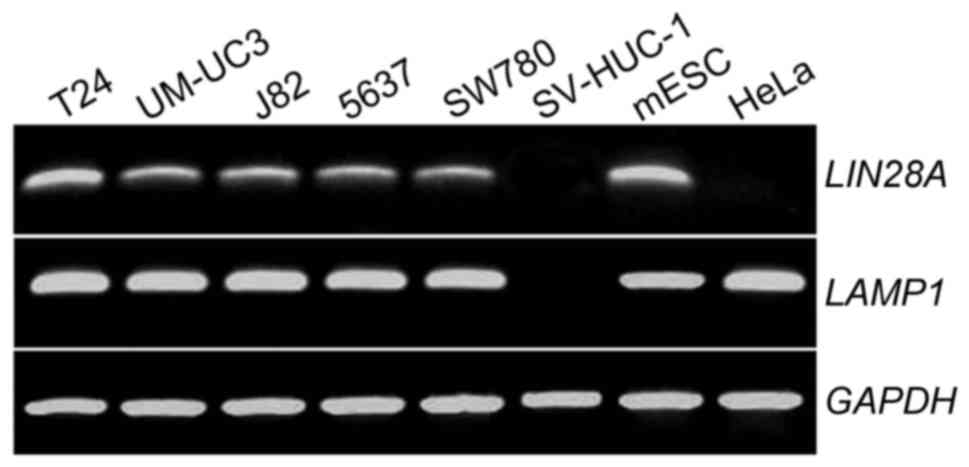
RT-PCR was used to determine if LAMP1 and LIN28A
mRNAs were expressed in five human bladder cell lines (T24, UMUC3,
J82, 5637 and SW780), one normal human bladder urothelial cell line
(SV-HUC-1) and a mESC line. mESCs were used as a positive control
for LIN28A and LAMP1 expression, SV-HUC-1 cells were used as
negative control for LAMP1 expression and HeLa cells were used as a
negative control for LIN28A (16,17).
The results demonstrated that LIN28A and LAMP1 were expressed in
all types of human bladder cancer cell lines and in mESCs (Fig. 1).
Three bladder cancer cell lines (UM-UC-3, 5637 and
SW780) were selected for further experimentation. Results from the
immunofluorescence expression assay revealed that LAMP1 and LIN28A
proteins were expressed in the cytoplasm of the three bladder
cancer cell lines, which means LAMP1 and LIN28A are expressed in
three malignant bladder cancer cells with different levels of
malignancy, but the relationship is not clear (Fig. 2).
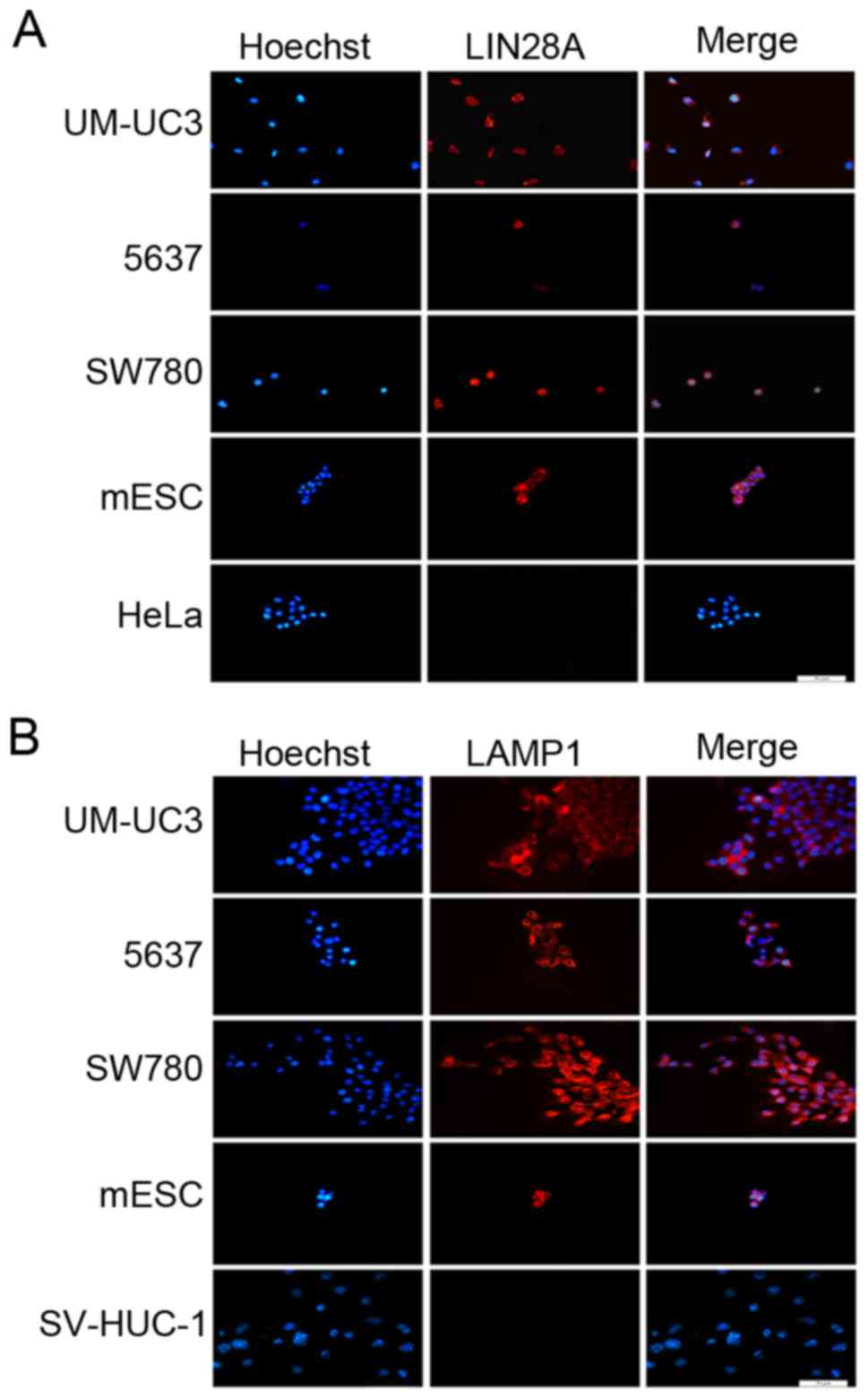 | Figure 2.LAMP1 and LIN28A proteins are
expressed in human bladder cancer cells and mESCs. Fluorescence
immunocytochemical analysis was used to determine the location of
(A) LIN28A protein expression in UM-UC3, 5637, SW780, mESCs and
HeLa cells and (B) LAMP1 protein expression in UM-UC3, 5637, SW780,
mESCs and SV-HUC-1 cells. mESCs were used as a positive control for
LIN28A and LAMP1 expression levels, HeLa cells were used as a
negative control for LIN28A expression and SV-HUC-1 cells were used
as a negative control for LAMP1 expression. Nuclei were stained
with Hoechst 33342; scale bar, 50 µm. LAMP1, lysosome-associated
membrane glycoprotein 1; mESCs, mouse embryonic stem cell. |
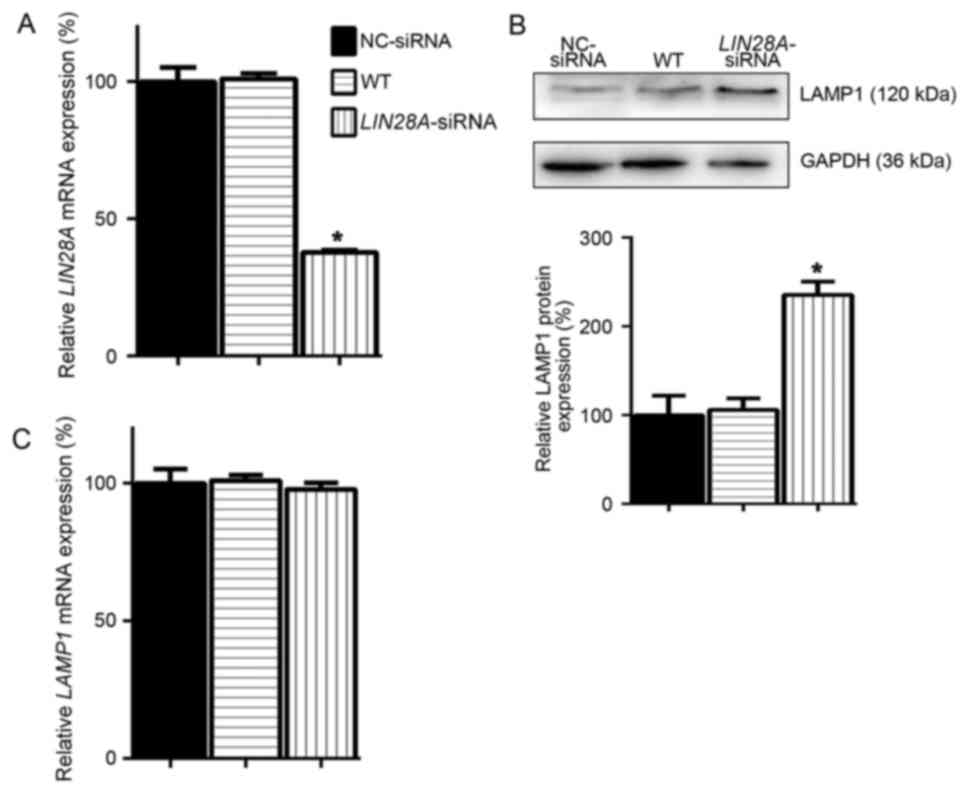
LIN28A inhibits LAMP1 protein
expression in mESCs
A previous study reported that LIN28A is a major
inhibitor of ER-related protein translation in ESCs (9). As an ER-related protein, the
expression of LAMP1 in ESCs may be regulated by LIN28A; therefore,
LAMP1 protein expression levels were examined, following the
successful knockdown of LIN28A mRNA expression in mESCs (Fig. 3A). In mESCs transfected with
LIN28A-siRNA the protein expression level of LAMP was demonstrated
to be significantly increased by 1.35-fold compared with expression
in NC-siRNA transfected cells (Fig.
3B). As the effects of LIN28A on its target gene LET7 are at
the post-transcriptional level, the expression levels of LAMP1 mRNA
were examined. No significant differences were identified in the
expression of LAMP1 mRNA in LIN28A-siRNA treated cells compared
with WT and NC-siRNA transfected cells (Fig. 3C), which suggested that LIN28A
inhibition of LAMP1 protein expression may be independent of LAMP1
mRNA levels in mESCs.
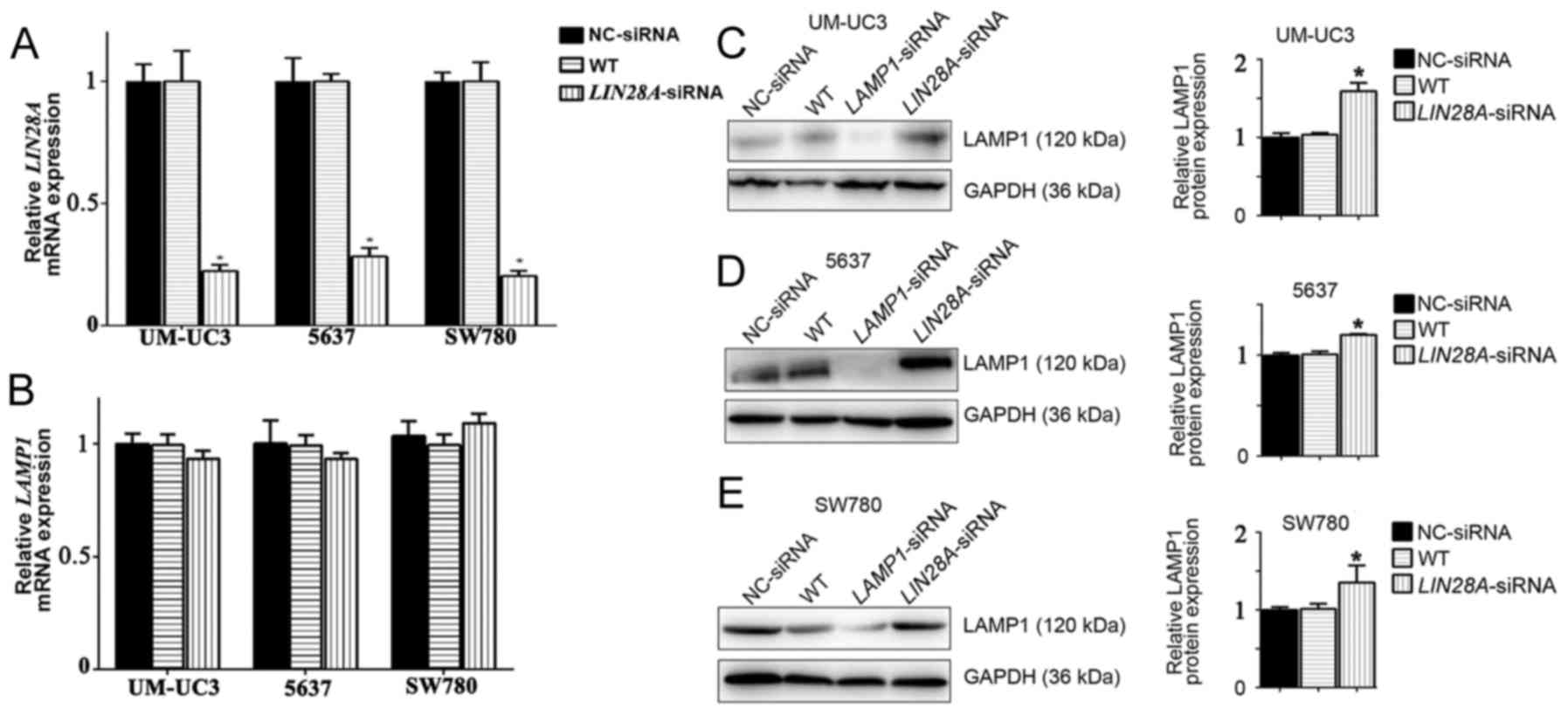
LIN28A inhibits LAMP1 protein
translation in human bladder cancer cells
As LIN28A was revealed to affect the protein
expression levels of LAMP1 in mESCs, human bladder cancer cells
were subsequently used in LIN28A knockdown experiments to determine
if a similar process occurred. Following successful transfection
with LIN28A-siRNA (Fig. 4A), LAMP1
mRNA expression levels were unaffected compared with NC-siRNA
transfected cells (Fig. 4B). In
addition, following knockdown of LIN28A, LAMP1 protein expression
levels were significantly increased in UM-UC3, 5637 and SW780 cells
by 1.59, 1.2 and 1.35-fold compared with NC-siRNA cells,
respectively (Fig. 4C-E). These
results indicated that LIN28A may inhibit the translation of LAMP1
protein in ESCs and in human cancer cells.
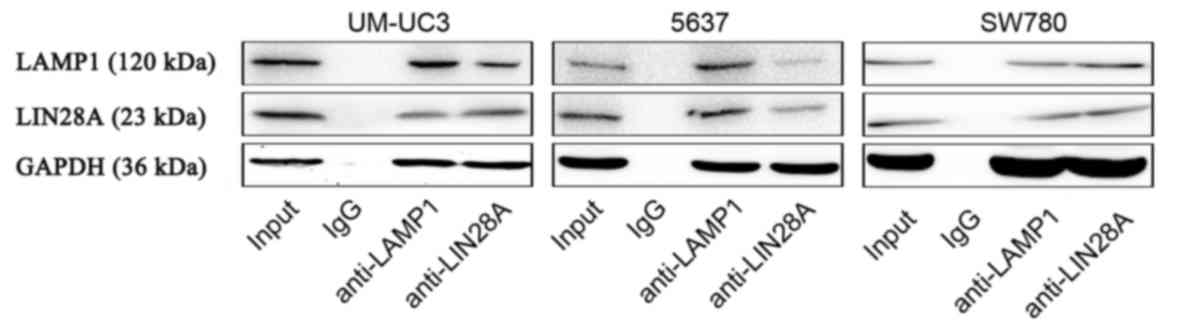
To further demonstrate the relationship between
LIN28A and LAMP1, Co-IP experiments were conducted, which
demonstrated that LIN28A protein interacts with LAMP1 protein
(Fig. 5). These data indicated
that LIN28A may serve a direct role in the regulation of LAMP1
protein expression.
Effects of LIN28A on cell migration,
proliferation and invasion of human bladder cancer cells in
vitro
To investigate the effects of LIN28A on cancer
cells, LIN28A knockdown was conducted in three types of bladder
cancer cells; the migration, invasion and proliferative capacities
were measured by wound-healing, invasion and CCK-8 assays,
respectively. The data revealed the three types of bladder cancer
cells (UM-UC3, 5637, SW780) markedly reduced the rate of cell
migration, invasion and proliferation after the knockout of LIN28A
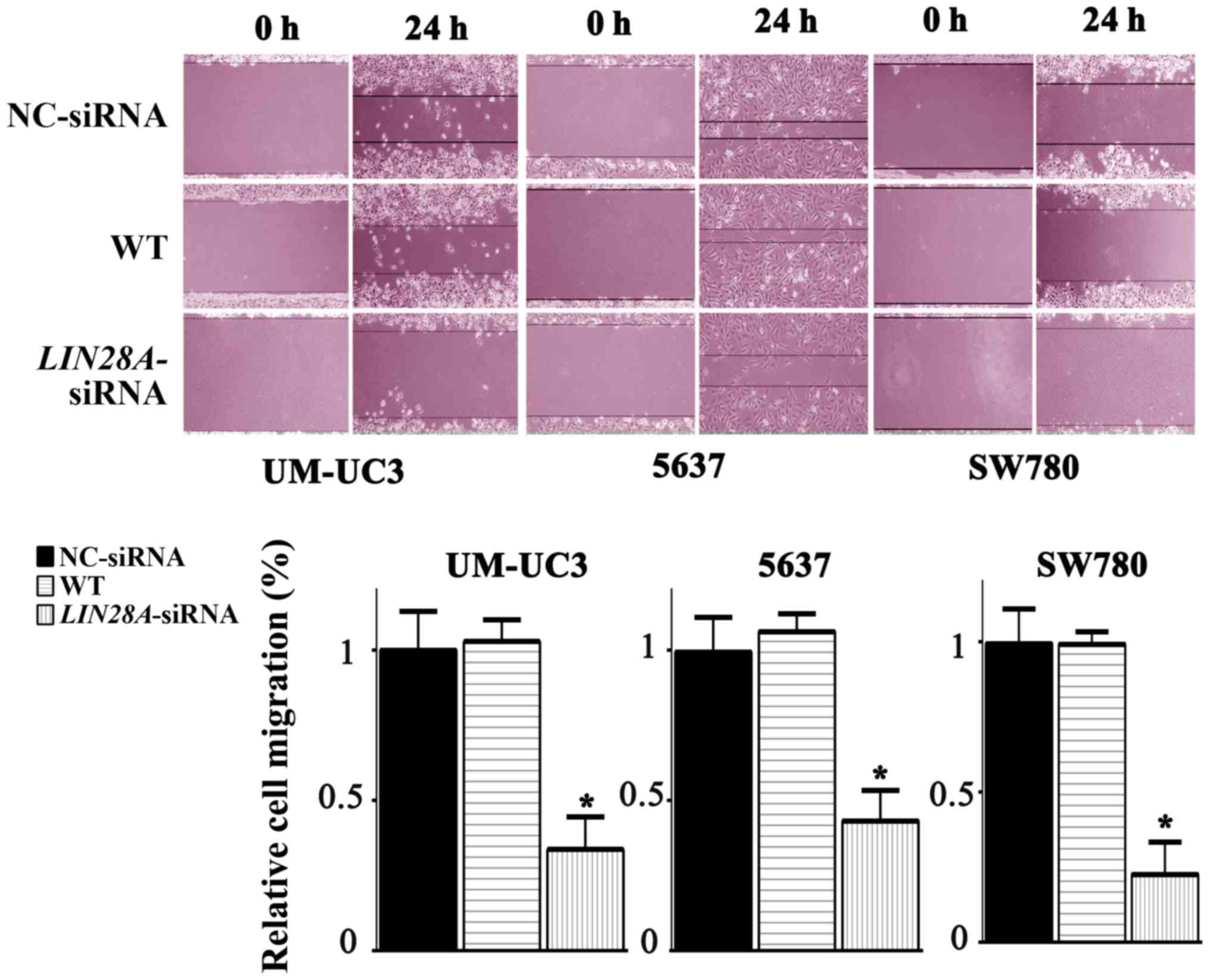
in bladder cancer cells. The migration distance exhibited by
si-LIN28A cells in the UM-UC3, 5637 and SW780 cell lines was
significantly decreased by 0.72, 0.66 and 0.75 fold compared with
the NC group, respectively (Fig.
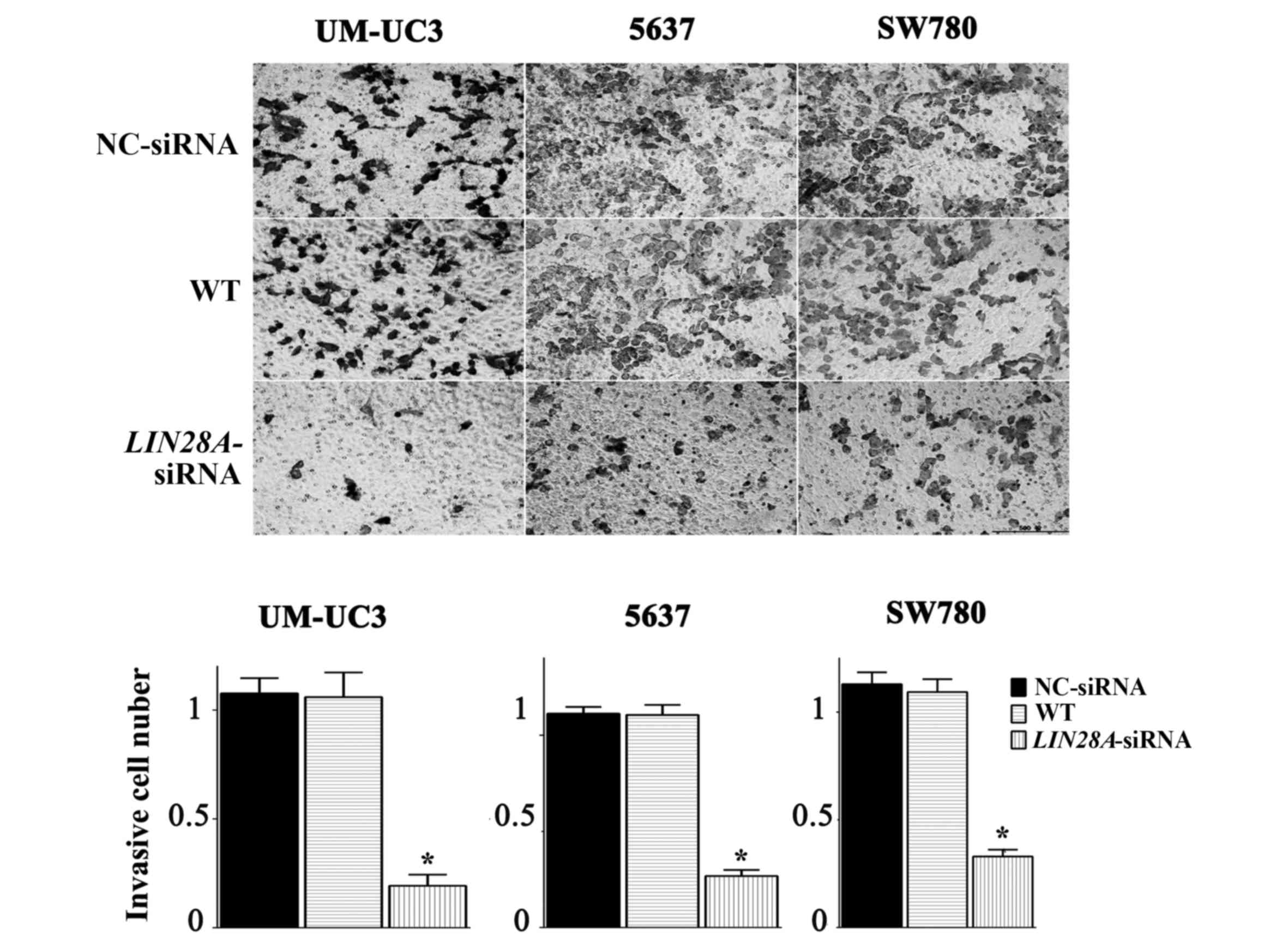
6). In addition, the invasive ability of si-LIN28A cells in the
UM-UC3, 5637 and SW780 cell lines was significantly decreased by
0.75, 0.53 and 0.63 fold compared with the NC group, respectively
(Fig. 7). Furthermore, the
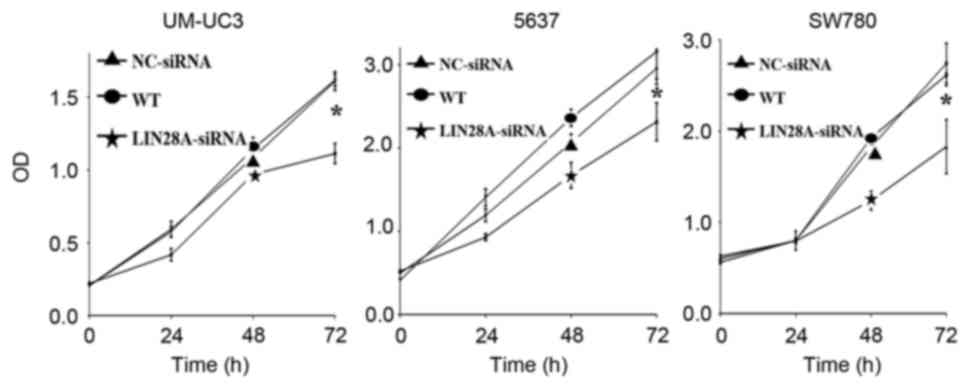
proliferative ability of si-LIN28A cells in the UM-UC3, 5637 and
SW780 cell lines at the 72 h time interval was significantly
suppressed by 0.52, 0.56 and 0.45 fold compared with the NC group,
respectively (Fig. 8). The results
demonstrated that LIN28A may suppress the migration, invasion and
proliferation of bladder cancer cells.
Discussion
LIN28A was previously reported to promote protein
translation in stem cells (14).
In addition, LIN28A was also reported to reduce the translation of
proteins in the ER and Golgi lumen (11).
Crosslinking-immunoprecipitation-sequencing technology was
previously used to reveal that LIN28A inhibits the expression of
ER-related proteins in ESCs (11).
The present study used western blotting to demonstrate that
expression of the ER-related protein LAMP1 was increased following
reduced LIN28A expression in mESCs and human bladder cancer cells.
Based on these results, it was speculated that the development of
ESCs and human tumor cells was comparable.
A number of genes have been previously demonstrated
to serve a key role in stem cells, some of which may exhibit
oncogene-like functions, such as LIN28A and OCT4
(8,9). In addition, cancer stem cells (CSCs)
have been reported to accelerate the process of tumor malignancy
(18). Epithelial-mesenchymal
transition has also been reported to serve an important role in ESC
differentiation (19,20) and in carcinoma development
(21,22).
The present study results suggested that the
inhibitory effect of LIN28A on LAMP1 protein expression was at the
post-transcriptional level and did not affect lamp1 mRNA
production in either mESCs or in human bladder cancer cells in
vitro. But the expression of LAMP1 protein increased following
knockdown of LIN28A, indicating that LIN28A may downregulated the
protein expression levels of LAMP1. It has been reported that
LIN28A may alter the ribosome structure of some protein synthesis;
LIN28A may inhibit the formation of LAMP1 protein without affecting
its mRNA expression levels in this way (23). LIN28A was previously reported to
post-transcriptionally regulate the expression of let7 miRNA
precursors (24). Recent studies
have revealed that, in addition to let7, LIN28A also
modifies a number of mRNAs and miRNAs at the post-transcriptional
level without affecting RNA synthesis, such as insulin-like growth
factor 2 and members of the miRNA-106 family (25,26).
These results are consistent with those from the present study.
LIN28A also serves an important role in human tumor
cells; an increasing number of studies have reported that the
overexpression of LIN28A is one of the biomarkers of CSCs, such as
in hepatocellular carcinoma (6),
and in ovarian (7), colon
(8) and prostate (27) cancers. The present study results
demonstrated that LIN28A was expressed in the human bladder cancer
cell lines UM-UC3, SW780 and 5637, but not in healthy SV-HUC-1
urothelial cells, and suggested that LIN28A may serve an important
role in the development of tumor cells by increasing the
proliferation, migration and invasion of bladder cancer cells.
It has been reported that LIN28A affects
mitochondrial localization in the cell cytoplasm, which suggested
an involvement in oxidative metabolism and oxidative
phosphorylation (OXPHOS) (28).
Previous studies have also reported that LIN28A is enriched in the
intracellular region, including the inner mitochondrial membrane,
thus inhibiting cellular oxidative metabolism (29,30).
LIN28A was reported to simultaneously repress cell OXPHOS and
promote cell anaerobic glycolysis (30). In addition, mitochondrial
localization in the cytoplasm is altered following LAMP1 knockdown,
which further suggested that LAMP1 may participate in mitochondrial
functions (28). Providing the
importance of mitochondria in cellular glucose metabolism, LAMP1
may participate in the process of OXPHOS (31). Inhibition of LAMP1 protein
translation may be one of the ways in which LIN28A regulates cell
metabolism. LIN28A may inhibit the expression of OXPHOS-associated
proteins; LAMP1 is an OXPHOS-associated protein (32). The ER-associated protein LAMP1 is a
mitochondria and OXPHOS-related protein that is inhibited by
LIN28A.
In conclusion, the results of the present study
demonstrated that LIN28A reduced LAMP1 protein expression without
inhibiting the level of LAMP1 mRNA expression. The results of the
present study, in agreement with Kim et al (33), suggested that embryonic stem cells
and tumor cells have similar transcription mechanisms, metabolic
pathways and protein synthesis pathways, which may provide a novel
treatment approach for the treatment of tumors. Furthermore, the
results also suggested that LIN28A may represent a novel target for
the treatment of bladder cancer.
Acknowledgements
We wish to thank Dr Liu for the provision of primer
sequences and Dr Chen for advanced experimental experience of
CO-IP. We also wish to thank the staff at MedSci Medicine for the
editing of our manuscript.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81270748).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PP, TC, GC and XG made substantial contributions to
the conception and design of the study; PP, TC, YZ and ZQ acquired
the data; PP, ZQ, JQ, GC and XG performed the analysis and
interpretation of data; PP, TC, YZ and JQ drafted and revised the
manuscript; and GC and XG provided the final approval of the
version of the manuscript to be published.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moss EG, Lee RC and Ambros V: The cold
shock domain protein LIN-28 controls developmental timing in C.
elegans and is regulated by the lin-4 RNA. Cell. 88:637–646. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Piskounova E, Polytarchou C, Thornton JE,
LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D and Gregory RI:
Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct
mechanisms. Cell. 147:1066–1079. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu J, Vodyanik MA, Smuga-Otto K,
Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA,
Ruotti V, Stewart R, et al: Induced pluripotent stem cell lines
derived from human somatic cells. Science. 318:1917–1920. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park IH, Zhao R, West JA, Yabuuchi A, Huo
H, Ince TA, Lerou PH, Lensch MW and Daley GQ: Reprogramming of
human somatic cells to pluripotency with defined factors. Nature.
451:141–146. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Korshunov A, Ryzhova M, Jones DT,
Northcott PA, van Sluis P, Volckmann R, Koster J, Versteeg R,
Cowdrey C, Perry A, et al: LIN28A immunoreactivity is a potent
diagnostic marker of embryonal tumor with multilayered rosettes
(ETMR). Acta Neuropathol. 124:875–881. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tian N, Han Z, Li Z, Zhou M and Fan C:
Lin28/let-7/Bcl-xL pathway: The underlying mechanism of drug
resistance in Hep3B cells. Oncol Rep. 32:1050–1056. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Enriquez VA, Cleys ER, Da Silveira JC,
Spillman MA, Winger QA and Bouma GJ: High LIN28A expressing ovarian
cancer cells secrete exosomes that induce invasion and migration in
HEK293 Cells. Biomed Res Int. 2015:7013902015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paz EA, LaFleur B and Gerner EW:
Polyamines are oncometabolites that regulate the LIN28/let-7
pathway in colorectal cancer cells. Mol Carcinog. 1 53
Suppl:E96–E106. 2014. View
Article : Google Scholar
|
|
9
|
Murray MJ, Saini HK, Siegler CA, Hanning
JE, Barker EM, van Dongen S, Ward DM, Raby KL, Groves IJ, Scarpini
CG, et al: LIN28 Expression in malignant germ cell tumors
downregulates let-7 and increases oncogene levels. Cancer Res.
73:4872–4884. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roos M, Rebhan MA, Lucic M, Pavlicek D,
Pradere U, Towbin H, Civenni G, Catapano CV and Hall J: Short
loop-targeting oligoribonucleotides antagonize Lin28 and enable
pre-let-7 processing and suppression of cell growth in
let-7-deficient cancer cells. Nucleic Acids Res. 43:e92015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cho J, Chang H, Kwon SC, Kim B, Kim Y,
Choe J, Ha M, Kim YK and Kim VN: LIN28A is a suppressor of
ER-associated translation in embryonic stem cells. Cell.
151:765–777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Saftig P and Klumperman J: Lysosome
biogenesis and lysosomal membrane proteins: Trafficking meets
function. Nat Rev Mol Cell Biol. 10:623–635. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eskelinen EL: Roles of LAMP-1 and LAMP-2
in lysosome biogenesis and autophagy. Mol Aspects Med. 27:495–502.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herreros-Villanueva M, Bujanda L,
Billadeau DD and Zhang J: embryonic stem cell factors and
pancreatic cancer. World J Gastroenterol. 20:2247–2254. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li MA, Amaral PP, Cheung P, Bergmann JH,
Kinoshita M, Kalkan T, Ralser M, Robson S, von Meyenn F, Paramor M,
et al: A lncRNA fine tunes the dynamics of a cell state transition
involving Lin28, let-7 and de novo DNA methylation. Elife. 6:pii:
e23468. 2017. View Article : Google Scholar
|
|
17
|
Cohnen A, Chiang SC, Stojanovic A, Schmidt
H, Claus M, Saftig P, Janßen O, Cerwenka A, Bryceson YT and Watzl
C: Surface CD107a/LAMP-1 protects natural killer cells from
degranulation-associated damage. Blood. 122:1411–1418. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chaffer CL, Marjanovic ND, Lee T, Bell G,
Kleer CG, Reinhardt F, D'Alessio AC, Young RA and Weinberg RA:
Poised chromatin at the ZEB1 promoter enables breast cancer cell
plasticity and enhances tumorigenicity. Cell. 154:61–74. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Stryjewska A, Dries R, Pieters T,
Verstappen G, Conidi A, Coddens K, Francis A, Umans L, van IJcken
WF, Berx G, et al: Zeb2 regulates cell fate at the exit from
epiblast state in mouse embryonic stem cells. Stem Cells.
35:611–625. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cheung LY, Davis SW, Brinkmeier ML, Camper
SA and Pérez-Millán MI: Regulation of pituitary stem cells by
epithelial to mesenchymal transition events and signaling pathways.
Mol Cell Endocrinol. 445:14–26. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cheng SW, Tsai HW, Lin YJ, Cheng PN, Chang
YC, Yen CJ, Huang HP, Chuang YP, Chang TT, Lee CT, et al: Lin28B is
an oncofetal circulating cancer stem cell-like marker associated
with recurrence of hepatocellular carcinoma. PLoS One.
8:e800532013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yuan WT, Peng CY, Lee SS, Chou MY, Yu CC
and Chang YC: Acquisition cancer stemness, mesenchymal
transdifferentiation, and chemoresistance properties by chronic
exposure of oral epithelial cells to arecoline. Oncotarget.
7:84072–84081. 2016.PubMed/NCBI
|
|
23
|
Shyh-Chang N and Daley GQ: Lin28: Primal
regulator of growth and metabolism in stem cells. Cell Stem Cell.
12:395–406. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Seltzer J, Scotton TC, Kang K, Zada G and
Carmichael JD: Gene expression in prolactinomas: A systematic
review. Pituitary. 19:93–104. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Thakar NY, Ovchinnikov DA, Hastie ML,
Gorman J and Wolvetang EJ: RELB alters proliferation of human
pluripotent stem cells via IMP3- and LIN28-mediated modulation of
the expression of IGF2 and other cell-cycle regulators. Stem Cells
Dev. 24:1888–1900. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Warrander F, Faas L, Kovalevskiy O, Peters
D, Coles M, Antson AA, Genever P and Isaacs HV: lin28 proteins
promote expression of 17~92 family miRNAs during amphibian
development. Dev Dyn. 245:34–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Culig Z: Words of wisdom: Re: Lin28
promotes growth of prostate cancer cells and activates the androgen
receptor. Eur Urol. 65:10132014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rajapakshe AR, Podyma-Inoue KA, Terasawa
K, Hasegawa K, Namba T, Kumei Y, Yanagishita M and Hara-Yokoyama M:
Lysosome-associated membrane proteins (LAMPs) regulate
intracellular positioning of mitochondria in MC3T3-E1 cells. Exp
Cell Res. 331:211–222. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu H, Shyh-Chang N, Segrè AV, Shinoda G,
Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG,
et al: The Lin28/let-7 axis regulates glucose metabolism. Cell.
147:81–94. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang J, Ratanasirintrawoot S,
Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D,
Xia Q, Seligson M, et al: LIN28 regulates Stem Cell metabolism and
conversion to primed pluripotency. Cell Stem Cell. 19:66–80. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Andrzejewski S, Gravel SP, Pollak M and
St-Pierre J: Metformin directly acts on mitochondria to alter
cellular bioenergetics. Cancer Metab. 2:122014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Docherty CK, Salt IP and Mercer JR: Lin28A
induces energetic switching to glycolytic metabolism in human
embryonic kidney cells. Stem Cell Res Ther. 7:782016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim J, Woo AJ, Chu J, Snow JW, Fujiwara Y,
Kim CG, Cantor AB and Orkin SH: A Myc network accounts for
similarities between embryonic stem and cancer cell transcription
programs. Cell. 143:313–324. 2010. View Article : Google Scholar : PubMed/NCBI
|