Introduction
Breast cancer is one of the most common malignancies
in women and demonstrates an increasing incident rate (1). Advances in neoadjuvant therapy have
greatly altered the treatment of patients with breast cancer,
particularly patients with Her-2-positive breast cancer.
Neoadjuvant therapy increases the opportunity for breast-conserving
surgery in patients that were originally candidates for mastectomy
and allows a potentially more defined prognosis (2). Trastuzumab, a Her-2-targeting
therapeutic antibody, has increased the survival of patients
suffering from Her-2-positive breast cancer. Various randomized
trials have demonstrated that trastuzumab significantly improved
the efficacy of adjuvant and neoadjuvant chemotherapy (3–6). At
present, trastuzumab is considered the standard primary therapy for
Her-2-positive breast cancer in the National Comprehensive Cancer
Network guidelines (https://www.nccn.org). Although the efficacy of
trastuzumab in the neoadjuvant settings for Her-2-positive breast
cancer is remarkable, the drug resistance limits its potential.
Around two-thirds of patients with Her-2-positive breast cancer
cannot benefit from Her-2-targeted therapy (7). Trastuzumab resistance may lead to
delays in treatment as well as unnecessary costs and
trastuzumab-associated side effects, therefore, it is imperative to
identify which patients are unlikely to benefit from trastuzumab
treatment. In the last decade, a number of studies have aimed to
investigate the mechanisms of resistance to Her-2-targeted
therapies and to identify molecular targets for
resistance-conferring factors.
The most commonly recognized anti-cancer mechanism
of trastuzumab is the targeting of the extracellular domain of the
Her-2 receptor and the inhibition of the downstream
phosphoinositide 3-kinase (PI3K)/Akt pathway; therefore, PIK3CA, a
mutation of the PI3K gene, was considered to be an important reason
for trastuzumab resistance (8),
while several other studies demonstrated that the PIK3CA gene is
common in Her-2-positive breast cancer but demonstrated no
significant association between the PIK3CA mutation and trastuzumab
resistance (9–11). Various studies have also
investigated the transcriptome of trastuzumab-resistant
Her-2-positive breast cancer, but these studies were based on
limited samples and demonstrated great heterogeneity (12–14).
In the present study, the microarray transcriptome
data of trastuzumab-resistant breast cancer and
trastuzumab-sensitive breast cancer in previously reported
neoadjuvant therapy studies were compared to identify potential
biomarkers for trastuzumab-sensitive Her-2-positive breast cancer
and investigate the potential molecular mechanisms of trastuzumab
resistance.
Materials and methods
Microarray data
The Gene Expression Omnibus (GEO) database
(http://www.ncbi.nlm.nih.gov/geo) was
searched for mRNA expression microarrays of Her-2-positive breast
cancer patients who received trastuzumab-based neoadjuvant
chemotherapy with the key terms ‘breast cancer AND (trastuzumab OR
herceptin)’, and 24 GEO series were identified. Nine were excluded
for not reporting neoadjuvant trastuzumab therapy, five were
excluded for not reporting data of human samples, three were
excluded for poor data quality, two were excluded for not reporting
mRNA microarray data and two were excluded for reporting no
complete response patients. As a result, three gene expression
profiles (GSE22358, GSE62327 and GSE66305) were finally obtained
from the GEO database. Microarray data of GSE22358 included 10
breast cancers with complete response and 13 breast cancers with no
complete response (12,14). GSE62327 consisted of 6 breast
cancers with complete response and 18 breast cancers with no
complete response (14). GSE66305
included 6 breast cancers with complete response and 17 breast
cancers with no complete response (13).
Data processing
The GEO database archives a large number of
high-throughput functional genomic studies that contain data that
are processed and normalized using various methods. GEO2R
(http://www.ncbi.nlm.nih.gov/geo/geo2r/) was applied to
screen differentially expressed mRNAs and genes between breast
cancer with partial response to trastuzumab-based neoadjuvant
chemotherapy and breast cancer with pathological complete response.
P<0.05 was set as the cut-off criteria for differently expressed
genes (DEGs). DEGs yielded by at least two datasets were selected
for further analysis. DEGs with P<0.05 and fold-change
(|FC|)>1.5 in all three datasets were selected as biomarkers of
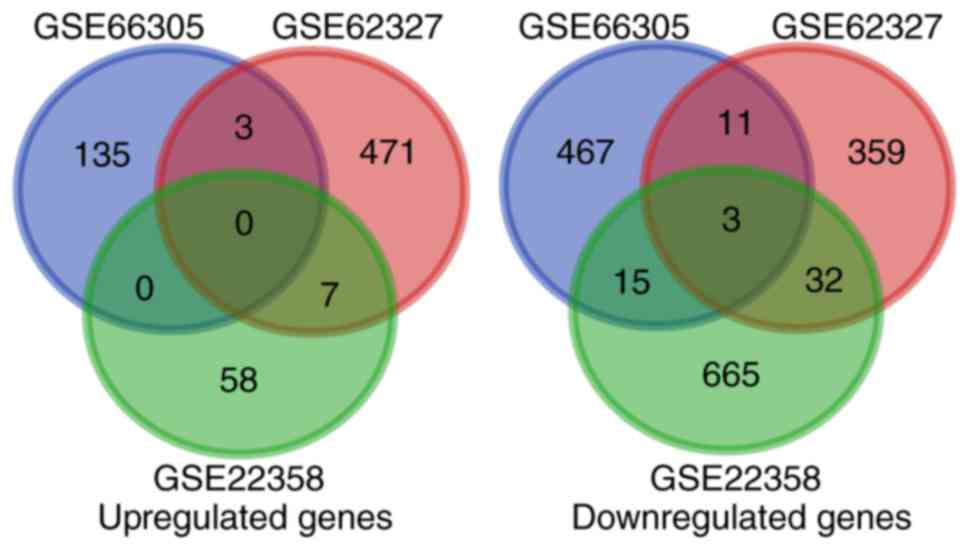
trastuzumab resistance. Venn Diagrams were created using the Venn
Diagrams software (http://bioinformatics.psb.ugent.be/webtools/Venn/) to
display the overlap of DEGs between the three datasets.
Functional and pathway enrichment
analysis
Upregulated and downregulated genes were subjected
to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway analysis using Database for Annotation,
Visualization and Integrated Discovery (DAVID, version 6.8;
http://david.abcc.ncifcrf.gov/)
software. GO and KEGG pathway enrichment analysis were performed
for identified DEGs using the DAVID database. P<0.05 was set as
the cut-off criterion.
Protein-protein interaction (PPI)
network construction and module selection
The identification of functional interactions
between proteins among the DEGs can provide context in the
molecular mechanism of cellular processing. In the present study, a
PPI network of DEGs was constructed using the Search Tool for the
Retrieval of Interacting Genes (STRING; http://string.embl.de/) database. The cut-off
criterion for PPI scores was set at 0.4. Subsequently, the PPI
network was analyzed using the Molecular Complete Detection (MCODE,
version 1.4.2) plug-in of the Cytoscape software (version 3.4.0)
(15) And enrichment analysis were
performed in the PPI network using the DAVID software.
Results
Identification of DEGs
Preliminary screening yielded a total of 2,376,
1,000 and 1,152 DEGs in the GSE22358, GSE62327 and GSE66305
datasets, respectively (1,305, 546 and 398 upregulated genes, and
1,071, 454 and 754 downregulated genes, respectively). Among them,
phosphate cytidylyltransferase 2 ethanolamine, estrogen receptor 1
(ESR1) and synaptogyrin 1 were upregulated in all three datasets,
and sex-determining region Y-box 11 (SOX11), ATPase phospholipid
transporting 8B2 (ATP8B2) and outer dense fiber of sperm tails
2-like (ODF2L) were downregulated in all three datasets.
Furthermore, when P<0.05 and |FC|>1.5 were used as the
criteria to further screen the DEGs with the most significant
difference of expression, a total of 981, 972 and 555 DEGs were
identified from the GSE22358, GSE62327 and GSE66305 datasets,
respectively (488, 548 and 105 upregulated genes, and 493, 424 and
450 downregulated genes, respectively). Among them, SOX11, ATP8B2
and ODF2L were downregulated with >1.5-fold alterations in all
three datasets (Fig. 1).
GO and pathway analysis
To assess the function of the DEGs, GO and KEGG
pathway enrichment analyses were performed using DAVID software.
The upregulated genes in partial response patients were primarily
enriched in ‘intracellular organelle lumen’, ‘transcription
activator activity’, ‘cytoskeletal protein binding’, ‘tissue
morphogenesis’ and ‘response to extracellular stimulus’.
Downregulated genes in partial response patients were primarily
enriched in ‘integral component of membrane’, ‘plasma membrane’,
‘extracellular exosome’, ‘signal transduction’ and ‘negative
regulation of apoptotic process’ (Table I). The upregulated KEGG pathways in
the partial response group were ‘pathways in cancer’, ‘basal cell
carcinoma’, ‘melanogenesis’ and ‘tight junction’. Downregulated
pathways in the partial response group were ‘PI3K-Akt signaling
pathway’, ‘FoxO signaling pathway’, ‘Rap1 signaling pathway’,
‘dopaminergic synapse’ and ‘cytokine-cytokine receptor
interactions’ (Table II).
 | Table I.Top 10 GO terms enriched in DEGs in
Her-2-positive breast cancer with partial response to
trastuzumab. |
Table I.
Top 10 GO terms enriched in DEGs in
Her-2-positive breast cancer with partial response to
trastuzumab.
| A, Upregulated
DEGs |
|---|
|
|---|
| GO term | Gene function
description | Count | P-value |
|---|
| GO:0070013 | Intracellular
organelle lumen | 11 | 0.026 |
| GO:0016563 | Transcription
activator activity | 5 | 0.026 |
| GO:0008092 | Cytoskeletal
protein binding | 5 | 0.050 |
| GO:0048729 | Tissue
morphogenesis | 4 | 0.016 |
| GO:0009991 | Response to
extracellular stimulus | 4 | 0.027 |
| GO:0033273 | Response to
vitamin | 3 | 0.016 |
| GO:0060562 | Epithelial tube
morphogenesis | 3 | 0.017 |
| GO:0046661 | Male sex
differentiation | 3 | 0.020 |
| GO:0043583 | Ear
development | 3 | 0.032 |
| GO:0043627 | Response to
estrogen stimulus | 3 | 0.038 |
|
| B, Downregulated
DEGs |
|
| GO term | Gene function
description | Count | P-value |
|
| GO:0016021 | Integral component
of membrane | 32 | 0.004 |
| GO:0005886 | Plasma
membrane | 26 | 0.011 |
| GO:0070062 | Extracellular
exosome | 21 | 0.004 |
| GO:0005887 | Integral component
of plasma membrane | 11 | 0.047 |
| GO:0007165 | Signal
transduction | 10 | 0.037 |
| GO:0043066 | Negative regulation
of apoptotic process | 7 | 0.009 |
| GO:0005925 | Focal adhesion | 6 | 0.018 |
| GO:0030424 | Axon | 5 | 0.011 |
| GO:0005913 | Cell-cell adherens
junction | 5 | 0.037 |
| GO:0010628 | Positive regulation
of gene expression | 5 | 0.020 |
 | Table II.KEGG pathways enriched in DEGs in
Her-2 positive breast cancer with partial response to
trastuzumab. |
Table II.
KEGG pathways enriched in DEGs in
Her-2 positive breast cancer with partial response to
trastuzumab.
| A, Upregulated
DEGs |
|---|
|
|---|
| KEGG term | Description | Count | P-value | Genes |
|---|
| hsa05200 | Pathways in
cancer | 5 | 0.008 | DVL3, WNT4, WNT3,
BCL2, RARA |
| hsa05217 | Basal cell
carcinoma | 3 | 0.008 | DVL3, WNT4,
WNT3 |
| hsa04916 | Melanogenesis | 3 | 0.025 | DVL3, WNT4,
WNT3 |
| hsa04530 | Tight junction | 3 | 0.044 | EPB41L1, MAGI1,
MYH14 |
|
| B, Downregulated
DEGs |
| KEGG
term |
Description | Count | P-value | Genes |
|
| hsa04151 | PI3K-Akt signaling
pathway | 8 | 0.004 | FLT1, OSMR, VEGFA,
CREB3L2, ITGB4, LPAR3, PRKAA1, BCL2L11 |
| hsa04068 | FoxO signaling
pathway | 5 | 0.008 | CDKN2B, MAPK14,
PRKAA1, STK4, BCL2L11 |
| hsa04015 | Rap1 signaling
pathway | 5 | 0.035 | FLT1, MAPK14,
VEGFA, LPAR3, CALML5 |
| hsa04728 | Dopaminergic
synapse | 4 | 0.040 | DDC, MAPK14,
CREB3L2, CALML5 |
| hsa04060 | Cytokine-cytokine
receptor interaction | 5 | 0.046 | TNFRSF21, FLT1,
OSMR, VEGFA, IL22RA2 |
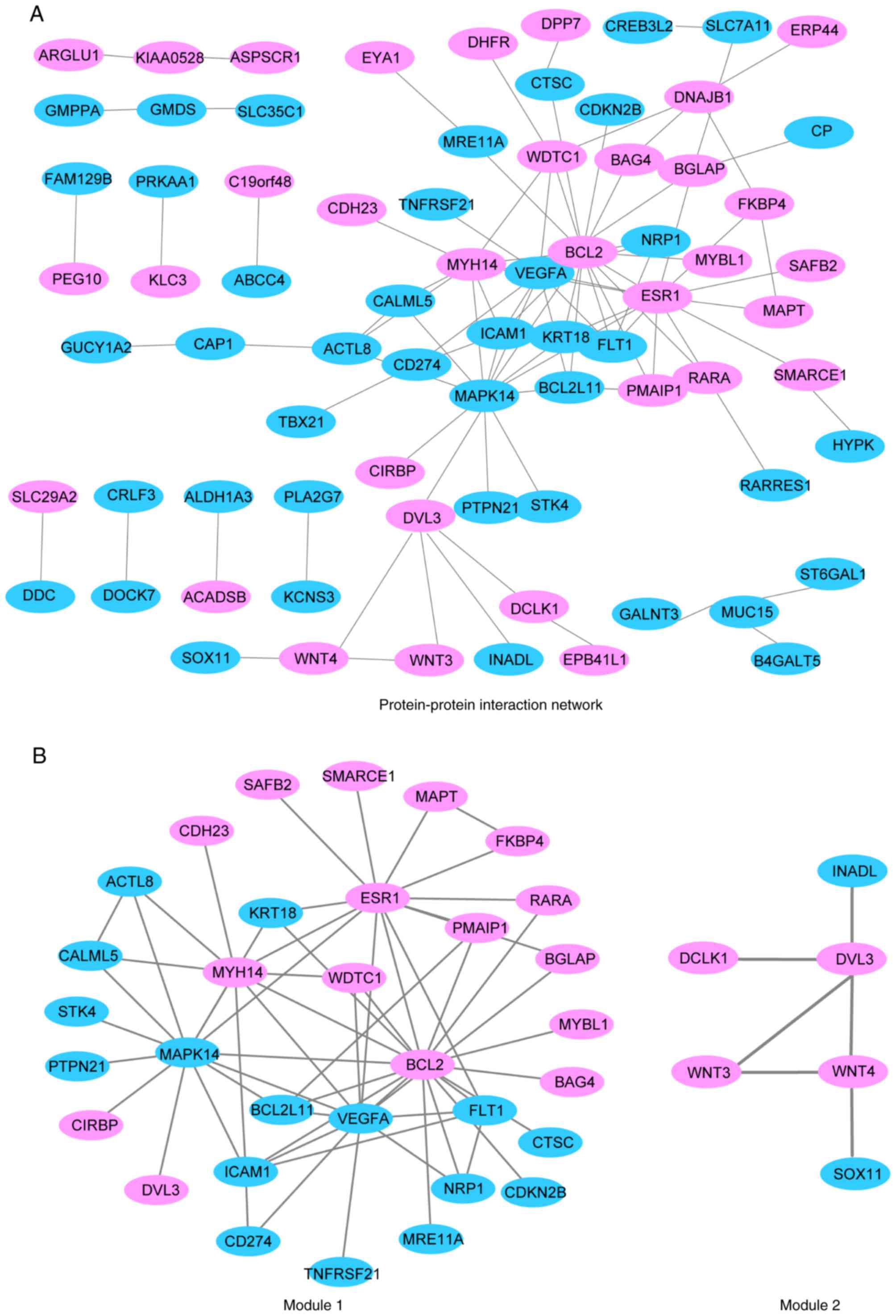
PPI network construction
Upregulated DEGs in the partial response group were
mapped with the STRING database. With a PPI score >0.4, a PPI
network with 96 nodes and 134 edges was constructed, as presented
in Fig. 2A. Two modules were
obtained from a PPI network of DEGs using MCODE, one with 32 nodes
and 60 edges, and the other with 6 nodes and 6 edges (Fig. 2B). GO term and KEGG pathway
enrichment analysis revealed that genes in module 1 were associated
with ‘protein binding’ and ‘cellular response to vascular
endothelial growth factor stimulus’ GO terms, while genes in module
2 were associated with ‘frizzled binding’, ‘canonical Wnt signaling
pathway’ and ‘neuron differentiation GO terms, and ‘basal cell
carcinoma’, ‘melanogenesis’, ‘Wnt signaling pathway’ and ‘signaling
pathways regulating pluripotency of stem cells’ KEGG pathways
(Table III).
 | Table III.Enriched GO terms and KEGG pathways
in the two protein-protein interaction network modules. |
Table III.
Enriched GO terms and KEGG pathways
in the two protein-protein interaction network modules.
| A, Module 1 |
|---|
|
|---|
| Category | GO/KEGG ID | Description | P-value |
|---|
|
GOTERM_MF_DIRECT | GO:0005515 | Protein
binding |
7.13×10−5 |
|
GOTERM_BP_DIRECT | GO:0035924 | Cellular response
to vascular endothelial growth factor stimulus |
8.90×10−6 |
|
| B, Module
2 |
|
Category | GO/KEGG
ID |
Description | P-value |
|
|
GOTERM_MF_DIRECT | GO:0005109 | Frizzled
binding |
2.65×10−5 |
|
GOTERM_BP_DIRECT | GO:0060070 | Canonical Wnt
signaling pathway |
1.44×10−4 |
|
GOTERM_BP_DIRECT | GO:0030182 | Neuron
differentiation |
1.89×10−4 |
| KEGG_PATHWAY | hsa05217 | Basal cell
carcinoma |
6.22×10−5 |
| KEGG_PATHWAY | hsa04916 | Melanogenesis |
2.07×10−4 |
| KEGG_PATHWAY | hsa04310 | Wnt signaling
pathway |
3.96×10−4 |
| KEGG_PATHWAY | hsa04550 | Signaling pathways
regulating pluripotency of stem cells |
4.08×10−4 |
Discussion
Utilization of trastuzumab has greatly improved the
prognosis of patients with Her-2-positive breast cancer.
Neoadjuvant regimens, including trastuzumab, have demonstrated
improved complete response rates compared with the previous
neoadjuvant regimens; however, given the high rate of trastuzumab
resistance and the high treatment expense, the effects of these
regimens are far from satisfying. The combined regimens of
trastuzumab and other targeted drugs are currently being tested
(16) and further knowledge of the
detailed mechanism of trastuzumab resistance is required. Several
studies have examined the gene expression profiles of patients
receiving neoadjuvant trastuzumab therapy using microarray gene
chips (12,13), but the majority of these studies
were performed with a small sample size and the results varied
between studies. In the present study, microarray data from three
GEO datasets were analyzed, which collectively included the gene
expression data of 22 complete response tumors and 48 partial
response tumors, in order to cast light on the mechanism of
trastuzumab resistance in Her-2-positive breast cancer.
Several molecular mechanisms have been proposed to
explain the action of the trastuzumab on Her-2, which are divided
into the following categories: Inhibiting the downstream PI3K/Akt
signaling pathway of Her-2 (17);
promoting the phosphorylation of phosphatase and tensin homolog
(18); reducing cancer cell
proliferation by reducing the expression of the cyclin D1 protein,
leading to G1 arrest; inhibiting angiogenesis by reducing the
production of the vascular endothelial growth factor (VEGF);
inhibiting Her-2 ectodomain cleavage (19); and binding the Fc-γ receptor III on
immune effector cells and inducing antibody dependent cell mediated
cytotoxicity (20). In the present
study, it was demonstrated that several targeted pathways of
trastuzumab exhibited a relatively lower activity in the partial
response group, which may have a role in trastuzumab
resistance.
Sustained excessive proliferative signaling is one
of the most fundamental hallmarks of cancer (21). It is reported that overexpression
of Her-2 may lead to the activation of the PI3K/Akt pathway, and
the excessively activated PI3K-Akt signaling pathway serves a
crucial role in the proliferation of breast cancer (22). Okutur et al (23) reported that 96% of Her-2-positive
breast cancer exhibited overexpression of the PI3K protein and
70.4% exhibited overexpression of the Akt protein. Inhibition of
the PI3K/Akt pathway through targeting Her-2 is considered to be
one of the key mechanisms underlying the anti-tumor effects of
trastuzumab (24); however, in the
present study, it was demonstrated that among the patients
receiving trastuzumab treatment, patients with partial responses
tended to have lower PI3K-Akt pathway activity prior to treatment,
compared with the patients with a complete response. In addition,
the present study revealed that genes associated with the
activation of the Wnt signaling pathway, including Wnt family
member 3 (WNT3), WNT4 and disheveled segment polarity protein 3,
were excessively expressed in the partial response group. The
expression of WNT3 was reported to activate the Wnt/β-catenin
pathway and promote a epithelial-mesenchymal transition-like
phenotype in trastuzumab-resistant Her-2-overexpressing breast
cancer cells, resulting in an increase in cell invasion and
proliferation (25). The results
of the current study indicated that a proportion of Her-2-positive
breast cancers may acquire trastuzumab resistance by downregulation
of the PI3K/Akt pathway and may maintain proliferative signaling by
the upregulation of the Wnt pathway. Therefore, combining
Wnt-targeted therapy with trastuzumab may help to enhance the
complete response rate of Her-2-targeted therapy.
Cancer cells have been demonstrated to acquire the
oxygen and nutrients required for their rapid proliferation by
inducing angiogenesis, which is also a key target of trastuzumab
(26). In the present study,
trastuzumab-resistant breast cancers exhibited downregulated
cytokine-cytokine receptor interaction pathways, including the
downregulation of VEGF and its receptor, Fms-related tyrosine
kinase 1. In addition, the PPI analysis demonstrated the
downregulation of a module primarily associated with the cellular
response to VEGF stimulus in the partial response group. These
results indicate that trastuzumab-resistant breast cancers may be
independent of tumor angiogenesis and therefore be resistant to the
antiangiogenic effect of trastuzumab. These results may also
suggest that trastuzumab-resistant breast cancers also tend to be
poorly vascularized even prior to neoadjuvant chemotherapy, which
may lead to low regional trastuzumab concentration in the tumor
foci.
In the present study, SOX11, ATP8B2 and ODF2L were
demonstrated to be downregulated in breast cancer with a partial
response to trastuzumab with >1.5 FC compared with the complete
response breast cancer cases. Enrichment and network analysis was
used in combination with DEGs to reduce the risk of type I errors
in the search for biomarkers. Although ATP8B2 and ODF2L were
demonstrated to be differentially expressed between the partial
response and complete response groups in all three datasets, the
pathways and GO terms associated with the two genes were not
enriched and no interactions were demonstrated between these two
genes and the other genes in the PPI network. SOX11 belongs to the
subgroup C of the SOX gene family, which encode transcription
factors with important roles in embryonic development and cell
differentiation, and may contribute to the development and
progression of central nervous system malignancies, solid tumors
and aggressive mantle cell lymphoma (27). SOX11 has been demonstrated to
directly activate the PI3K/Akt signaling pathway and suppress the
Wnt signaling pathway (27,28),
and promote tumor angiogenesis by regulation of platelet-derived
growth factor (27). As mentioned
above, the PI3K/Akt pathway and tumor angiogenesis were
demonstrated to be suppressed, and the Wnt pathway was activated,
in trastuzumab-resistant breast cancer. These results are
consistent with the function of SOX11, which indicates that
downregulation of SOX11 may serve a critical role in the
acquirement of trastuzumab resistance. Therefore, SOX11 may be used
as a potential biomarker for trastuzumab sensitivity and, more
importantly, it may also serve as a potential therapeutic target of
trastuzumab resistance in breast cancer.
The results of the current study demonstrated that
ESR1 was upregulated in the partial response group in all three
datasets, and DEGs upregulated in the partial response group were
enriched in the ‘response to estrogen stimulus’ GO term. In
addition, PPI analysis also highlighted an ER-associated PPI
network; ESR1, B-cell lymphoma 2 (BCL2), DnaJ homolog subfamily B
member 1, WD and tetratricopeptide repeats 1, and myosin heavy
chain 14, were the top 5 nodes in this network based on the
centrality degree. ESR1 protein, also termed ER, has been
extensively investigated in breast cancer and is one of the
defining features in classifying tumor subtype and assigning
therapeutic strategies in breast cancer (29). ER was considered to be an
alternative pathway to Her-2 blockade due to its ability to
activate certain Her-2 signaling members, including transforming
growth factor-a (30). ER-negative
breast cancers have been reported to exhibit an enhanced complete
response rate to trastuzumab-based chemotherapy in several clinical
studies, regardless of the treatment regimen (31,32).
Patients with ER positive and Her-2 positive breast cancer
demonstrated worse trastuzumab response compared with ER negative
and Her-2 positive patients, while hormone therapy targeting ER
following trastuzumab and chemotherapy resulted in increased
progression-free survival (33).
Therefore, Her-2 blockade combined with endocrine therapy may also
be a reasonable neoadjuvant regimen for Her-2-positive ER-positive
breast cancer.
Additionally, in the present study, the expression
of BCL2, an estrogen-associated protein, was demonstrated to be
increased in the partial response group (34). BCL2 has been demonstrated to
prevent apoptosis and inhibit proliferation (35). Paradoxical results have been
reported regarding the effect of BCL2 expression on the treatment
of breast cancer. Several studies reported that increased
expression of BCL2 may predict a good prognosis in breast cancer
(36,37), while other studies demonstrated
that the prognostic effect of BCL2 may differ between different
molecular subtypes of breast cancer (36,38,39).
Giuliano et al (40)
described the parallel upregulation of BCL2 as a mechanism of
survival entirely dependent on ER activity and potentially leading
to anti-Her-2 resistance (40,41).
In addition, an in vitro experiment performed on BT474
cells, a Her-2-overexpressing breast cancer cell line, demonstrated
that increased BCL2 expression contributed to trastuzumab
resistance (39). These results
indicate that Her-2-positive breast cancer may acquire trastuzumab
resistance via the ESR1/BCL2 pathway. Therefore, BCL2 may be used
as a novel molecular target for improving the response of breast
cancer to trastuzumab.
In conclusion, the present study demonstrated that
Her-2-positive breast cancer with trastuzumab resistance exhibited
low PI3K/Akt pathway, low tumor angiogenesis and high ER pathway
activity. Trastuzumab-resistant breast cancers may acquire
proliferation signaling by upregulating the Wnt signaling pathway.
Therefore, combination therapy consisting of trastuzumab and
anti-Wnt or hormone therapy may be a promising treatment modality
and should be investigated in further studies. Furthermore, low
SOX11 and high BCL2 expression may be employed as biomarkers for
neoadjuvant trastuzumab therapy of Her-2-positive breast
cancer.
Acknowledgements
The authors would like to thank the members of the
teams of Gluck S, De Cecco L and Guarneri V for the supplementary
microarray data used in the present study (GSE22358, GSE62327 and
GSE66305 datasets, respectively), and Dr Jianming Zeng, University
of Macau and Dr Guangchuang Yu (University of Hong Kong) for their
support in using bioinformatics tools.
Funding
The present study was supported by a grant from the
National Natural Science Foundation of China (grant no. 30901481)
and the Major Research Program of Shandong Province (grant no.
2017GSF221016).
Availability of data and materials
Microarray data used in this article can be
downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo) with the accession
number GSE22358, GSE62327 and GSE66305.
Authors' contributions
Data analysis was performed by BZ, HN, LL and YZ.
Manuscript was drafted by BZ and YZ. YS, LS and DH performed the
GEO and reference search. HN was in charge of language editing.
Ethics approval and consent to
participate
All data used in this study comes from the GEO
database and no clinical trial or animal experiment was included in
this study. This study was granted an exemption from requiring
ethics approval by the ethics committee of Qilu Hospital (Qingdao,
China).
Consent for publication
All data used in this study came from the GEO
database, and the data submitters have declared that their studies
comply with the NIH genomic data sharing policy and have the
appropriate consent/permission to submit the data to a public
database and these information does not compromise participant
privacy: (https://www.ncbi.nlm.nih.gov/geo/info/faq.html#patient).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal AL: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Guarneri V, Broglio K, Kau SW,
Cristofanilli M, Buzdar AU, Valero V, Buchholz T, Meric F,
Middleton L, Hortobagyi GN and Gonzalez-Angulo AM: Prognostic value
of pathologic complete response after primary chemotherapy in
relation to hormone receptor status and other factors. J Clin
Oncol. 24:1037–1044. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Romond EH, Perez EA, Bryant J, Suman VJ,
Geyer CE Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman
PA, et al: Trastuzumab plus adjuvant chemotherapy for operable
HER2-positive breast cancer. N Engl J Med. 353:1673–1684. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Slamon D, Eiermann W, Robert N, Pienkowski
T, Martin M, Press M, Mackey J, Glaspy J, Chan A, Pawlicki M, et
al: Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J
Med. 365:1273–1283. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gianni L, Eiermann W, Semiglazov V,
Manikhas A, Lluch A, Tjulandin S, Zambetti M, Vazquez F, Byakhow M,
Lichinitser M, et al: Neoadjuvant chemotherapy with trastuzumab
followed by adjuvant trastuzumab versus neoadjuvant chemotherapy
alone, in patients with HER2-positive locally advanced breast
cancer the NOAH trial): A randomised controlled superiority trial
with a parallel HER2-negative cohort. Lancet. 375:377–384. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Untch M, Rezai M, Loibl S, Fasching PA,
Huober J, Tesch H, Bauerfeind I, Hilfrich J, Eidtmann H, Gerber B,
et al: Neoadjuvant treatment with trastuzumab in HER2-positive
breast cancer: Results from the GeparQuattro study. J Clin Oncol.
28:2024–2031. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nahta R and Esteva FJ: HER2 therapy:
Molecular mechanisms of trastuzumab resistance. Breast Cancer Res.
8:2152006. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Berns K, Horlings HM, Hennessy BT,
Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM,
Stemke-Hale K, Hauptmann M, et al: A functional genetic approach
identifies the PI3K pathway as a major determinant of trastuzumab
resistance in breast cancer. Cancer Cell. 12:395–402. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Barbareschi M, Cuorvo LV, Girlando S,
Bragantini E, Eccher C, Leonardi E, Ferro A, Caldara A, Triolo R,
Cantaloni C, et al: PI3KCA mutations and/or PTEN loss in
Her2-positive breast carcinomas treated with trastuzumab are not
related to resistance to anti-Her2 therapy. Virchows Arch.
461:129–139. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
de Oliveira Taveira M, Nabavi S, Wang Y,
Tonellato P, Esteva FJ, Cantley LC and Wulf GM: Genomic
characteristics of trastuzumab-resistant Her2-positive metastatic
breast cancer. J Cancer Res Clin Oncol. 143:1255–1262. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bianchini G, Kiermaier A, Bianchi GV, Im
YH, Pienkowski T, Liu MC, Tseng LM, Dowsett M, Zabaglo L, Kirk S,
et al: Biomarker analysis of the NeoSphere study: Pertuzumab,
trastuzumab, and docetaxel versus trastuzumab plus docetaxel,
pertuzumab plus trastuzumab, or pertuzumab plus docetaxel for the
neoadjuvant treatment of HER2-positive breast cancer. Breast Cancer
Res. 19:162017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gluck S, Ross JS, Royce M, McKenna EF Jr,
Perou CM, Avisar E and Wu L: TP53 genomics predict higher clinical
and pathologic tumor response in operable early-stage breast cancer
treated with docetaxel-capecitabine ± trastuzumab. Breast Cancer
Res Treat. 132:781–791. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guarneri V, Dieci MV, Frassoldati A,
Maiorana A, Ficarra G, Bettelli S, Tagliafico E, Bicciato S,
Generali DG, Cagossi K, et al: Prospective biomarker analysis of
the randomized CHER-LOB study evaluating the dual anti-HER2
treatment with trastuzumab and lapatinib plus chemotherapy as
neoadjuvant therapy for HER2-positive breast cancer. Oncologist.
20:1001–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Triulzi T, De Cecco L, Sandri M, Prat A,
Giussani M, Paolini B, Carcangiu ML, Canevari S, Bottini A, Balsari
A, et al: Whole-transcriptome analysis links trastuzumab
sensitivity of breast tumors to both HER2 dependence and immune
cell infiltration. Oncotarget. 6:28173–28182. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Toomey S, Eustace AJ, Fay J, Sheehan KM,
Carr A, Milewska M, Madden SF, Teiserskiene A, Kay EW, O'Donovan N,
et al: Impact of somatic PI3K pathway and ERBB family mutations on
pathological complete response (pCR) in HER2-positive breast cancer
patients who received neoadjuvant HER2-targeted therapies. Breast
Cancer Res. 19:872017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yakes FM, Chinratanalab W, Ritter CA, King
W, Seelig S and Arteaga CL: Herceptin-induced inhibition of
phosphatidylinositol-3 kinase and Akt is required for
antibody-mediated effects on p27, cyclin D1, and antitumor action.
Cancer Res. 62:4132–4141. 2002.PubMed/NCBI
|
|
18
|
Nagata Y, Lan KH, Zhou X, Tan M, Esteva
FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, et al: PTEN
activation contributes to tumor inhibition by trastuzumab, and loss
of PTEN predicts trastuzumab resistance in patients. Cancer Cell.
6:117–127. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Molina MA, Codony-Servat J, Albanell J,
Rojo F, Arribas J and Baselga J: Trastuzumab (herceptin), a
humanized anti-Her2 receptor monoclonal antibody, inhibits basal
and activated Her2 ectodomain cleavage in breast cancer cells.
Cancer Res. 61:4744–4749. 2001.PubMed/NCBI
|
|
20
|
Clynes RA, Towers TL, Presta LG and
Ravetch JV: Inhibitory Fc receptors modulate in vivo cytotoxicity
against tumor targets. Nat Med. 6:443–446. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dey N, De P and Leyland-Jones B:
PI3K-AKT-mTOR inhibitors in breast cancers: From tumor cell
signaling to clinical trials. Pharmacol Ther. 175:91–106. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Okutur K, Bassulu N, Dalar L, Aydin K,
Bozkurt M, Pilanci KN, Dogusoy GB, Tecimer C, Mandel NM and Demir
G: Predictive and prognostic significance of p27, Akt, PTEN and
PI3K expression in HER2-positive metastatic breast cancer. Asian
Pac J Cancer Prev. 16:2645–2651. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lan KH, Lu CH and Yu D: Mechanisms of
trastuzumab resistance and their clinical implications. Ann N Y
Acad Sci. 1059:70–75. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Timmermans-Sprang EP, Gracanin A and Mol
JA: High basal Wnt signaling is further induced by PI3K/mTor
inhibition but sensitive to cSRC inhibition in mammary carcinoma
cell lines with HER2/3 overexpression. BMC Cancer. 15:5452015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Klos KS, Zhou X, Lee S, Zhang L, Yang W,
Nagata Y and Yu D: Combined trastuzumab and paclitaxel treatment
better inhibits ErbB-2-mediated angiogenesis in breast carcinoma
through a more effective inhibition of Akt than either treatment
alone. Cancer. 98:1377–1385. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Palomero J, Vegliante MC, Rodriguez ML,
Eguileor A, Castellano G, Planas-Rigol E, Jares P, Ribera-Cortada
I, Cid MC, Campo E and Amador V: SOX11 promotes tumor angiogenesis
through transcriptional regulation of PDGFA in mantle cell
lymphoma. Blood. 124:2235–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kuo PY, Leshchenko VV, Fazzari MJ, Perumal
D, Gellen T, He T, Iqbal J, Baumgartner-Wennerholm S, Nygren L,
Zhang F, et al: High-resolution chromatin immunoprecipitation ChIP)
sequencing reveals novel binding targets and prognostic role for
SOX11 in mantle cell lymphoma. Oncogene. 34:1231–1240. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Huang B, Warner M and Gustafsson JÅ:
Estrogen receptors in breast carcinogenesis and endocrine therapy.
Mol Cell Endocrinol. 418:240–244. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Menyhart O, Santarpia L and Győrffy B: A
comprehensive outline of trastuzumab resistance biomarkers in HER2
overexpressing breast cancer. Curr Cancer Drug Targets. 15:665–683.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gianni L, Pienkowski T, Im YH, Roman L,
Tseng LM, Liu MC, Lluch A, Staroslawska E, de la Haba-Rodriguez J,
Im SA, et al: Efficacy and safety of neoadjuvant pertuzumab and
trastuzumab in women with locally advanced, inflammatory, or early
HER2-positive breast cancer (NeoSphere): A randomised multicentre,
open-label, phase 2 trial. Lancet Oncol. 13:25–32. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rimawi MF, Mayer IA, Forero A, Nanda R,
Goetz MP, Rodriguez AA, Pavlick AC, Wang T, Hilsenbeck SG,
Gutierrez C, et al: Multicenter phase II study of neoadjuvant
lapatinib and trastuzumab with hormonal therapy and without
chemotherapy in patients with human epidermal growth factor
receptor 2-overexpressing breast cancer: TBCRC 006. J Clin Oncol.
31:1726–1731. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Montemurro F, Rossi V, Rocca Cossu M,
Martinello R, Verri E, Redana S, Adamoli L, Valabrega G, Sapino A,
Aglietta M, et al: Hormone-receptor expression and activity of
trastuzumab with chemotherapy in HER2-positive advanced breast
cancer patients. Cancer. 118:17–26. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Park SH, Kim H and Song BJ: Down
regulation of bcl2 expression in invasive ductal carcinomas is both
estrogen- and progesterone-receptor dependent and associated with
poor prognostic factors. Pathol Oncol Res. 8:26–30. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Aizawa K, Ueki K, Suzuki S, Yabusaki H,
Kanda T, Nishimaki T, Suzuki T and Hatakeyama K: Apoptosis and
Bbcl-2 expression in gastric carcinomas: Correlation
withclinicopathological variables, p53 expression, cell
proliferation and prognosis. Int J Oncol. 14:85–91. 1999.PubMed/NCBI
|
|
36
|
Eom YH, Kim HS, Lee A, Song BJ and Chae
BJ: BCL2 as a Subtype-specific prognostic marker for breast cancer.
J Breast Cancer. 19:252–260. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dawson SJ, Makretsov N, Blows FM, Driver
KE, Provenzano E, Le Quesne J, Baglietto L, Severi G, Giles GG,
McLean CA, et al: BCL2 in breast cancer: A favourable prognostic
marker across molecular subtypes and independent of adjuvant
therapy received. Br J Cancer. 103:668–675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hwang KT, Han W, Kim J, Moon HG, Oh S,
Song YS, Kim YA, Chang MS and Noh DY: Prognostic Influence of BCL2
on molecular subtypes of breast cancer. J Breast Cancer. 20:54–64.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Crawford A and Nahta R: Targeting Bcl-2 in
Herceptin-resistant breast cancer cell lines. Curr Pharmacogenomics
Person Med. 9:184–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Giuliano M, Hu H, Wang YC, Fu X, Nardone
A, Herrera S, Mao S, Contreras A, Gutierrez C, Wang T, et al:
Upregulation of ER signaling as an adaptive mechanism of cell
survival in HER2-positive breast tumors treated with Anti-HER2
therapy. Clin Cancer Res. 21:3995–4003. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang Q, Moran MS and Haffty BG: Bcl-2
expression predicts local relapse for early-stage breast cancer
receiving conserving surgery and radiotherapy. Breast Cancer Res
Treat. 115:343–348. 2009. View Article : Google Scholar : PubMed/NCBI
|