Introduction
Endothelial dysfunction, which is closely associated
with vascular endothelial cell injury, is involved in an array of
pathophysiological processes, including coronary artery diseases,
diabetes and hypertension (1–4).
Oxidized low-density lipoprotein (oxLDL)-induced injury to vascular
endothelial cells has been revealed to be an important initial
event in atherosclerosis (5–7). It
has been reported that the activation of endothelial cells by oxLDL
may occur via the induction of the expression of various cell
surface adhesion molecules, which mediate the rolling adhesion of
leukocytes, including monocytes and T cells (8,9).
Following adhesion to the endothelium, leukocytes have been
demonstrated to migrate into the intima where monocytes then
differentiate into macrophages and induce atherosclerotic lesions
(6–11). OxLDL has been reported to induce
the onset of atherogenesis involving endothelial cells and
macrophages (12). However, the
mechanisms underlying the activation of macrophage differentiation
by oxLDL-treated endothelial cells remain unclear.
Macrophages are heterogeneous and polarize into
specific subsets, such as classically activated proinflammatory
macrophage M1 (M1)-like and alternatively activated macrophage M2
(M2)-like macrophages (13).
Different markers are employed for the identification of M1 and M2
macrophages. Markers for M1 macrophages include tumor necrosis
factor-α, inducible nitric oxide synthase, interleukin (IL)-6 and
integrin-α-X (also termed CD11c), while markers for M2 macrophages
include macrophage mannose receptor 1 (also termed CD206),
arginase-1 and resistin-like-α (14,15).
In human atherosclerotic plaques, markers for M1 and M2 macrophages
are present in the early and advanced stages of plaque development
(16,17). Previous studies in
hypercholesterolemic mice have demonstrated that M2 macrophages
reduce atherosclerosis, whereas suppression of M2 polarization
enhances plaque progression (18–20).
Therefore, it may be concluded that macrophage polarization is
important for plaque progression in atherosclerosis.
Exosomes are nano-sized membrane vesicles released
by cells, with a diameter of 30–100 nm (21–23).
Exosomes are important inter-cellular transporters, carrying bulk
biomaterials to regulate the cellular functions of endothelial
cells, vascular smooth muscle cells, fibroblasts and leukocytes
(24–27). However, the function of exosomes
secreted by oxLDL-treated endothelial cells during atherosclerosis
remains to be determined.
Long non-coding RNAs (lncRNAs) are non-protein
coding transcripts that are composed of >200 nucleotides, and
studies have demonstrated that certain lncRNAs have important
functions in the regulation of numerous biological processes,
including cell proliferation, differentiation and death (28–30).
Metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is
a widely expressed single-exon lncRNA. MALAT1 was initially
identified in non-small cell lung cancer and is reported to be
involved in the regulation of various biological processes by
functioning as a competing endogenous RNA (31–33).
Therefore, the aim of the present study was to
investigate the function of exosomal lncRNA-MALAT1 in the process
of oxLDL-treated endothelial cell-activated macrophage
differentiation.
Materials and methods
Cell culture
THP-1 (American Type Culture Collection, Manassas,
VA, USA) human monocyte cells were maintained in RPMI-1640 (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and human umbilical vein
endothelial cells immortalized with telomerase reverse
transcriptase (HUVEC/TERT2; American Type Culture Collection) were
maintained in Dulbecco's Modified Eagle's Medium/F12 (Thermo Fisher
Scientific, Inc.), and each were supplemented with 10%
heat-inactivated fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.), penicillin (100 U/ml) and streptomycin (100
µg/ml) in a humidified atmosphere of 5% CO2 at 37°C. All
cells were confirmed to be free of mycoplasma contamination with
the MycoFluor™ Mycoplasma Detection kit (Thermo Fisher Scientific,
Inc.).
Exosome isolation and co-culture with
THP-1 cells
In order to isolate exosomes, HUVECs
(2×107) were treated with 50 µg/ml oxLDL (Shanghai Luwen
Biotechnology Co., Ltd., Shanghai, China) for 48 h at 37°C and the
supernatant was collected. The supernatant was centrifuged twice
(1,000 × g for 10 min and 3,000 × g for 30 min at 4 C) to remove
the cells and fragments, added to the Total Exosome Isolation kit
(Thermo Fisher Scientific, Inc.) overnight and further centrifuged
at 10,000 × g for 1 h at 4°C. Exosomes were resuspended in PBS and
stored at −80°C. The concentration of exosomes was determined using
a bicinchoninic acid assay. Exosomes (50 ng/ml) were subsequently
added to THP-1 cells (1×105) in culture medium for 24 h
at 37°C. Exosomes were stained with PHK67 (cat. no. MINI67;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) to observe
endocytosis, according to the manufacturer's protocol.
Transmission electron microscopic
observation of exosomes
The exosome suspension was added to an equal volume
of 4% paraformaldehyde at 4°C for 30 min and applied to a
Formvar/Carbon film-coated transmission electron microscope grid
(Alliance Biosystems, Inc., Osaka, Japan). Subsequently, the sample
was fixed by incubation with 1% glutaraldehyde for 5 min at 25°C,
washed with PBS and incubated with 1% uranyl acetate for 5 min at
25°C. The sample was subsequently observed under a transmission
electron microscope (Hitachi H7650; Hitachi, Ltd., Tokyo,
Japan).
Knockdown of MALAT1 in THP-1
cells
The knockdown sequence for MALAT1 was as follows:
5′-ACGGAAGTAATTCAAGATCAA-3′. Lentiviral vectors for human MALAT1
knockdown were constructed by Shanghai Hanyin Industrial Co., Ltd.
(Shanghai, China). The recombinant lentivirus and the empty vector
negative control (NC) lentivirus (Shanghai Hanyin Industrial Co.,
Ltd.) were prepared and titrated to 109 transfection
U/ml. To isolate stably transfected cell lines, THP-1 cells were
seeded in 6-well plates at a density of 2×105 cells/well
and infected with the same titer of virus (Shanghai Hanyin
Industrial Co., Ltd., Shanghai, China) with 8 µg/ml polybrene
(Shanghai Hanyin Industrial Co., Ltd.) on the following day at
37°C. At ~72 h post-viral infection, the RPMI-1640 culture medium
was replaced with selection medium containing 4 µg/ml puromycin and
the cells were cultured for ≥14 days at 37°C. The
puromycin-resistant cells were amplified in medium containing 2
µg/ml puromycin for 7–9 days 37°C and then transferred to medium
without puromycin. The knockdown efficiency was confirmed via
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) 24 h after co-culture, as described below.
Western blot analysis
Protein concentration was determined using the
bicinchoninic acid protein assay method. Membranes were blocked in
5% non-fat milk at 25°C for 30 min. To analyze the expression of
the markers CD9, CD63 and CD206 in exosomes (control or
oxLDL-treated) or THP-1 cells that were co-cultured with exosomes
(control or oxLDL-treated), western blot assays were performed
using the following primary antibodies: Mouse anti-human CD63 (cat.
no. ab59479; 1:1,000; Abcam, Cambridge, UK), rabbit anti-human CD9
(cat. no. ab92726; 1:1,000; Abcam), rabbit anti-human CD206 (cat.
no. 18704-1-AP; 1:1,000; ProteinTech Group, Inc., Chicago, IL, USA)
and mouse anti-actin (cat. no. MAB1501; 1:10,000; EMD Millipore,
Billerica, MA, USA). Briefly, exosomes or cells were lysed with
radioimmunoprecipitation assay buffer [50 mM Tris-HCl (pH 7.5), 150
mM NaCl, 1% Triton X-100 and 0.5% Na-deoxycholate] containing
protease inhibitors (Complete™, Mini protease inhibitor cocktail;
Sigma-Aldrich; Merck KGaA). A total of 20–30 µg lysate were
separated on 8–12% SDS-PAGE gels and transferred to polyvinylidene
fluoride membranes. The membranes were subsequently incubated with
the aforementioned primary antibodies overnight at 4°C. The primary
antibody incubation was followed by incubation with the following
secondary antibodies at room temperature for 1 h: Horseradish
peroxidase (HRP)-conjugated anti-rabbit (cat. no. 7074; 1:10,000;
CST Biological Reagents Co., Ltd., Shanghai, China) and
HRP-conjugated anti-mouse (cat. no. 7076; 1:10,000; CST Biological
Reagents Co., Ltd.). The bound antibodies were detected using an
enhanced chemiluminescence kit (cat. no. PI32209; Pierce; Thermo
Fisher Scientific, Inc.). Quantity One analysis software (version
4.6.9; Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to
quantify the relative band intensities from western blot
images.
RT-qPCR
Total RNA was extracted from exosomes (control or
oxLDL-treated) or THP-1 cells that were co-cultured with exosomes
(control or oxLDL-treated) using TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.). An equal quantity of total RNA was used for
first-strand cDNA synthesis using the reverse transcription system
(cat. no. A3500; Promega Corporation, Madison, WI, USA) according
to the manufacturer's protocol. The synthesized first-strand cDNA
(2 µl) was used for each qPCR assay. A SYBR-Green PCR Master Mix
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for
qPCR experiments. GAPDH was used as an internal reference control.
The primers sequences were as follows: MALAT1 forward,
5′-TTGTAGACTGGAGAAGATAGG-3′ and reverse, 5′-ACTGAAGAGCATTGGAGAT-3′;
CD206 forward, 5′-GGGTTGCTATCACTCTCTATGC-3′ and reverse,
5′-TTTCTTGTCTGTTGCCGTAGTT-3′; Arg-1 forward,
5′-TGGACAGACTAGGAATTGGCA-3′ and reverse,
5′-CCAGTCCGTCAACATCAAAACT-3′; GAPDH forward,
5′-CATGGCCTTCCGTGTTCCTA-3′ and reverse 5′-GCGGCACGTCAGATCCA-3′.
qPCR cycling conditions consisted of initial denaturation for 3 min
at 95°C, followed by 45 cycles of 95°C for 10 sec and 58°C for 45
sec in a LightCycler® 480 instrument (Roche Diagnostics,
Basel, Switzerland). Melt curve analysis was performed at the end
of each run between 58–95°C. Data were analyzed using Microsoft
Excel 2013 (Microsoft Corporation, Redmond, WA, USA). The
2−ΔΔCq relative quantification method was used to
analyze quantitative RT-qPCR data using the housekeeping gene GAPDH
for normalization (34).
ELISA
ELISAs were performed on THP-1 cells that had been
co-cultured with exosomes from control or oxLDL-treated HUVECs to
determine the effect on IL-10 and IL-12 levels. IL-10 and IL-12
ELISA kits were purchased from Abcam (cat. nos. ab185986 and
ab46035, respectively), and the analysis was performed according to
the manufacturer's protocol.
Statistical analysis
Statistical differences between two groups were
determined using the Student's t-test. Experiments were repeated
three times. Data are presented as the mean ± standard error of the
mean. SPSS software was used to perform statistical analysis
(version 17.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Exosomal MALAT1 is increased in
oxLDL-treated HUVECs
To investigate the functions of oxLDL-treated
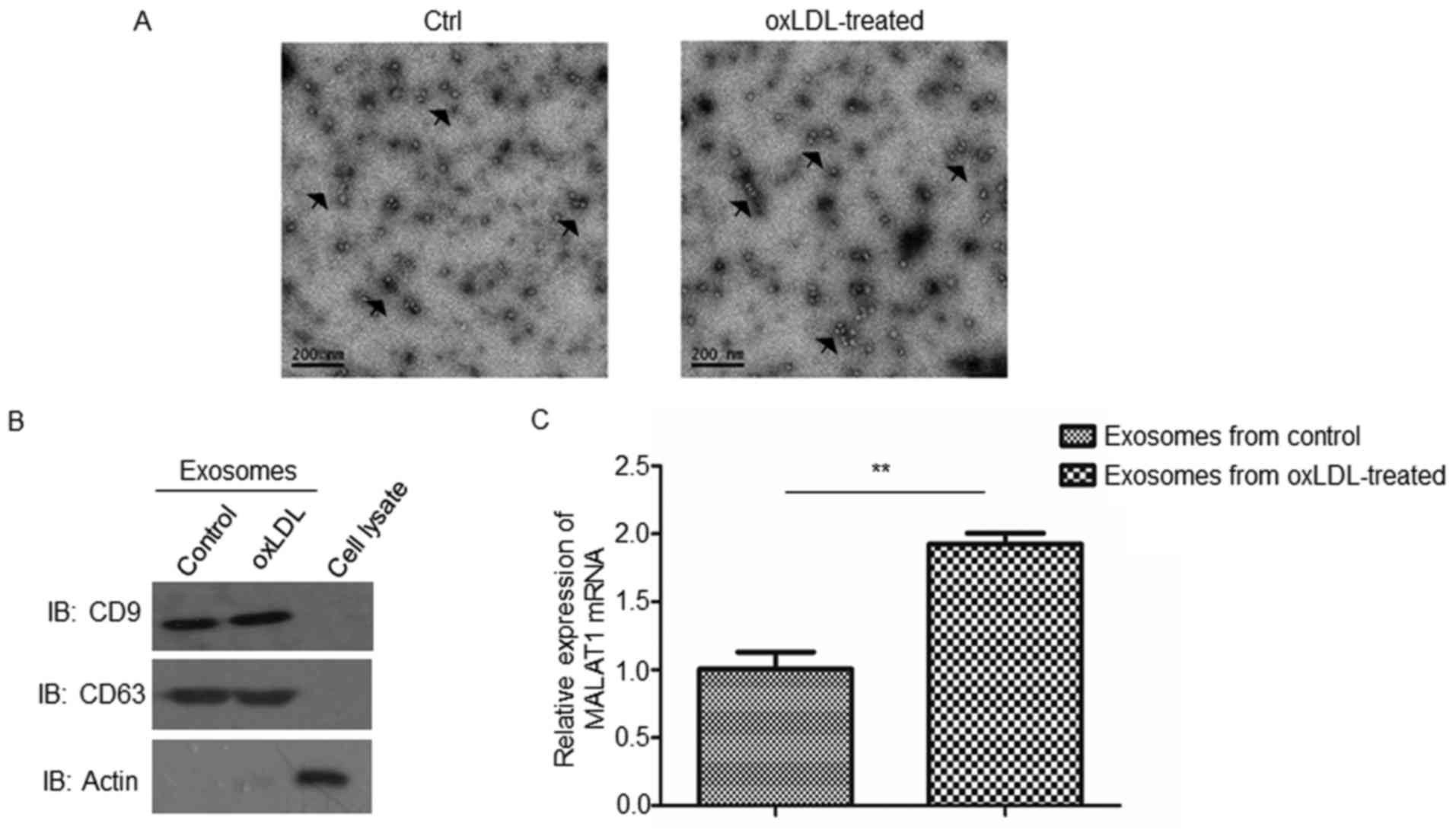
HUVEC-derived exosomes in atherosclerosis, exosomes were isolated
from oxLDL-treated or untreated HUVECs. The morphology of isolated
exosomes was observed under transmission electron microscopy
(Fig. 1A). As revealed in Fig. 1A, the diameters of the exosomes
ranged from 30–120 nm. Furthermore, the results of western blot
analysis demonstrated that, compared with cell lysates, exosomes
were enriched with CD9 and CD63 exosomal markers (Fig. 1B), thus confirming the effective
isolation of exosomes.
Following this, levels of MALAT1 mRNA expression
were investigated in isolated exosomes via RT-qPCR, and the results
revealed that exosomes secreted by oxLDL-treated HUVECs exhibited
significantly enhanced levels of MALAT1 mRNA compared with those
excreted by untreated controls (Fig.
1C).
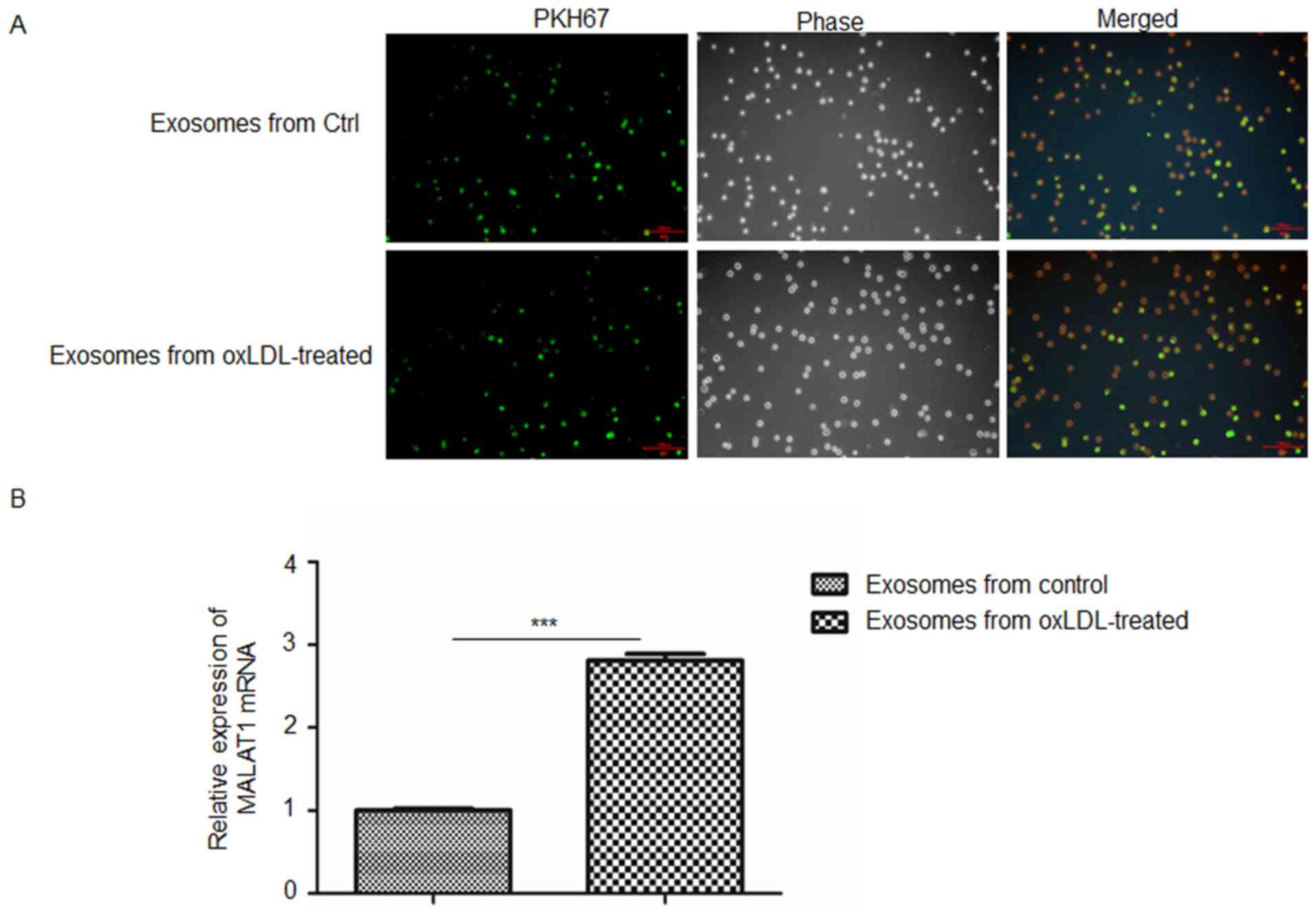
Monocytes endocytose exosomes isolated
from oxLDL-treated HUVECs
The present study investigated whether exosomes
released from oxLDL-treated HUVECs could be taken up by monocytes.
Exosomes labeled with PHK67 (green) were co-cultured with THP-1
cells. Following 24 h of co-culture, THP-1 cells were fixed and
observed by fluorescence microscopy. As revealed in Fig. 2A, exosomes were observed in
co-cultured THP-1 cells, thus indicating that monocytes
successfully endocytosed exosomes derived from untreated and
oxLDL-treated HUVECs. Furthermore, the levels of MALAT1 mRNA in
co-cultured THP-1 cells were investigated via RT-qPCR. The RT-qPCR
results demonstrated that THP-1 cells co-cultured with exosomes
secreted by oxLDL-treated HUVECs exhibited significantly enhanced
levels of MALAT1 mRNA compared with those co-cultured with exosomes
excreted by untreated control HUVECs (Fig. 2B).
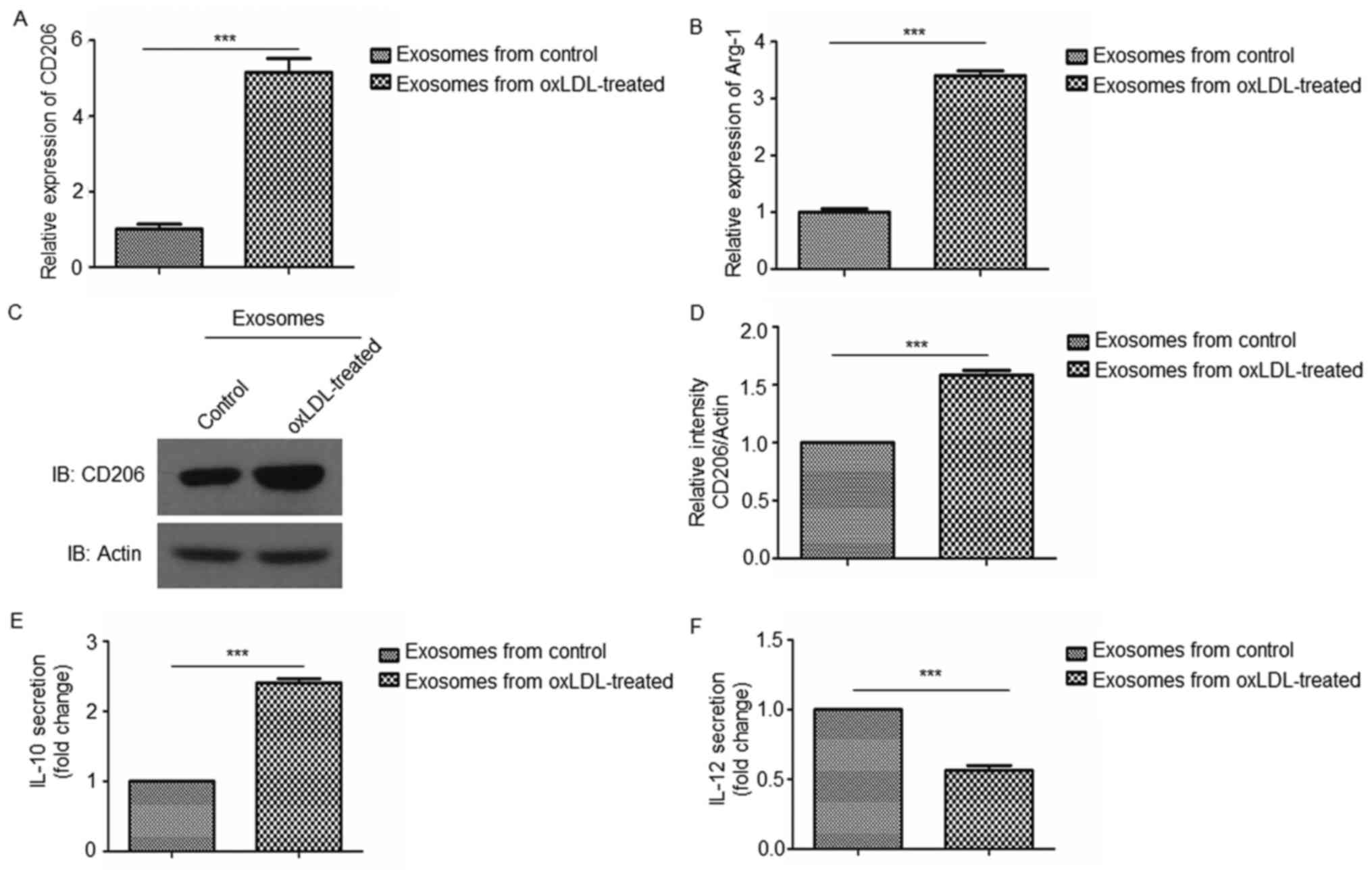
Exosomes derived from oxLDL-treated
HUVECs promote M2 macrophage polarization
In order to investigate the effects of oxLDL-treated
HUVEC-derived exosomes on macrophage polarization, THP-1 cells were
co-cultured with exosomes isolated from oxLDL-treated or untreated
HUVECs and the expression of M1 and M2 macrophage markers were
investigated by RT-qPCR, western blotting and ELISA analyses. The
results demonstrated that oxLDL-treated HUVEC-derived exosomes
enhanced the expression of the M2 macrophage markers (CD206, Arg-1
and IL-10; Fig. 3A-E) compared
with oxLDL-untreated HUVEC-derived exosomes, whereas the expression
of the M1 macrophage marker (IL-12; Fig. 3F) was reduced compared with
oxLDL-untreated HUVEC-derived exosomes. These results indicate that
exosomes derived from oxLDL-treated endothelial cells promoted M2
macrophage polarization.
Suppression of MALAT1 expression
reverses exosome-mediated M2 macrophage polarization
To further verify the effect of exosomal MALAT1 on
macrophage polarization, MALAT1 expression was knocked down in
THP-1 cells and successful knockdown was confirmed by RT-qPCR
(Fig. 4A). oxLDL-treated
HUVEC-derived exosomes were co-cultured with THP-1 cells with or
without MALAT1 knockdown. The results of subsequent RT-qPCR,
western blotting and ELISA analyses revealed that knockdown of
MALAT1 in THP-1 cells co-cultured with oxLDL-treated HUVEC-derived
exosomes led to the reduced expression of M2 macrophage markers
(CD206, Arg-1 and IL-10; Fig.
4B-F) and increased levels of IL-12, an M1 macrophage marker
(Fig. 4G), compared with those
co-cultured with oxLDL-treated HUVEC-derived exosomes without
knockdown of MALAT1. These results indicate that the suppression of
MALAT1 expression reversed M2 macrophage polarization mediated by
oxLDL-treated HUVEC-derived exosomes.
Discussion
Recent studies have demonstrated that exosomes
mediate intercellular communication via the transfer of RNAs and
proteins, which have important roles in numerous physiological and
pathological processes, including cardiovascular disease (21–27).
Chen et al (35) reported
that exosomal lncRNA growth arrest-specific 5 regulated the
apoptosis of macrophages and vascular endothelial cells in
atherosclerosis. In the present study, it was revealed that the
expression of exosomal MALAT1 was enhanced in oxLDL-treated HUVECs.
Furthermore, the results of the present study revealed that,
following co-culture with monocytes, these exosomes were
endocytosed and promoted M2 macrophage polarization via enhanced
expression of the M2 macrophage markers CD206, Arg-1 and IL-10, and
reduced expression of the M1 macrophage marker, IL-12.
Additionally, it was demonstrated that the suppression of MALAT1
expression in monocytes reversed M2 macrophage polarization
mediated by oxLDL-treated HUVEC-derived exosomes.
OxLDL contributes to atherosclerotic progression via
numerous mechanisms, including the induction of endothelial cell
dysfunction and macrophage foam cell formation (12). In addition, oxLDL-induced injury in
the retinal pigment epithelium was reported to enhance the exosomal
and apoptotic bleb release of CD46 and CD59, which are membrane
complement regulatory factors, thus indicating that oxLDL
stimulation may affect exosomal release (36). Reciprocal interactions between
endothelial cells and macrophages have been revealed in angiogenic
vascular niches via secreted microvesicles, such as exosomes
(37). In the present study, it
was demonstrated that oxLDL-stimulated HUVEC-derived exosomes
mediated the polarization of macrophages. In addition, in recent
studies, exosomes from numerous cell types, including stem cells
and cancer cells, have been demonstrated to be involved in
macrophage polarization (38–40).
However, the underlying molecular mechanisms of this process remain
unknown and require further investigation.
It has previously been established that exosomes
contain mRNAs, non-coding RNAs and proteins (23–25).
It was previously reported that exosomes derived from hypoxic
epithelial ovarian cancer deliver microRNA-940 for the induction of
M2 macrophage polarization (39).
Additionally, exosomal MALAT1 has been reported to have important
roles in the regulation of numerous pathological processes,
including those involved in cancer and endocrinology (41–43).
Serum MALAT-1 exosomal expression was upregulated and demonstrated
to promote cell proliferation and migration in non-small cell lung
cancer (42). Huangfu et al
(44) demonstrated that oxLDL
induced MALAT1 transcription through the nuclear factor-κB pathway.
Furthermore, Tang et al (45) revealed that MALAT1 protects the
endothelium from ox-LDL-induced endothelial dysfunction, in part
through competition with miR-22-3p for endogenous CXCR2 and AKT
mRNA. The results of the present study indicate a novel mechanism
for the onset of atherogenesis involving endothelial cells and
macrophages. M2 macrophages have atheroprotective function by
preventing foam cell formation (17). Thus, the results of the present
study demonstrate that exosomal MALAT1 derived from oxLDL-treated
endothelial cells may promote M2 macrophage polarization, and this
may provide a novel scientific basis for the understanding of
atherosclerosis progression.
Acknowledgements
Not applicable.
Funding
This research was supported by the Natural Sciences
Foundation of Zhejiang Province (grant nos. LY17H020003 and
LY17H020006) and Zhejiang Medical Science and Technology Project
(grant no. 2015KYA070).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CH and JH designed the study, acquired, analyzed and
interpreted the data, and wrote the manuscript. YW, SL, QW and WL
collected and analyzed the data. JZ was involved in designing the
experiments, interpreting data, and drafting and revising the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gkaliagkousi E, Gavriilaki E,
Triantafyllou A and Douma S: Clinical significance of endothelial
dysfunction in essential hypertension. Curr Hypertens Rep.
17:852015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Prieto D, Contreras C and Sánchez A:
Endothelial dysfunction, obesity and insulin resistance. Curr Vasc
Pharmacol. 12:412–426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Jager SCA, Meeuwsen JAL, van Pijpen FM,
Zoet GA, Barendrecht AD, Franx A, Pasterkamp G, van Rijn BB,
Goumans MJ and den Ruijter HM: Preeclampsia and coronary plaque
erosion: Manifestations of endothelial dysfunction resulting in
cardiovascular events in women. Eur J Pharmacol. 816:129–137. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang X, Li Y, Li Y, Ren X, Zhang X, Hu D,
Gao Y, Xing Y and Shang H: Oxidative stress-mediated
atherosclerosis: Mechanisms and therapies. Front Physiol.
8:6002017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen M, Ren L, Meng Y, Shi L, Chen L, Yu
B, Wu Q and Qi G: The protease inhibitor E64d improves
ox-LDL-induced endothelial dysfunction in human aortic endothelial
cells. Can J Physiol Pharmacol. 96:120–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li J, Liang X, Wang Y, Xu Z and Li G:
Investigation of highly expressed PCSK9 in atherosclerotic plaques
and ox-LDL-induced endothelial cell apoptosis. Mol Med Rep.
16:1817–1825. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li Z, Huang H, Huang L, Du L, Sun Y and
Duan Y: Prevention of oxidized low density lipoprotein-induced
endothelial cell injury by DA-PLGA-PEG-cRGD nanoparticles combined
with ultrasound. Int J Mol Sci. 18:pii: E815. 2017.
|
|
8
|
Amberger A, Maczek C, Jürgens G, Michaelis
D, Schett G, Trieb K, Eberl T, Jindal S, Xu Q and Wick G:
Co-expression of ICAM-1, VCAM-1, ELAM-1 and Hsp60 in human arterial
and venous endothelial cells in response to cytokines and oxidized
low-density lipoproteins. Cell Stress Chaperones. 2:94–103. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Weber C, Erl W, Weber KS and Weber PC:
Effects of oxidized low density lipoprotein, lipid mediators and
statins on vascular cell interactions. Clin Chem Lab Med.
37:243–251. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Di Pietro N, Formoso G and Pandolfi A:
Physiology and pathophysiology of oxLDL uptake by vascular wall
cells in atherosclerosis. Vascul Pharmacol. 84:1–7. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Westhorpe CL, Dufour EM, Maisa A,
Jaworowski A, Crowe SM and Muller WA: Endothelial cell activation
promotes foam cell formation by monocytes following
transendothelial migration in an in vitro model. Exp Mol Pathol.
93:220–226. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pirillo A, Norata GD and Catapano AL:
LOX-1, OxLDL, and atherosclerosis. Mediators Inflamm.
2013:1527862013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang N, Liang H and Zen K: Molecular
mechanisms that influence the macrophage m1-m2 polarization
balance. Front Immunol. 5:6142014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bories GFP and Leitinger N: Macrophage
metabolism in atherosclerosis. FEBS Lett. 591:3042–3060. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liberale L, Dallegri F, Montecucco F and
Carbone F: Pathophysiological relevance of macrophage subsets in
atherogenesis. Thromb Haemost. 117:7–18. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chistiakov DA, Bobryshev YV, Nikiforov NG,
Elizova NV, Sobenin IA and Orekhov AN: Macrophage phenotypic
plasticity in atherosclerosis: The associated features and the
peculiarities of the expression of inflammatory genes. Int J
Cardiol. 184:436–445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Colin S, Chinetti-Gbaguidi G and Staels B:
Macrophage phenotypes in atherosclerosis. Immunol Rev. 262:153–166.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fadini GP, Simoni F, Cappellari R, Vitturi
N, Galasso S, de Kreutzenberg Vigili S, Previato L and Avogaro A:
Pro-inflammatory monocyte-macrophage polarization imbalance in
human hypercholesterolemia and atherosclerosis. Atherosclerosis.
237:805–808. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu R, Li C, Wu Y, Shen L, Ma J, Qian J and
Ge J: Role of KCa3.1 channels in macrophage polarization and its
relevance in atherosclerotic plaque instability. Arterioscler
Thromb Vasc Biol. 37:226–236. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin Y, Chen Z and Kato S:
Receptor-selective IL-4 mutein modulates inflammatory vascular cell
phenotypes and attenuates atherogenesis in apolipoprotein
E-knockout mice. Exp Mol Pathol. 99:116–127. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martínez MC and Andriantsitohaina R:
Extracellular vesicles in metabolic syndrome. Circ Res.
120:1674–1686. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Todorova D, Simoncini S, Lacroix R,
Sabatier F and Dignat-George F: Extracellular vesicles in
angiogenesis. Circ Res. 120:1658–1673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nomura S: Extracellular vesicles and blood
diseases. Int J Hematol. 105:392–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goetzl EJ, Schwartz JB, Mustapic M, Lobach
IV, Daneman R, Abner EL and Jicha GA: Altered cargo proteins of
human plasma endothelial cell-derived exosomes in atherosclerotic
cerebrovascular disease. FASEB J. 31:3689–3694. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lu X: The role of exosomes and
exosome-derived microRNAs in atherosclerosis. Curr Pharm Des. Apr
13–2017.(Epub ahead of print).
|
|
26
|
Niu C, Wang X, Zhao M, Cai T, Liu P, Li J,
Willard B, Zu L, Zhou E, Li Y, et al: Macrophage foam cell-derived
extracellular vesicles promote vascular smooth muscle cell
migration and adhesion. J Am Heart Assoc. 5:pii: e00409. 2016.
View Article : Google Scholar
|
|
27
|
Cai J, Wu G, Jose PA and Zeng C:
Functional transferred DNA within extracellular vesicles. Exp Cell
Res. 349:179–183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Y, Zheng L, Wang Q and Hu YW: Emerging
roles and mechanisms of long noncoding RNAs in atherosclerosis. Int
J Cardiol. 228:570–582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li H, Zhu H and Ge J: Long noncoding RNA:
Recent updates in atherosclerosis. Int J Biol Sci. 12:898–910.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou T, Ding JW, Wang XA and Zheng XX:
Long noncoding RNAs and atherosclerosis. Atherosclerosis.
248:51–61. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang X, Hamblin MH and Yin KJ: The long
noncoding RNA malat1: Its physiological and pathophysiological
functions. RNA Biol. 14:1705–1714. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Uchida S and Dimmeler S: Long noncoding
RNAs in cardiovascular diseases. Circ Res. 116:737–750. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yoshimoto R, Mayeda A, Yoshida M and
Nakagawa S: MALAT1 long non-coding RNA in cancer. Biochim Biophys
Acta. 1859:192–199. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen L, Yang W, Guo Y, Chen W, Zheng P,
Zeng J and Tong W: Exosomal lncRNA GAS5 regulates the apoptosis of
macrophages and vascular endothelial cells in atherosclerosis. PLoS
One. 12:e01854062017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ebrahimi KB, Fijalkowski N, Cano M and
Handa JT: Oxidized low-density-lipoprotein-induced injury in
retinal pigment epithelium alters expression of the membrane
complement regulatory factors CD46 and CD59 through exosomal and
apoptotic bleb release. Adv Exp Med Biol. 801:259–265. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Baer C, Squadrito ML, Iruela-Arispe ML and
De Palma M: Reciprocal interactions between endothelial cells and
macrophages in angiogenic vascular niches. Exp Cell Res.
319:1626–1634. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao H, Shang Q, Pan Z, Bai Y, Li Z, Zhang
H, Zhang Q, Guo C, Zhang L and Wang Q: Exosomes from
adipose-derived stem cells attenuate adipose inflammation and
obesity through polarizing M2 macrophages and beiging in white
adipose tissues. Diabetes. 67:235–247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen X, Ying X and Wang X, Wu X, Zhu Q and
Wang X: Exosomes derived from hypoxic epithelial ovarian cancer
deliver microRNA-940 to induce macrophage M2 polarization. Oncol
Rep. 38:522–528. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Song Y, Dou H, Li X, Zhao X, Li Y, Liu D,
Ji J, Liu F, Ding L, Ni Y and Hou Y: Exosomal miR-146a contributes
to the enhanced therapeutic efficacy of interleukin-1β-primed
mesenchymal stem cells against sepsis. Stem Cells. 35:1208–1221.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gezer U, Özgür E, Cetinkaya M, Isin M and
Dalay N: Long non-coding RNAs with low expression levels in cells
are enriched in secreted exosomes. Cell Biol Int. 38:1076–1079.
2014.PubMed/NCBI
|
|
42
|
Zhang R, Xia Y, Wang Z, Zheng J, Chen Y,
Li X, Wang Y and Ming H: Serum long non coding RNA MALAT-1
protected by exosomes is up-regulated and promotes cell
proliferation and migration in non-small cell lung cancer. Biochem
Biophys Res Commun. 490:406–414. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
El Bassit G, Patel RS, Carter G, Shibu V,
Patel AA, Song S, Murr M, Cooper DR, Bickford PC and Patel NA:
MALAT1 in human adipose stem cells modulates survival and
alternative splicing of PKCδII in HT22 cells. Endocrinology.
158:183–195. 2017.PubMed/NCBI
|
|
44
|
Huangfu N, Xu Z, Zheng W, Wang Y, Cheng J
and Chen X: LncRNA MALAT1 regulates oxLDL-induced CD36 expression
via activating β-catenin. Biochem Biophys Res Commun.
495:2111–2117. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang Y, Jin X, Xiang Y, Chen Y, Shen CX,
Zhang YC and Li YG: The lncRNA MALAT1 protects the endothelium
against ox-LDL-induced dysfunction via upregulating the expression
of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett.
589:3189–3196. 2015. View Article : Google Scholar : PubMed/NCBI
|