Introduction
Osteoporosis (OP) is a common bone disorder that
increases the risk of fractures and is characterized by low bone
mass (1,2). OP can be classified into primary,
secondary and idiopathic types. Primary OP typically affects
postmenopausal women and senile men, and is therefore further
divided into postmenopausal OP and senile OP (3). OP that occurs in young males with
absence of secondary causes of bone loss is termed male idiopathic
osteoporosis (MIO) (4). OP results
from an imbalance between bone formation by osteoblasts and bone
absorption by osteoclasts (5).
Decreased recruitment of osteoblasts and failure of vitamin D
activation were demonstrated to be associated with senile OP
(6). Estrogen deprivation is the
primary cause of postmenopausal OP and also serves a role in senile
OP (7). Although MIO is an
important public health problem worldwide (8), the pathogenesis of MIO differs from
the other two types of OP and has not yet been elucidated. However,
evidence indicates that osteoblastic defects may lead to MIO
(9) and that the insulin-like
growth factor I (IGF-I) pathway, and alterations in sex hormones
and sclerostin production, also serve roles in the pathogenesis of
MIO (9–11). To develop accurate and effective
diagnosis and treatment strategies for MIO, the molecular
mechanisms underlying the development of this disease requires
investigation.
Differentially expressed genes (DEGs) were
previously reported to be associated with the pathogenesis of OP,
and RNA sequencing is an approach that has been used to identify
DEGs in numerous diseases, including OP (12). Using RNA sequencing, the DEGs in
patients with MIO, and senile and postmenopausal OP, compared with
normal controls (NCs), were identified. DEGs that were common among
these three types of OP and unique DEGs only present in patients
with MIO were further analyzed. Functional annotation was performed
and a protein-protein interaction (PPI) network was constructed to
further investigate the biological functions of DEGs in OP. To the
best of our knowledge, the present study is the first to identify
DEGs in patients with MIO by RNA-sequencing. The results of the
present study may be used in the future for identification of genes
and pathways associated with MIO and aid in elucidating the
pathogenesis of this disease.
Materials and methods
Patients
Patients with OP (2 patients with senile OP, 2
patients with postmenopausal OP and 2 patients with MIO) and two NC
patients were recruited from June to August 2016 at Jining No. 1
People's Hospital (Jining, China). Characteristics of patients with
MIO and healthy NC patients are presented in Table I. Written informed consent was
obtained from all participants and the present study was approved
by the Ethics Committee of Jining No. 1 People's Hospital (Jining,
China).
 | Table I.Patient characteristics. |
Table I.
Patient characteristics.
|
| MIO | Postmenopausal
OP | Senile OP | Normal control |
|---|
| No. of
patients | 2 | 2 | 2 | 2 |
| Male/female | 2/0 | 0/2 | 2/0 | 2/0 |
| Age (years) | 38.0±1.0 | 73±5.0 | 89±0.0 | 33.5±1.5 |
| Smoking history
(n) | 2 | 0 | 0 | 0 |
| Alcohol consumption
(n) | 2 | 0 | 0 | 2 |
RNA isolation and sequencing
Whole blood samples were obtained from all eight
participants. Total RNA was isolated from whole blood samples and
purified using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and an RNeasy Mini kit (Qiagen,
Inc., Valencia, CA, USA), respectively. The quantity and integrity
of purified RNA was verified using an Agilent 2100 Bioanalyzer
(Agilent Technologies, Inc., Santa Clara, CA, USA). mRNA that
passed quality control (RNA integrity number >7) was used to
construct libraries with a TruSeq RNA Sample Prep kit v2 (Illumina,
Inc., San Diego, CA, USA), as described below. Initially, mRNA was
fragmented at 95°C for 8 min with fragmentation buffer.
Subsequently, the fragmented mRNA samples were used as templates
for the synthesis of the first cDNA strand using First Strand
Master Mix and Super Script II reverse transcriptase (Invitrogen;
Thermo Fisher Scientific, Inc.) at 25°C for 10 min, 42°C for 50 min
and 70°C for 15 min. The second cDNA strand was synthesized
following the addition of Second Strand Master Mix and incubation
at 16°C for 1 h. Finally, end repair, adenylate 3′ ends and adapter
ligation were performed according to the manufacturer's protocol of
the TruSeq RNA Sample Prep kit v2. Polymerase chain reaction (PCR)
was performed with PCR Master Mix and RNA PCR primer cocktail in
the TruSeq RNA Sample Prep kit v2 (Illumina, Inc., San Diego, CA,
USA) to amplify the libraries according to the manufacturer's
instructions. The following thermocycling conditions were used for
the PCR: Initial denaturation at 98°C for 30 sec; 11 cycles of 98°C
for 10 sec, 65°C for 30 sec and 72°C for 15 sec, followed by a
final extension step of 72°C for 10 min. The enriched cDNA
libraries were subsequently sequenced using the Illumina HiSeq 2500
(Illumina, Inc.) sequencing platform.
Identification of DEGs
Read QC tool in FastQC version 0.11.4 software
(13) was used for the quality
control of FASTQ data. Trimming of raw data was performed with
cutadapt version 1.9.1 (14). Low
quality reads, including adaptor sequences and reads with ratios of
N base >10%, were removed and clean reads were obtained. TopHat
version 2.1.1 (15) was used to
align the clean reads to the human genome (GRCh38.p7 assembly,
https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.38)
(16). The quantification of mRNA
and standardized output was performed using Cuffquant and Cuffdiff
in Cufflinks version 2.2.1 (15).
Fragments per kilobase of exon per million fragments mapped were
used to determine the transcript abundance of each gene. P<0.05
and abs (count_1-count_2) >100 served as the thresholds for the
identification of DEGs, where abs indicated the absolute value, and
count_1 and count_2 indicated the cases of OP (senile,
postmenopausal and MIO) and the control group, respectively. The
DEGs in MIO, and postmenopausal and senile OP, compared with NCs
were identified. DEGs that were common among the three types of OP
were identified. A heat map of DEGs in MIO and common DEGs in the
three types of OP were generated using the heat map.2 function in
the gplots package in R version 3.3.3 (17).
Functional annotation
Gene ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway enrichment analyses were performed using
the online software GeneCodis 3 (http://genecodis.cnb.csic.es/analysis) (18) to interpret the biological function
of DEGs in MIO, postmenopausal OP and senile OP. A false discovery
rate (FDR) <0.05 was considered to indicate a statistically
significant difference.
PPI network construction
To determine disease-associated pathways and
functions of proteins at the molecular level, the PPI networks of
DEGs in MIO, and postmenopausal and senile OP, were constructed
using Biological General Repository for Interaction Datasets
(http://thebiogrid.org/) and Cytoscape version
3.3.3 (19). Nodes were used to
represent proteins and edges to represent interactions between
proteins. Based on Cytoscape version 3.3.3, proteins with a degree
of >160 were defined as hub proteins of the PPI network.
Results
Identification of DEGs
Compared with NCs, a total of 519 (185 upregulated
and 334 downregulated), 368 (272 upregulated and 96 downregulated)
and 1,472 (646 upregulated and 826 downregulated) DEGs were
identified in MIO, senile OP and postmenopausal OP, respectively.
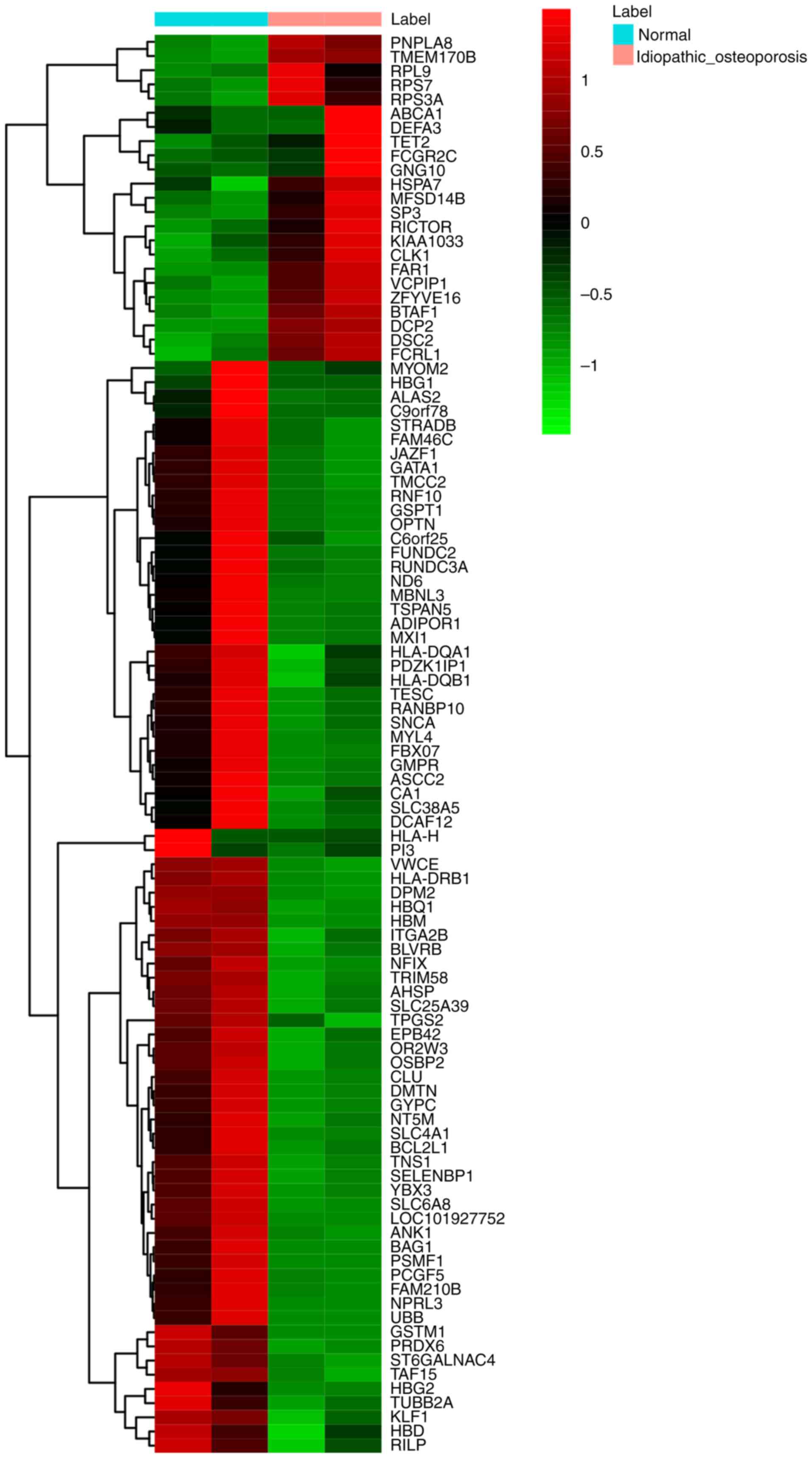
The top 20 upregulated and downregulated DEGs in MIO are presented
in Table II.
 | Table II.Top 20 downregulated and upregulated
differentially expressed genes in patients with male idiopathic
osteoporosis compared with normal controls. |
Table II.
Top 20 downregulated and upregulated
differentially expressed genes in patients with male idiopathic
osteoporosis compared with normal controls.
| A, Downregulated
genes |
|---|
|
|---|
| Gene ID (NCBI) | Gene | P-value |
|---|
| 212 | ALAS2 |
5.00×10−5 |
| 2038 | EPB42 |
5.00×10−5 |
| 25793 | FBXO7 |
5.00×10−5 |
| 2766 | GMPR |
5.00×10−5 |
| 3047 | HBG1 |
5.00×10−5 |
| 3048 | HBG2 |
5.00×10−5 |
| 3117 | HLA-DQA1 |
5.00×10−5 |
| 3123 | HLA-DRB1 |
5.00×10−5 |
| 3136 | HLA-H |
5.00×10−5 |
| 55796 | MBNL3 |
5.00×10−5 |
| 9172 | MYOM2 |
5.00×10−5 |
| 8991 | SELENBP1 |
5.00×10−5 |
| 6521 | SLC4A1 |
5.00×10−5 |
| 6535 | SLC6A8 |
5.00×10−5 |
| 6622 | SNCA |
5.00×10−5 |
| 7145 | TNS1 |
5.00×10−5 |
| 25893 | TRIM58 |
5.00×10−5 |
| 10098 | TSPAN5 |
5.00×10−5 |
| 7280 | TUBB2A |
5.00×10−5 |
| 220001 | VWCE |
5.00×10−5 |
|
| B, Upregulated
genes |
|
| Gene ID | Gene | P-value |
|
| 84188 | FAR1 |
5.00×10−5 |
| 9103 | FCGR2C |
5.00×10−5 |
| 84641 | MFSD14B |
5.00×10−5 |
| 253260 | RICTOR |
5.00×10−5 |
| 9765 | ZFYVE16 |
5.00×10−5 |
| 1824 | DSC2 |
1.00×10−4 |
| 3311 | HSPA7 |
1.00×10−4 |
| 2790 | GNG10 |
1.50×10−4 |
| 6670 | SP3 |
2.00×10−4 |
| 23325 | KIAA1033 |
2.50×10−4 |
| 50640 | PNPLA8 |
2.50×10−4 |
| 6201 | RPS7 |
2.50×10−4 |
| 19 | ABCA1 |
3.50×10−4 |
| 167227 | DCP2 |
4.00×10−4 |
| 1195 | CLK1 |
5.00×10−4 |
| 1668 | DEFA3 |
5.00×10−4 |
| 100113407 | TMEM170B |
5.00×10−4 |
| 80124 | VCPIP1 |
5.00×10−4 |
| 6189 | RPS3A |
5.50×10−4 |
| 54790 | TET2 |
5.50×10−4 |
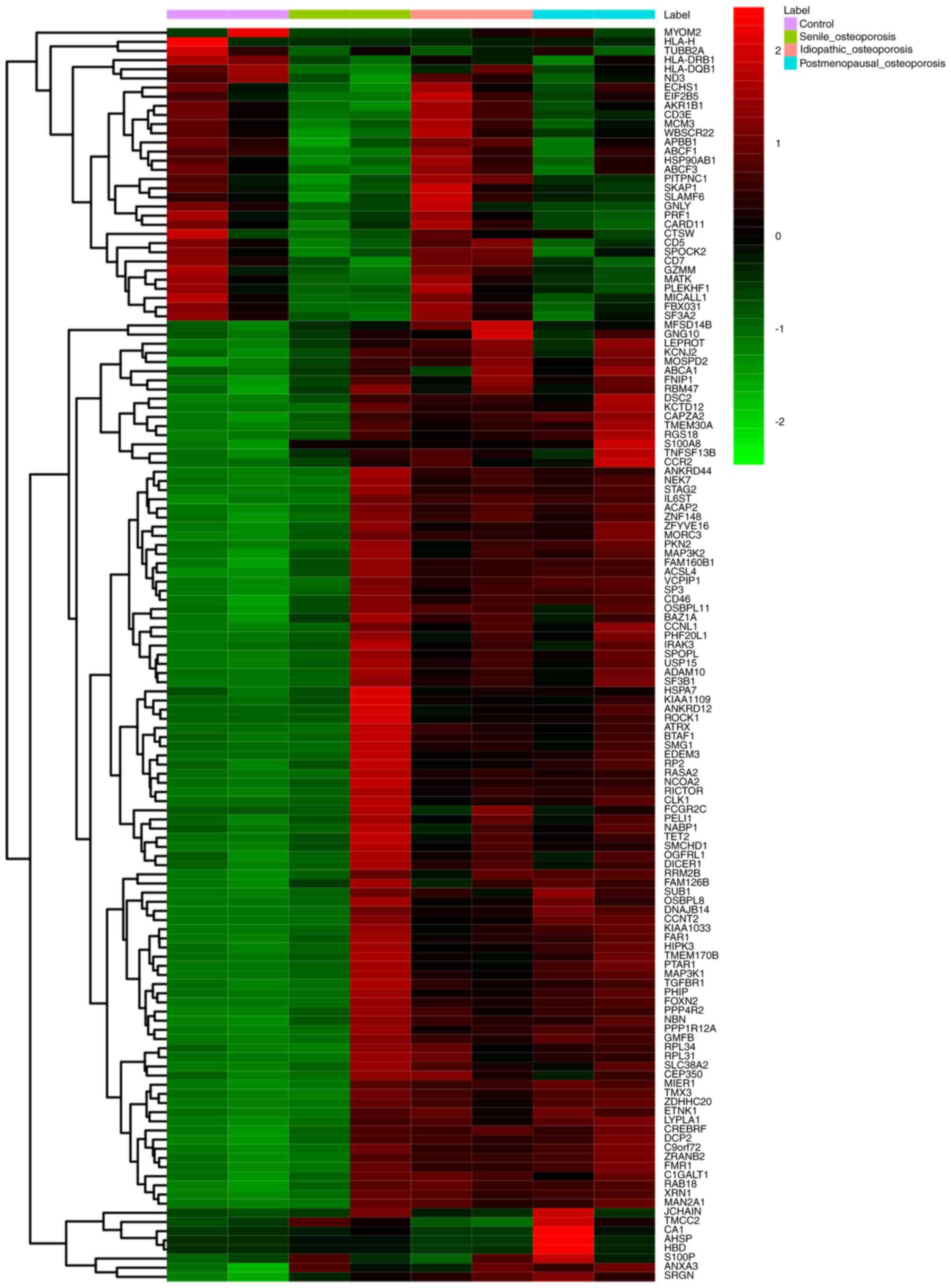
A total of 138 shared DEGs were identified in the
three types of OP compared with NCs. A total of 7 DEGs, including
α-hemoglobin stabilizing protein (AHSP), carbonic anhydrase 1
(CA1), hemoglobin subunit δ (HBD), transmembrane and coiled-coil
domain family 2 (TMCC2), joining chain of multimeric IgA and IgM
(JCHAIN), RP2 ARL3 GTPase-activating protein (RP2) and S100
calcium-binding protein P (S100P), were differentially expressed in
MIO compared with both senile and postmenopausal OP (Table III). Certain DEGs, including
AHSP, CA1, HBD and TMCC2, were downregulated in MIO but upregulated
in senile and postmenopausal OP, compared with NCs. A heat map of
the top 100 DEGs in MIO is presented in Fig. 1, while a heat map of the 138 common
DEGs among the three types of OP is presented in Fig. 2.
 | Table III.Differentially expressed genes in MIO
compared with NCs and senile and postmenopausal OP. |
Table III.
Differentially expressed genes in MIO
compared with NCs and senile and postmenopausal OP.
| Gene ID (NCBI) | Gene | P-value 1 | P-value 2 | P-value 3 | Differential
expression |
|---|
| 51327 | AHSP |
5.00×10−5 |
5.00×10−5 |
5.00×10−5 | Downregulated |
| 759 | CA1 |
1.00×10−4 |
5.00×10−5 |
5.00×10−5 | Downregulated |
| 3045 | HBD |
1.50×10−4 |
5.00×10−5 |
5.00×10−5 | Downregulated |
| 3512 | JCHAIN |
1.12×10−2 |
4.15×10−3 |
1.50×10−4 | Upregulated |
| 6102 | RP2 |
9.45×10−3 |
3.48×10−2 |
2.20×10−2 | Upregulated |
| 6286 | S100P |
2.95×10−3 |
4.59×10−2 |
6.00×10−4 | Upregulated |
| 9911 | TMCC2 |
1.00×10−4 |
2.50×10−4 |
5.00×10−5 | Downregulated |
Functional annotation
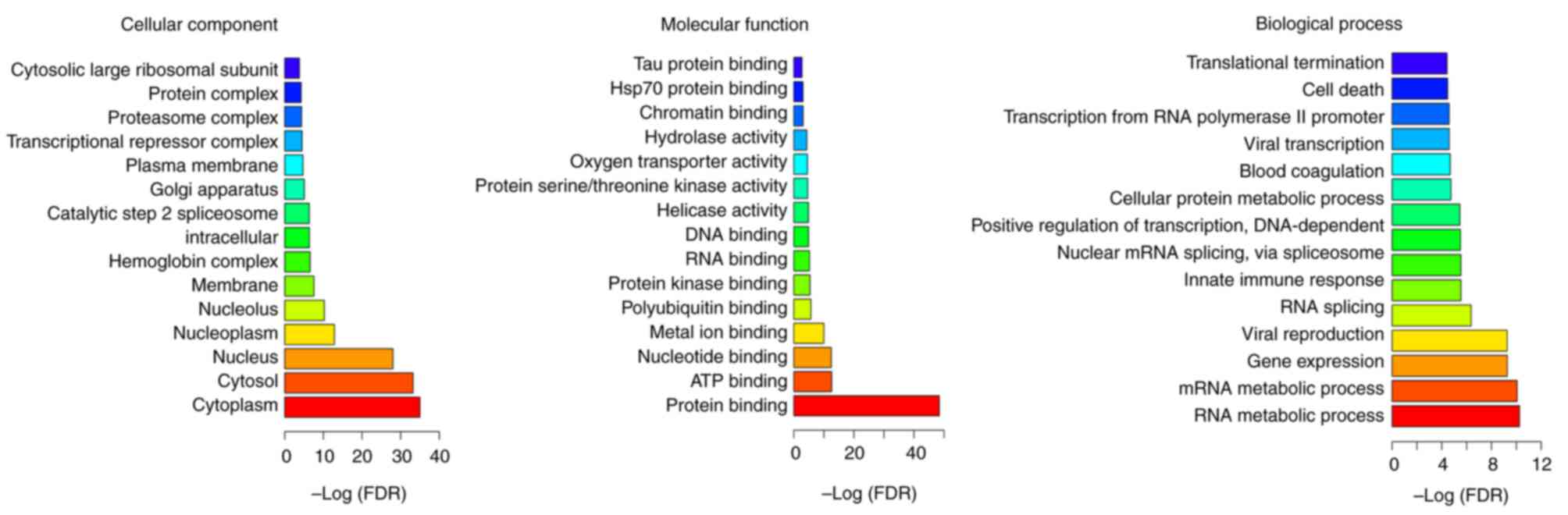
According to the GO enrichment analysis, ‘RNA
metabolic process’ (FDR=5.60×10−11), ‘mRNA metabolic
process’ (FDR=8.59×10−11), ‘cytoplasm’
(FDR=9.85×10−36), ‘cytosol’ (FDR=5.98×10−34),
‘protein binding’ (FDR=4.02×10−49) and ‘ATP binding’
(FDR=2.55×10−13) were the most significantly enriched GO
terms of DEGs in MIO (Fig. 3).
‘Peptidyl-serine phosphorylation’ (FDR=1.37×10−5),
‘protein phosphorylation’ (FDR=2.43×10−5), ‘cytoplasm’
(FDR=1.82×10−8), ‘nucleus’ (FDR=8.03×10−7),
‘protein binding’ (FDR=4.30×10−11) and ‘ATP binding’
(FDR=3.33×10−8) were the most significantly enriched GO
terms of the 138 common DEGs among the three types of OP (data not
shown).
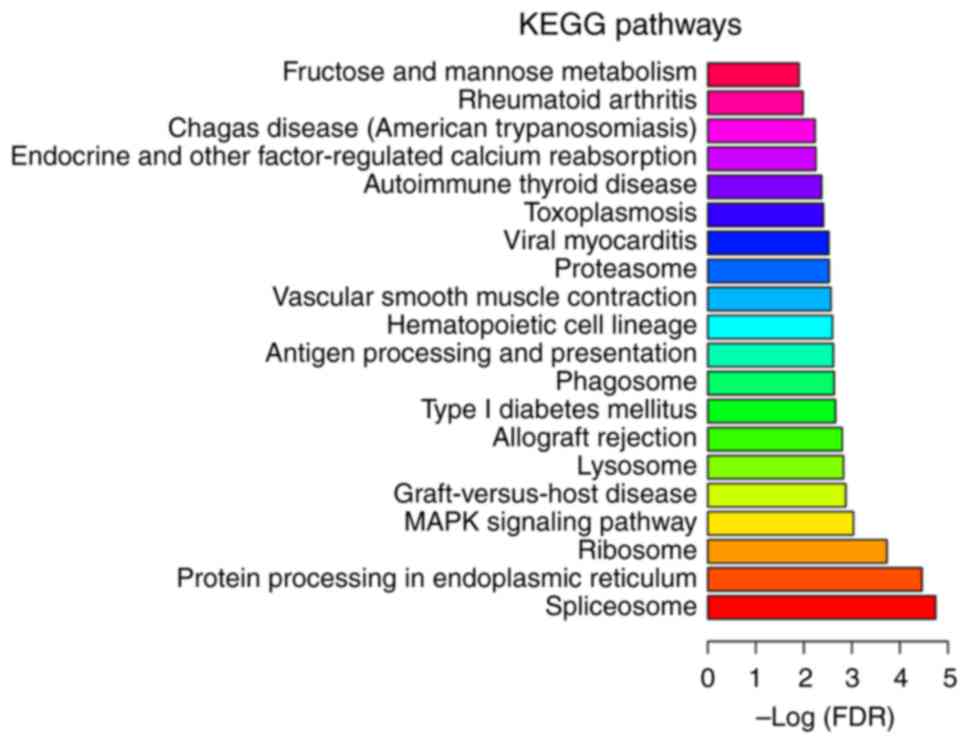
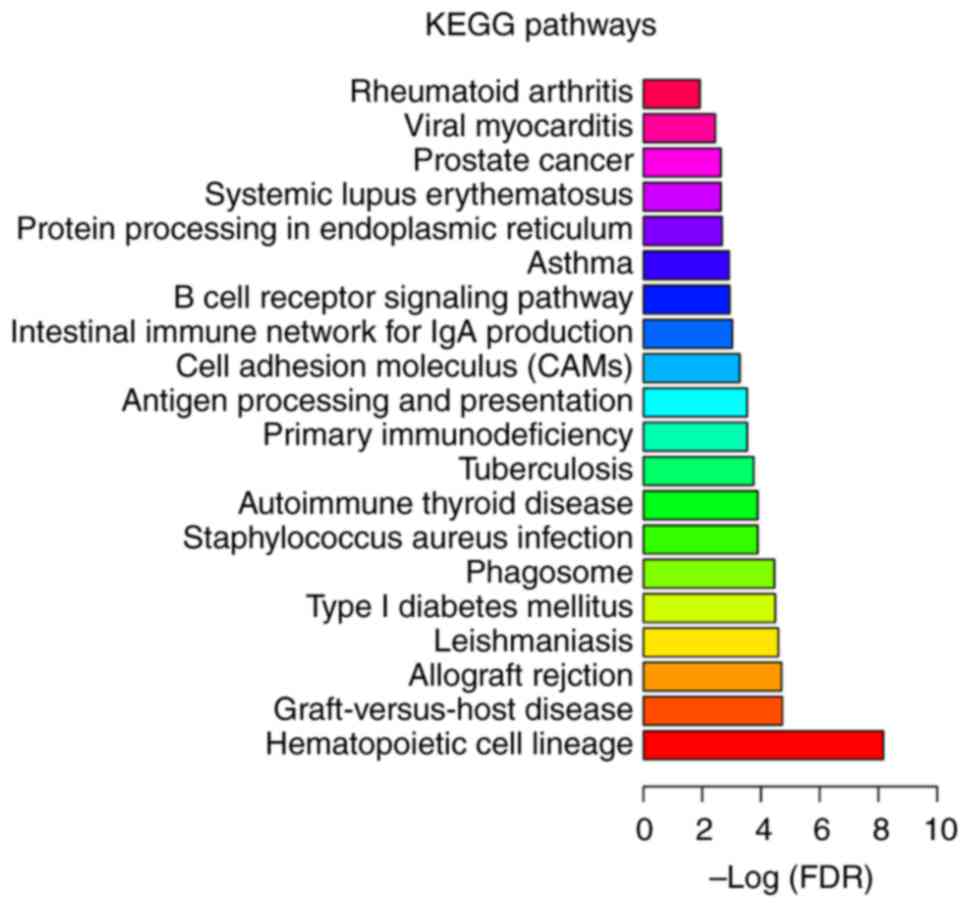
Following KEGG enrichment analysis, ‘MAPK signaling
pathway’ (FDR=9.25×10−4), ‘type I diabetes mellitus’
(FDR=2.19×10−3), ‘antigen processing and presentation’
(FDR=2.42 ×10−3) and ‘hematopoietic cell lineage’
(FDR=2.53×10−3) were determined to be the most
significantly enriched pathways in MIO (Fig. 4). Four DEGs including T-cell
antigen CD7 (CD7), T-cell surface glycoprotein CD3 ε chain (CD3E),
T-cell surface glycoprotein CD5 (CD5) and major histocompatibility
complex class II antigen DR β 1 (HLA-DRB1) in MIO were
significantly enriched in the ‘hematopoietic cell lineage’ pathway.
In addition, ‘hematopoietic cell lineage’
(FDR=6.71×10−9), ‘graft-versus-host disease’
(FDR=1.84×10−5) and ‘allograft rejection’
(FDR=1.98×10−5) were the most significantly enriched in
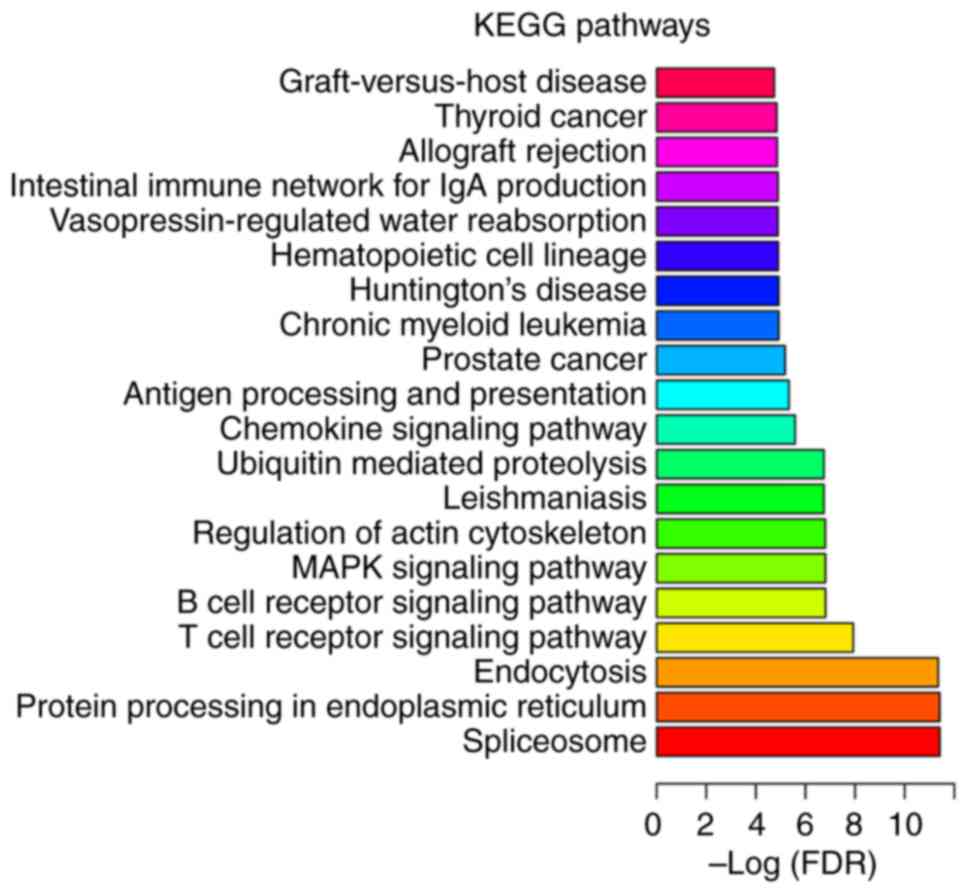
senile OP (Fig. 5), while
‘spliceosome’ (FDR=3.79×10−12), ‘protein processing in
endoplasmic reticulum’ (FDR=3.83×10−12) and
‘endocytosis’ (FDR=4.41×10−12) were the most
significantly enriched pathways in postmenopausal OP (Fig. 6). ‘Hematopoietic cell lineage’ was
a significantly enriched pathway that was common among all three
types of OP (Figs. 4–6).
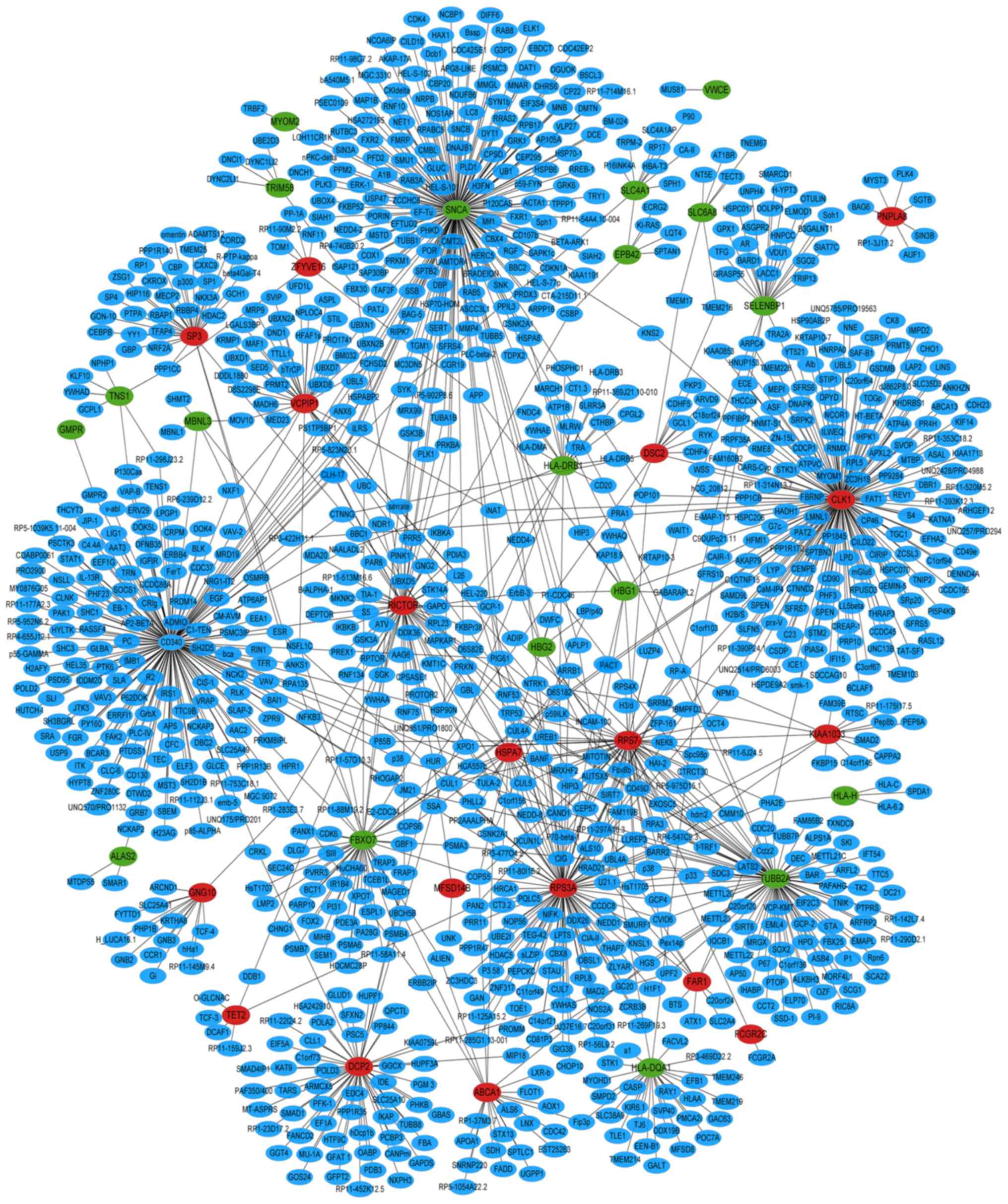
PPI network construction
The PPI network of the top 20 upregulated and
downregulated DEGs in MIO was constructed, which included 1,066
nodes and 1,246 edges (Fig. 7).
According to the PPI network, α-synuclein (SNCA; degree, 179;
clustering coefficient, 0), CDC-like kinase 1 (CLK1; degree, 162;
clustering coefficient, 0) and receptor tyrosine-protein kinase
erbB-2; degree, 160; clustering coefficient, 0) were the 3 hub
proteins in MIO. The PPI network of the 138 common DEGs among the
three types of OP consisted of 2,680 nodes and 3,957 edges (data
not shown). Heat shock protein 90 α family class B member 1
(degree, 233; clustering coefficient, 2.22×10−4),
capping actin protein of muscle Z-line α subunit 2 (degree, 229;
clustering coefficient, 6.51×10−4) and CLK1 (degree,
162; clustering coefficient, 1.53×10−4) were the 3 hub
proteins. CLK1 was a common hub protein in the MIO-specific PPI
network and OP-specific PPI network.
Discussion
MIO is an important public health problem worldwide
(8) and the mechanism underlying
this disease remains to be elucidated. The present study identified
DEGs in MIO, and senile and postmenopausal OP, compared with NCs.
The common DEGs among these three types of OP and unique DEGs only
identified in patients with MIO were investigated. The primary
symptom of OP is bone loss induced by an imbalance between bone
resorption and bone formation. Factors that regulate this balance
may be involved in the pathogenesis of OP. Osteoblastic defects
have been reported to be associated with the mechanism underlying
the development of MIO, and regulators of osteoblasts and bone
formation may therefore be associated with MIO.
‘MAPK signaling pathway’ was a significantly
enriched pathway of DEGs in patients with MIO included in the
present study. The mitogen-activated protein kinase (MAPK) pathway
regulates numerous cellular activities, including cell
proliferation, differentiation, survival, death and transformation
(20). The MAPK signaling pathway
primarily consists of an extracellular signal-regulated kinase, p38
and c-Jun NH2-terminal protein kinase (JNK) (21). Furthermore, the JNK signaling
pathway was reported to be associated with the differentiation and
proliferation of osteoblasts (22–25).
The MAPK signaling pathway and the DEGs enriched in this pathway
may serve a role in MIO by regulating bone formation.
Previous studies have demonstrated that IGF-I, which
has a similar function and structure to insulin, is crucial for the
progression of MIO (10,26). In the present study, DEGs in
patients with MIO were significantly enriched in ‘type I diabetes
mellitus’. As a deficiency in insulin can impair bone formation
induced by osteoblasts, type I diabetes mellitus (TIDM) with low
serum IGF-I levels has been reported to be a risk factor for OP
(27). Therefore, we hypothesized
that the TIDM pathway and its associated DEGs in MIO, including
perforin 1, major histocompatibility complex (HLA) class I C, HLA
class II DQα1, HLA class II DRβ1, HLA class II DQβ1, may serve a
role in MIO by regulating bone formation via IGF-I. Furthermore,
HLA alleles have been reported to be associated with OP by
regulating bone mineral density (28).
In addition to genes involved in the regulation of
bone formation by osteoblasts, genes involved in the process of
bone resorption by osteoclasts may also be associated with the
regulation of bone formation and the mechanism of MIO. Osteoclasts
originate from pluripotent hematopoietic cells (5). In the present study, four DEGs (CD7,
CD3E, CD5 and HLA-DRB1) in MIO were significantly enriched in the
‘hematopoietic cell lineage’ pathway, which supported the
hypothesis that the number and activity of osteoclasts may be
associated with MIO. Furthermore, ‘hematopoietic cell lineage’ was
a common pathway enriched in the other two types of OP, which
emphasizes the potential importance of this pathway in OP.
Tetraspanin (TSPAN)5 is a member of the TSPAN family
of proteins and was a unique DEG in MIO that was not differentially
expressed in the other two types of OP. TSPAN5 was previously
reported to be an inhibitor of osteoclastogenesis (24). In the present study, downregulated
TSPAN5 was detected in the blood of patients with MIO, which
indicated that TSPAN5 may be involved in the process of MIO as
reduced TSPAN5 levels leads to the promotion of
osteoclastogenesis.
SNCA was another unique DEG in the present study,
which was identified only in patients with MIO. SNCA has been
demonstrated to be associated with osteoclastogenesis and bone
resorption (28) by interacting
with the bone morphogenetic protein pathway and modulating bone
loss induced by deficiency of estrogen (29–31).
In the present study, SNCA was downregulated in patients with MIO
and may serve a role in the development of this disease by
regulating bone resorption. Furthermore, SNCA was a hub gene in the
MIO-specific PPI network, which emphasizes the potential that this
gene may be associated with the pathogenesis of MIO.
In the present study, CLK1 was a hub protein of both
the MIO- and OP-specific PPI networks. CLK1 is a dual specificity
kinase (31) and was upregulated
in all three types of OP, which indicates that CLK1 may regulate
the process of OP. However, to the best of our knowledge, an
association between CLK1 and OP has not been previously reported.
Further studies are required to validate the function of CLK1 in
the pathogenesis of OP.
Of the 138 common DEGs in all three types of OP, 7
DEGs (AHSP, CA1, HBD, TMCC2, JCHAIN, RP2 and S100P) were
significantly differentially expressed in MIO compared with the
other two types of OP. Compared with NCs, CA1 was significantly
downregulated in MIO, while it was upregulated in both senile and
postmenopausal OP. As a member of the CA family, CA1 has been
reported to function in bone formation, bone remodeling and
calcification (32). CA1 catalyzes
the hydration and dehydration reactions of
CO2/H2CO3. As CaCO3 is
associated with initial bone formation, CA1 contributes to bone
formation and biomineralization by forming a CaCO3
precipitate (32,33). Increased CA1 activity was
demonstrated to promote bone resorption (34). Although the mechanism remains to be
elucidated, the hydrogen ions produced by CA1 can dissolve the
mineral components of bones and may enhance bone resorption
(34). By regulating
calcification, CA1 serves different roles in bone resorption and
formation. As the expression of CA1 in MIO was significantly
different compared with the other two types of OP in the present
study, it may be hypothesized that CA1 may be involved in the
process of MIO by regulating bone mass and that the mechanism may
be different compared with that in senile and postmenopausal OP.
Further experiments are required to verify this hypothesis.
In the current study, S100P was upregulated in all
three types of OP compared with NC. However, the expression of
S100P in MIO was markedly decreased compared with senile and
postmenopausal OP. S100P is a member of the S100 family of
calcium-binding proteins and is a target of bone morphogenetic
protein 4 (35). Furthermore,
abnormal calcium binding was previously reported to be associated
with OP (36), which indicates
that S100P may serve a unique role in MIO.
In conclusion, the MAPK signaling pathway and the
TIDM pathway may be associated with the process of MIO by
regulating bone formation. Furthermore, the hematopoietic cell
lineage pathway and two unique DEGs in MIO (SNCA and TSPAN5) may be
associated with bone resorption. CA1 and S100P may regulate the
process of MIO via functions in calcification and dysregulation of
calcium binding. In the present study, CA1 was downregulated in MIO
and upregulated in senile and postmenopausal OP, which indicates a
unique role for this protein in MIO. However, a lack of biological
validation of the results of the current study was a limitation.
Further validation with a larger sample sizes, as well as in
vitro and in vivo experiments are required to confirm
these results.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LF and BX made substantial contributions the
conceptualization and design of the study. LF, YW, JZ and BT
collected and analyzed the data. LF and YW interpreted the data.
All authors were involved in drafting and revising the manuscript
and gave final approval of the manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
participants and the present study was approved by the Ethics
Committee of Jining No.1 People's Hospital (Jining, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CA1
|
carbonic anhydrase 1
|
|
CLK1
|
CDC-like kinase 1
|
|
DEGs
|
differentially expressed genes
|
|
GO
|
gene ontology
|
|
HLA
|
human leukocyte antigen
|
|
IGF-I
|
insulin-like growth factor I
|
|
JNK
|
c-Jun NH2-terminal kinase
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
MAPK
|
mitogen-activated protein kinase
|
|
OP
|
osteoporosis
|
|
MIO
|
male idiopathic osteoporosis
|
|
NC
|
normal control
|
|
PPI
|
protein-protein interaction
|
|
SNCA
|
α-synuclein
|
|
TIDM
|
type I diabetes mellitus
|
|
TSPAN5
|
tetraspanin 5
|
References
|
1
|
Karasik D, Rivadeneira F and Johnson ML:
The genetics of bone mass and susceptibility to bone diseases. Nat
Rev Rheumatol. 12:4962016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Arjmandi BH, Johnson SA, Pourafshar S,
Navaei N, George KS, Hooshmand S, Chai SC and Akhavan NS:
Bone-protective effects of dried plum in postmenopausal women:
Efficacy and possible mechanisms. Nutrients. 9:pii: E496. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Usman J and Siddiqui H: Osteoporosis in
family practice. J Pak Med Assoc. 53:433–436. 2003.PubMed/NCBI
|
|
4
|
Brumsen C, Papapoulos SE, Lentjes EG,
Kluin PM and Hamdy NA: A potential role for the mast cell in the
pathogenesis of idiopathic osteoporosis in men. Bone. 31:556–561.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dudakovic A, Camilleri ET, Riester SM,
Paradise CR, Gluscevic M, O'Toole TM, Thaler R, Evans JM, Yan H,
Subramaniam M, et al: Enhancer of Zeste homolog 2 inhibition
stimulates bone formation and mitigates bone loss caused by
ovariectomy in skeletally mature mice. J Biol Chem.
291:24594–24606. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee YH, Kim JE, Roh YH, Choi HR, Rhee Y,
Kang DR and Lim SK: The combination of vitamin D deficiency and
mild to moderate chronic kidney disease is associated with low bone
mineral density and deteriorated femoral microarchitecture: Results
from the KNHANES 2008–2011. J Clin Endocrinol Metab. 99:3879–3888.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Klein-Nulend J, van Oers RF, Bakker AD and
Bacabac RG: Bone cell mechanosensitivity, estrogen deficiency, and
osteoporosis. J Biomech. 48:855–865. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gielen E, Vanderschueren D, Callewaert F
and Boonen S: Osteoporosis in men. Best Pract Res Clin Endocrinol
Metab. 25:321–335. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Patsch JM, Kohler T, Berzlanovich A,
Muschitz C, Bieglmayr C, Roschger P, Resch H and Pietschmann P:
Trabecular bone microstructure and local gene expression in iliac
crest biopsies of men with idiopathic osteoporosis. J Bone Miner
Res. 26:1584–1592. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Paccou J, Dewailly J and Cortet B: Reduced
levels of serum IGF-1 is related to the presence of osteoporotic
fractures in male idiopathic osteoporosis. Joint Bone Spine.
79:78–82. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Muschitz C, Kocijan R, Pahr D, Patsch JM,
Amrein K, Misof BM, Kaider A, Resch H and Pietschmann P:
Ibandronate increases sclerostin levels and bone strength in male
patients with idiopathic osteoporosis. Calcif Tissue Int.
96:477–489. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mohammadi Z, Keshtkar A, Fayyazbakhsh F,
Ebrahimi M, Amoli MM, Ghorbani M, Khashayar P, Dini M, Ebrahimi-Rad
M and Larijani B: Prevalence of osteoporosis and vitamin D receptor
gene polymorphisms (FokI) in an Iranian general population based
study (Kurdistan) (IMOS). Med J Islam Repub Iran. 29:2382016.
|
|
13
|
Andrews S: FastQC A Quality Control tool
for High Throughput Sequence Data. 2014.
|
|
14
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet J. 17:2011.
View Article : Google Scholar
|
|
15
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ai H, Ai Y and Meng F: GenomeLandscaper:
Landscape analysis of genome-fingerprints maps assessing chromosome
architecture. Sci Rep. 8:10262018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gheinani Hashemi A, Burkhard FC, Rehrauer
H, Fournier Aquino C and Monastyrskaya K: MicroRNA MiR-199a-5p
regulates smooth muscle cell proliferation and morphology by
targeting WNT2 signaling pathway. J Biol Chem. 290:7067–7086. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tabas-Madrid D, Nogales-Cadenas R and
Pascual-Montano A: GeneCodis3: A non-redundant and modular
enrichment analysis tool for functional genomics. Nucleic Acids
Res. 40:(Web Server Issue). W478–W483. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Demchak B, Hull T, Reich M, Liefeld T,
Smoot M, Ideker T and Mesirov JP: Cytoscape: The network
visualization tool for GenomeSpace workflows. F1000Res.
3:1512014.PubMed/NCBI
|
|
20
|
Kim EK and Choi EJ: Pathological roles of
MAPK signaling pathways in human diseases. Biochim Biophys Acta.
1802:396–405. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Che J P, Li W, Yan Y, Liu M, Wang GC, Li
QY, Yang B, Yao XD and Zheng JH: Expression and clinical
significance of the nin one binding protein and p38 MAPK in
prostate carcinoma. Int J Clin Exp Pathol. 6:2300–2311.
2013.PubMed/NCBI
|
|
22
|
Kim HK, Kim MG and Leem KH: Osteogenic
activity of collagen peptide via ERK/MAPK pathway mediated boosting
of collagen synthesis and its therapeutic efficacy in osteoporotic
bone by back-scattered electron imaging and microarchitecture
analysis. Molecules. 18:15474–15489. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hah YS, Kang HG, Cho HY, Shin SH, Kim UK,
Park BW, Lee SI, Rho GJ, Kim JR and Byun JH: JNK signaling plays an
important role in the effects of TNF-α and IL-1β on in vitro
osteoblastic differentiation of cultured human periosteal-derived
cells. Mol Biol Rep. 40:4869–4881. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou J, Fujiwara T, Ye S, Li X and Zhao H:
Downregulation of Notch modulators, tetraspanin 5 and 10, inhibits
osteoclastogenesis in vitro. Calcif Tissue Int. 95:209–217. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim DY, Kim GW and Chung SH: Nectandrin a
enhances the BMP-induced osteoblastic differentiation and
mineralization by activation of p38 MAPK-Smad signaling pathway.
Korean J Physiol Pharmacol. 17:447–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Zhang L, Zhou L, Yu ZP, Qi F, Liu B,
Zi SX, Li L, Li Y, Wang SB, et al: Beneficial effects of
non-matched allogeneic cord blood mononuclear cells upon patients
with idiopathic osteoporosis. J Transl Med. 10:1022012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yong L, Chengjun H, Jinhua SH and Paul J:
possible linkage between osteoporosis and genes associated with
type 1 diabetes mellitus. Med One. 1:e1700012017.
|
|
28
|
Calabrese G, Mesner LD, Foley PL, Rosen CJ
and Farber CR: Network analysis implicates alpha-synuclein (Snca)
in the regulation of ovariectomy-induced bone loss. Sci Rep.
6:294752016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bohaty I, Jurásné-Lukovics M,
Somogyiné-Vári É, Kozma L, Dankó K and Molnár I: HLA class I, Cw*01
and Cw*15 alleles can play a preventing role in serum IL17
elevation associated with postmenopausal osteoporosis in Hungary.
Endocrine. Apr 27–2013.DOI: 10.1530/endoabs.32.P85.
|
|
30
|
Alam I, Carr LG, Liang T, Liu Y, Edenberg
HJ, Econs MJ and Turner CH: Identification of genes influencing
skeletal phenotypes in congenic P/NP rats. J Bone Mineral Res.
25:1314–1325. 2010. View
Article : Google Scholar
|
|
31
|
Jain P, Karthikeyan C, Moorthy NS, Waiker
DK, Jain AK and Trivedi P: Human CDC2-like kinase 1 (CLK1): a novel
target for Alzheimer's disease. Curr Drug Targets. 15:539–550.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chang X, Zheng Y, Yang Q, Wang L, Pan J,
Xia Y, Yan X and Han J: Carbonic anhydrase I (CA1) is involved in
the process of bone formation and is susceptible to ankylosing
spondylitis. Arthrit Res Ther. 14:R1762012. View Article : Google Scholar
|
|
33
|
Li W, Chen WS, Zhou PP, Cao L and Yu LJ:
Influence of initial pH on the precipitation and crystal morphology
of calcium carbonate induced by microbial carbonic anhydrase.
Colloids Surf B Biointerfaces. 102:281–287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chang X, Han J, Zhao Y, Yan X, Sun S and
Cui Y: Increased expression of carbonic anhydrase I in the synovium
of patients with ankylosing spondylitis. BMC Musculoskelet Disord.
11:2792010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tóthová V and Gibadulinová A: S100P, a
peculiar member of S100 family of calcium-binding proteins
implicated in cancer. Acta Virol. 57:238–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li XL, Wu MS, Zhu XW, Deng YC, Ye YY, Zhao
SZ, Ren LZ and Li B: Effects of flavonoids from Cuscuta chinensis
on intestinal calcium-binding protein mRNA expression in
ovariectomized osteoporosis model rats. Chin J Tissue Eng Res.
4271–4276. 2014.
|