Introduction
Endometriosis is a common gynecological disease
characterized by the growth of endometrial tissue outside of the
uterine cavity (1,2). It can be subdivided into three main
subtypes according to the site of the growth, namely, ovarian
endometriosis, pelvic endometriosis and deep infiltrating
endometriosis (3,4). The main clinical features of
endometriosis include pelvic pain, dysmenorrhea and infertility,
and this condition may affect up to 15% of women of reproductive
age (5–7).
Despite extensive investigation, the detailed
molecular etiology of endometriosis is still not fully understood
(8,9). It has been determined that genetic
factors are involved in the development of endometriosis based on
observations of the familial occurrence of this disorder, as the
first-degree relatives of individuals with endometriosis exhibit a
higher risk (10,11). Alternatively, endometriosis has
been proposed to be a precancerous lesion, since certain subtypes
of ovarian and endometrial cancers have been implicated as being
derived from this disease (12–14).
Furthermore, somatic mutations in multiple oncogenes and tumor
suppressor genes, including KRAS, protein phosphatase 2 scaffold
subunit A alpha (PPP2R1A), phosphatidylinositol-4,5-bisphosphate
3-kinase catalytic subunit alpha (PIK3CA) and AT-rich interaction
domain 1A (ARID1A), have been identified in certain subtypes of
endometriosis via large-scale genomic analysis (15) and candidate gene strategy (16).
CTCF is an essential epigenetic regulator that
affects chromatin structure organization to control gene
transcription via facilitating or preventing promoter-enhancer
interactions (17,18). Previous studies have identified
frequent CTCF mutations in diverse human cancer types, including
endometrial cancer (19–21), and CTCF mutations have been shown
to promote the progress of endometrial cancer (19). Although a previous exome-sequencing
study identified CTCF somatic mutations in 16 ovarian endometriosis
samples (22), due to the great
heterogeneity of this disease, the detailed CTCF mutation spectrum
of ovarian endometriosis samples remains largely undetermined
(23,24). Therefore, in the present study, a
cohort of 92 ovarian endometriosis samples was collected in order
to identify the somatic CTCF mutations of the endometriotic
lesions.
Patients and methods
Patients
The present study included 92 Chinese patients with
ovarian endometriosis enrolled at the Jiangxi Provincial Maternal
and Child Health Hospital from June 2013 to July 2014. Following
assessment by two independent pathologists, paired endometriotic
lesions and EDTA-anticoagulated blood samples were simultaneously
collected from each patient. Samples of healthy eutopic endometrial
tissues from 67 control patients without endometriosis, and of
healthy ovarian tissues from 46 patients with ovarian cysts
(without endometriosis) were also collected. This study was
performed in accordance with the Declaration of Helsinki. The
Institutional Review Board of the Jiangxi Provincial Maternal and
Child Health Hospital approved the study. Informed consent was
obtained from all participants prior to the study commencing.
Clinical data
The current age and the age at the time of menarche
of each patient was recorded (Table
I). The serum estrogen, progesterone, cancer antigen 125
(CA125), thyroid stimulating hormone (TSH), free triiodothyronine
(FT3), free thyroxine (FT4), carcino embryonic antigen (CEA),
α-fetoprotein (AFP) and squamous cell carcinoma antigen (SCCA)
levels were determined with a radioimmunoassay method, as
previously described (25).
 | Table I.Association of CTCF mutation with
clinical features in 92 patients with ovarian endometriosis. |
Table I.
Association of CTCF mutation with
clinical features in 92 patients with ovarian endometriosis.
| Feature | Wild type
(n=90) | Mutant type
(n=2) | P-value |
|---|
| Age (years) | 33.46±6.62 | 31.50±7.78 | 0.35 |
| Age of menarche
(years) | 13.23±1.23 | 13.50±2.12 | 0.81 |
| E2 (pg/ml) | 122.65±86.23 | 75.61±53.25 | 0.56 |
| P (ng/ml) | 1.48±3.21 | 0.85±0.49 | 0.86 |
| CA125 (µ/ml) | 112.38±198.23 | 85.03±18.60 | 0.62 |
| TSH (mIU/ml) | 2.34±1.12 | 1.82±0.63 | 0.20 |
| FT3 (pg/ml) | 2.95±0.26 | 3.07±0.08 | 0.65 |
| FT4 (ng/dl) | 1.27±0.08 | 1.29±0.07 | 0.49 |
| CEA (ng/ml) | 1.13±0.38 | 0.92±0.16 | 0.40 |
| AFP (ng/ml) | 2.89±1.82 | 2.96±0.89 | 0.18 |
| SCCA (ng/ml) | 1.43±1.08 | 1.28±1.06 | 0.46 |
Mutational analysis of the CTCF
gene
Total DNA was extracted from the endometriotic
lesions and paired blood samples using the TIANamp Blood DNA kit
(Tiangen Biotech Co., Ltd., Beijing, China). The DNA was quantified
using a SmartSpec Plus spectrophotometer (Bio-Rad Laboratories,
Inc., Hercules, CA, USA). The entire coding regions of the CTCF
gene were PCR amplified with a series of primer pairs (Table II). Briefly, ~50 ng total DNA was
used for each PCR amplicon in a final volume of 30 µl with rTaq DNA
polymerase (Takara Biotechnology, Dalian, China). The PCR
conditions were: One denaturation cycle (94°C for 3 min), 35 PCR
cycles of denaturation, annealing and product extension (94°C for
30 sec; 50–60°C as specified in Table
II for 30 sec; 72°C for 30 sec) and a final extension step
(72°C for 10 min). The PCR products were visualized on a 1.5%
agarose gel stained with ethidium bromide and purified with a
TIANgel Midi DNA Purification kit (Tiangen Biotechnology, Beijing,
China). The purified PCR products were sequenced with an ABI Prism
3730 DNA sequencer (Applied Biosystems; Thermo Fisher Scientific,
Inc., Waltham, MA, USA). The potential CTCF somatic mutations were
verified by comparison with the paired blood samples. The
identified mutations were searched in the HGMD (www.hgmd.cf.ac.uk/ac/index.php), dbSNP
(www.ncbi.nlm.nih.gov/snp), ExAC
(exac.broadinstitute.org) or 1000G
(www.ncbi.nlm.nih.gov/variation/tools/1000genomes)
databases.
 | Table II.PCR primer sequences for the
amplification of CTCF. |
Table II.
PCR primer sequences for the
amplification of CTCF.
| Exon | Forward primer
(5′→3′) | Reverse primer
(5′→3′) | Annealing
temperature (°C) | Amplicon size
(bp) |
|---|
| 3-1 |
TGCTTTAAATAACAATCT |
TCTGAAGAAGGGTGGGGTC | 56 | 263 |
| 3-2 |
AACAGCAGTGTACAGATG |
CTTCTACTGAAGTGGTAGC | 52 | 235 |
| 3-3 |
CAGCTTGTTCAAGTACCT |
TGCCAACTAGGATCTTCC | 52 | 236 |
| 3–4 |
GAGGTGGAGACACTAGAAC |
CACTATGGATAAACTCGT | 58 | 285 |
| 4 |
ACTCTGCAGCAAGTAAGT |
ACATTCTTATCCAGCAC | 50 | 224 |
| 5 |
TTCCTGTTACTCCATCCT |
CTGCCTAAGAGAGATACCA | 55 | 199 |
| 6 |
CTCTTGTTACAGTCTGTG |
GAGTGGAGAAGTCCTAC | 57 | 194 |
| 7 |
AATTACAGTATTTATTCA |
CACTAGTTAATCTACTTA | 60 | 223 |
| 8 |
GGCTTTTTACTGTGCTT |
ACACCAGACACCGAGAA | 55 | 230 |
| 9 |
CCCTATGCCGTTTCAGGA |
AGGCAAAGTGAAGTTCTG | 50 | 225 |
| 10 |
AGTGGTGTGAAAGAGGAT |
TCAAGGAACAAGTCACT | 56 | 191 |
| 11 |
TGCTTCCTGATTTCATGA |
GAGATGAACAACTTACGC | 58 | 200 |
| 12 |
CTGTGCTCTTCTTTGCCAG |
GCACAAGGCTCCGCCATC | 54 | 220 |
Evolutionary conservation
analysis
Evolutionary conservation analysis was used to
evaluate the potential pathogenicity of the identified CTCF
mutations. A total of 20 different vertebrate species sequences
were retrieved from the GenBank database (www.ncbi.nlm.nih.gov/genbank), including Homo
sapiens (NP_006556), Pan troglodytes (XP_009429318),
Mus musculus (NP_851839), Rattus norvegicus
(NP_114012), Heterocephalus glaber (XP_021112509),
Mesocricetus auratus (XP_012973364), Cricetulus
griseus (XP_003508601), Bos taurus (NP_001069216),
Bison bison (XP_010849335), Tursiops truncatus
(XP_004317606), Canis lupus familiaris (XP_005620876),
Sus scrofa (NP_001231589), Ovis aries (XP_012045353),
Equus caballus (XP_001497859), Pteropus alecto
(XP_015443188), Hipposideros armiger (XP_019511268),
Pygoscelis adeliae (XP_009328626), Gallus gallus
(NP_990663), Columba livia (XP_021157070) and Danio
rerio (NP_001001844). Sequence alignment was performed with
MEGA software (version 4.0) (26).
In silico analysis of the CTCF
mutations
Two online bioinformatics programs were used to
predict the pathogenic potential of the identified CTCF mutations,
including MutationTaster (www.mutationtaster.org) (27) and PolyPhen-2 (genetics.bwh.harvard.edu/pph2) (28). These programs predict whether each
mutation is likely to be to be benign or pathogenic, according to
the probability score.
Statistical analysis
SPSS 17.0 statistical package (SPSS, Inc., Chicago,
IL, USA) was used to calculate statistical significance. The
Fisher's exact test was used to assess the potential association
between nominal variables and CTCF mutation, whereas the
Mann-Whitney method was used to analyze the potential association
between continuous variables and CTCF mutations. P<0.05 was
considered to indicate a statistically significant difference and
all P-values were two-tailed.
Results
CTCF mutation in ovarian
endometriosis
The study consisted of 92 patients with ovarian
endometriosis; the age range was 21–50 years, the age of menarche
range was 10–18 years, and the serum estrogen, progesterone, CA125,
TSH, FT3, FT4, CEA, AFP and SCCA levels are summarized in Table I. In total, 2 different somatic
heterozygote CTCF mutations were identified in the endometriotic
lesions in 2/92 (2.2%) ovarian endometriosis tissue samples,
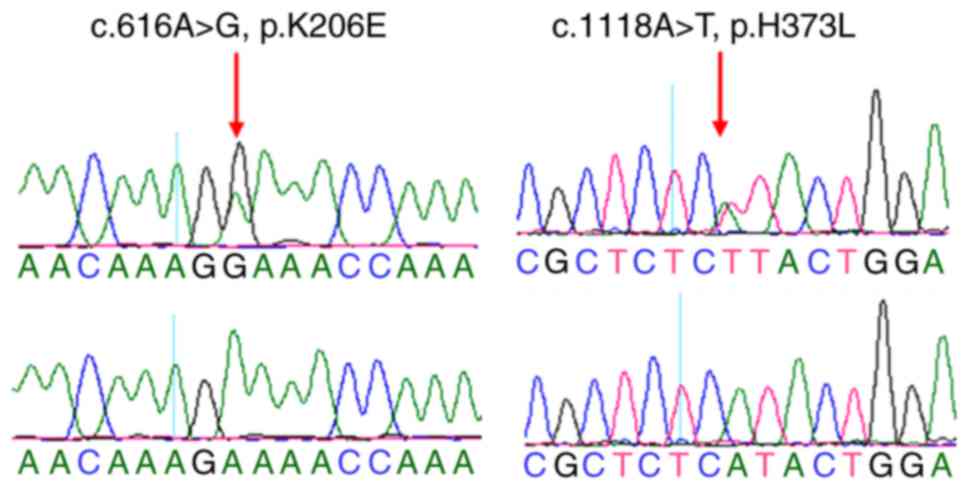
including a p.K206E (c.616A>G) and a p.H373L (c.1118A>T)
mutation. These mutations were not identified in the paired blood
samples, indicating that they were somatic (Fig. 1). The p.K206E-mutated sample was
from a 26-year-old woman who also experienced primary infertility,
while the p.H373L-mutated individual was a 37-year-old who was also
diagnosed with uterine leiomyoma. The p.H373L mutation was
identified in 1/699 endometrial cancer samples in a previous study
(29), while the p.K206E mutation
was not previously reported either in the HGMD or dbSNP databases.
In addition, somatic CTCF mutations were not detected in the
remaining 90 ovarian endometriosis samples or in the 67 healthy
control eutopic endometrial tissues and 46 healthy ovarian tissue
samples from patients with ovarian cysts.
Association of CTCF mutations with
clinical features
The potential associations between CTCF mutations
and clinical features were analyzed. This analysis did not identify
any associations between CTCF mutations and the clinical features
considered (Table I).
The causative potential of the CTCF
mutations
The potential pathogenicity of the identified CTCF
mutations was evaluated by MutationTaster and PolyPhen-2 programs.
The p.K206E and p.H373L mutations scored 56 and 99 on
MutationTaster, respectively, and were predicted to be ‘disease
causing’ mutations. The Poly-Phen2 program predicted that the
p.K206E mutation was ‘possibly damaging’ and the p.H373L was
‘probably damaging’, with a score of 0.956 for p.K206E
(sensitivity, 0.79; specificity, 0.95) and 0.997 for p.H373L
(sensitivity, 0.41; specificity, 0.98), with mutations considered
‘probably damaging’ when the prediction score value was >0.95
(28). The two identified
mutations were not reported in either the ExAC or 1000G
databases.
Evolutionary conservation analysis of
the CTCF mutations
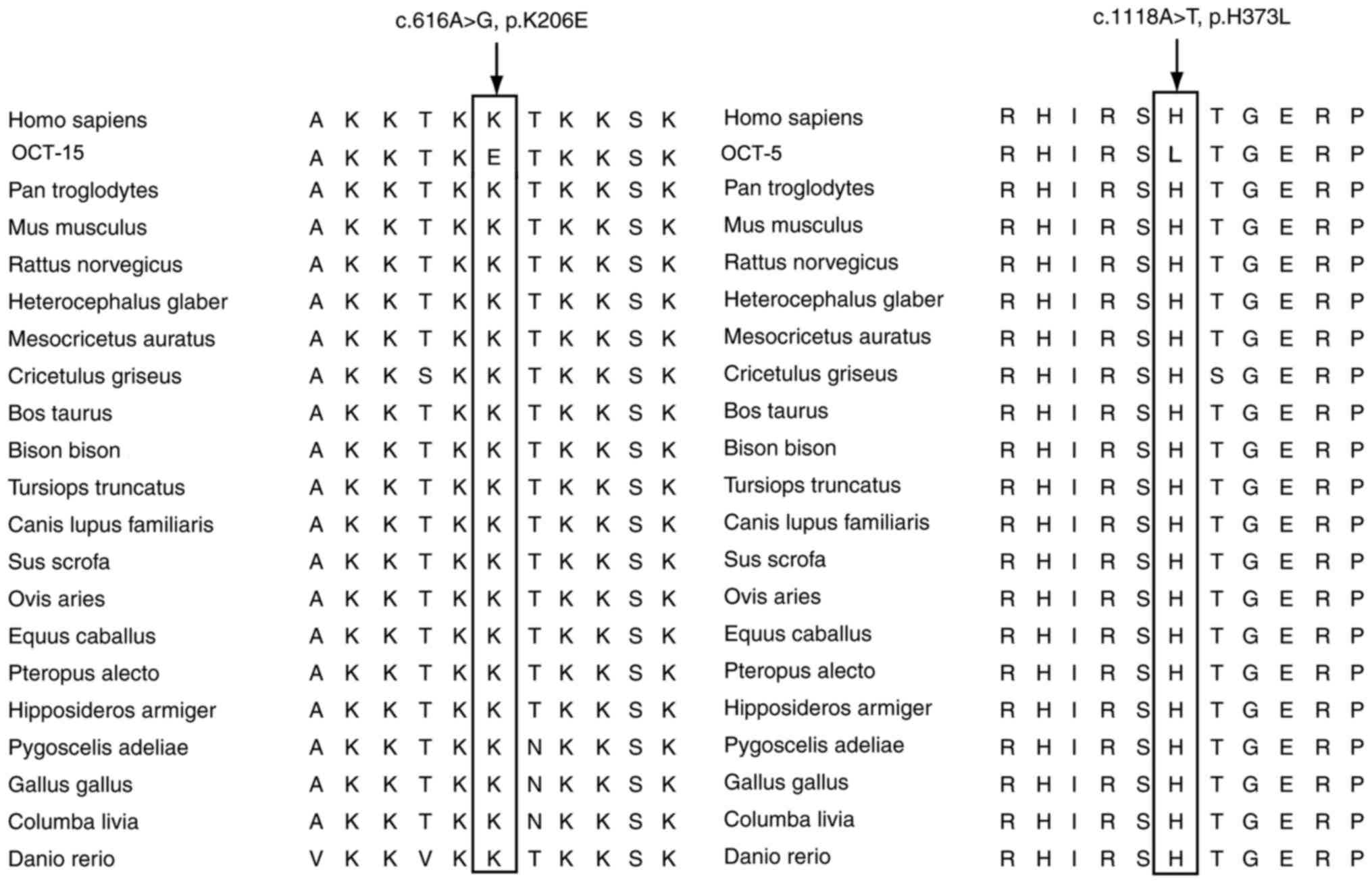
The conservation trends of the CTCF amino acids were
investigated based on sequence alignment with the human CTCF
protein sequence. The alignment results showed that the two
potentially pathogenic mutations (p.K206E and p.H373L) were at
highly conserved sites in the 20 vertebrate species, from Homo
sapiens to Danio rerio (Fig. 2).
Discussion
As a critical chromatin-organizing factor, CTCF
serves an important role in chromatin insulation, enhancer blocking
and transcriptional regulation (17,18,30,31).
CTCF mutations are frequently identified in endometrial cancer
(20,21,29),
while they are less common in several other cancer types, including
liver (32) and breast (33) cancer. As endometriosis, a
potentially premalignant condition, shares some genetic features
with CTCF-mutated endometrial cancer, including frequent KRAS and
PIK3CA mutations (20,34,35),
the present study hypothesized that there may also be CTCF
mutations in endometriotic lesions.
In the present study, 2 heterozygous somatic
mutations in CTCF, p.K206E (c.616A>G) and p.H373L
(c.1118A>T), were identified in 2/92 ovarian endometriosis
samples. The association of CTCF mutations with endometriosis
remains largely uncharacterized, with the exception of two previous
large-scale genomic analyses profiling global somatic mutations in
endometriosis, including a study of 16 Chinese patients with
ovarian endometriosis (22), and
of 24 Euro-American and Japanese patients with deeply infiltrating
endometriosis (15). The mutation
frequency of CTCF in the samples of the present study was notably
different from these two previous studies; the previous exome-based
sequencing effort identified a CTCF mutation frequency of 25%
(4/16; P=0.004) in Chinese patients with ovarian endometriosis
(22). It can be hypothesized that
the high sensitivity for the detection of mutations using the exome
sequencing technique may be the main reason for the differential
mutation frequency between the present study and the previous study
(36). The identified CTCF
mutations in the previous study (22) were not subject to Sanger sequencing
verification, which has a detection threshold of 6.6–20% mutant
alleles (37–39). The other genomic analysis of 24
patients with deeply infiltrating endometriosis failed to identify
any CTCF mutations (15); one
possible explanation is that CTCF mutations may be specific to
ovarian endometriosis. However, this hypothesis should be treated
with caution due to the relatively small sample size of the
previous study (15).
Additionally, CTCF mutations were not identified in the 67 healthy
eutopic endometrial tissues samples and 46 health ovarian tissue
samples from patients with ovarian cysts, implying that CTCF
mutations may participate in the development of ovarian
endometriosis.
The present study failed to identify any association
between CTCF mutations and the clinical characteristics of the
sample cohort; however, this conclusion should also be treated with
caution, as the sample size of patients with CTCF mutation was too
small (n=2). To overcome this potential statistical bias, the
association should be further analyzed with a larger sample size in
a future study.
The in silico prediction results indicated
that the two CTCF mutations were ‘probably damaging’, while the
evolutionary conservation analysis results showed the two mutated
amino acids were evolutionarily highly conserved across 20
vertebrate species, from Homo sapiens to Danio rerio.
Furthermore, the p.H373L mutation identified in the present study
was previously detected in 1 of 699 patients with endometrial
cancer (29). These results imply
that the CTCF mutations identified in the patients with ovarian
endometriosis may be pathogenic. Combined with the high frequency
of CTCF mutations in ovarian endometriosis in the previous study
(22), it can be hypothesized that
CTCF mutations may contribute to the pathogenesis of ovarian
endometriosis.
In conclusion, the present study identified two CTCF
somatic mutations in the endometriotic lesions of Chinese patients
with ovarian endometriosis. In silico prediction and
evolutionary conservation analysis implied that these mutations may
be pathogenic. The findings of the present study, together with
previous studies, suggest that CTCF mutations may contribute to the
development of ovarian endometriosis.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Natural Science Foundation of Jiangxi Province (grant no.
20151BAB205012).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JG performed the experiment. BC and XX performed
data analysis. FW collected the sample and clinical data. BZ
designed the study and prepared the manuscript.
Ethics approval and consent to
participate
The Institutional Review Board of the Jiangxi
Provincial Maternal and Child Health Hospital approved the study.
Informed consent was obtained from all participants prior to the
study commencing.
Consent for publication
Informed consent was obtained from all participants
prior to the study commencing.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bedaiwy MA, Allaire C, Yong P and Alfaraj
S: Medical management of endometriosis in patients with chronic
pelvic pain. Semin Reprod Med. 35:38–53. 2017.PubMed/NCBI
|
|
2
|
Cakmak H, Seval-Celik Y, Arlier S,
Guzeloglu-Kayisli O, Schatz F, Arici A and Kayisli UA: p38
Mitogen-activated protein kinase is involved in the pathogenesis of
endometriosis by modulating inflammation, but not cell survival.
Reprod Sci. 25:587–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Exacoustos C, Zupi E and Piccione E:
Ultrasound Imaging for ovarian and deep infiltrating endometriosis.
Semin Reprod Med. 35:5–24. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gordts S, Koninckx P and Brosens I:
Pathogenesis of deep endometriosis. Fertil Steril. 108(872–885):
e12017.PubMed/NCBI
|
|
5
|
Tanbo T and Fedorcsak P:
Endometriosis-associated infertility: Aspects of pathophysiological
mechanisms and treatment options. Acta Obstet Gynecol Scand.
96:659–667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chui MH, Wang TL and Shih IM:
Endometriosis: Benign, malignant, or something in between?
Oncotarget. 8:78263–78264. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schwartz Kohl AS, Wölfler MM, Mitter V,
Rauchfuss M, Haeberlin F, Eberhard M, von Orelli S, Imthurn B,
Imesch P, Fink D and Leeners B: Endometriosis, especially mild
disease: A risk factor for miscarriages. Fertil Steril.
108(806–814): e22017.
|
|
8
|
Uimari O, Rahmioglu N, Nyholt DR, Vincent
K, Missmer SA, Becker C, Morris AP, Montgomery GW and Zondervan KT:
Genome-wide genetic analyses highlight mitogen-activated protein
kinase (MAPK) signaling in the pathogenesis of endometriosis. Hum
Reprod. 32:780–793. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Burghaus S, Fasching PA, Häberle L, Rübner
M, Büchner K, Blum S, Engel A, Ekici AB, Hartmann A, Hein A, et al:
Genetic risk factors for ovarian cancer and their role for
endometriosis risk. Gynecol Oncol. 145:142–147. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matalliotakis IM, Arici A, Cakmak H,
Goumenou AG, Koumantakis G and Mahutte NG: Familial aggregation of
endometriosis in the Yale series. Arch Gynecol Obstet. 278:507–511.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McLeod BS and Retzloff MG: Epidemiology of
endometriosis: An assessment of risk factors. Clin Obstet Gynecol.
53:389–396. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wiegand KC, Shah SP, Al-Agha OM, Zhao Y,
Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, et
al: ARID1A mutations in endometriosis-associated ovarian
carcinomas. N Engl J Med. 363:1532–1543. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Mang M, Wang Y, Wang L, Klein R,
Kong B and Zheng W: Tubal origin of ovarian endometriosis and clear
cell and endometrioid carcinoma. Am J Cancer Res. 5:869–879.
2015.PubMed/NCBI
|
|
14
|
Cochrane DR, Tessier-Cloutier B, Lawrence
KM, Nazeran T, Karnezis AN, Salamanca C, Cheng AS, McAlpine JN,
Hoang LN, Gilks CB and Huntsman DG: Clear cell and endometrioid
carcinomas: Are their differences attributable to distinct cells of
origin? J Pathol. 243:26–36. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Anglesio MS, Papadopoulos N, Ayhan A,
Nazeran TM, Noë M, Horlings HM, Lum A, Jones S, Senz J, Seckin T,
et al: Cancer-associated mutations in endometriosis without cancer.
N Engl J Med. 376:1835–1848. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vestergaard AL, Thorup K, Knudsen UB, Munk
T, Rosbach H, Poulsen JB, Guldberg P and Martensen PM: Oncogenic
events associated with endometrial and ovarian cancers are rare in
endometriosis. Mol Hum Reprod. 17:758–761. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansen AS, Pustova I, Cattoglio C, Tjian R
and Darzacq X: CTCF and cohesin regulate chromatin loop stability
with distinct dynamics. Elife. 6:pii: e25776. 2017. View Article : Google Scholar
|
|
18
|
Pérez-García A, Marina-Zárate E,
Álvarez-Prado ÁF, Ligos JM, Galjart N and Ramiro AR: CTCF
orchestrates the germinal centre transcriptional program and
prevents premature plasma cell differentiation. Nat Commun.
8:160672017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marshall AD, Bailey CG, Champ K, Vellozzi
M, O'Young P, Metierre C, Feng Y, Thoeng A, Richards AM, Schmitz U,
et al: CTCF genetic alterations in endometrial carcinoma are
pro-tumorigenic. Oncogene. 36:4100–4110. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cancer Genome Atlas Research Network, .
Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H,
Robertson AG, Pashtan I, Shen R, et al: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Walker CJ, Miranda MA, O'Hern MJ, McElroy
JP, Coombes KR, Bundschuh R, Cohn DE, Mutch DG and Goodfellow PJ:
Patterns of CTCF and ZFHX3 mutation and associated outcomes in
endometrial cancer. J Natl Cancer Inst. 107:pii: djv249. 2015.
View Article : Google Scholar
|
|
22
|
Li X, Zhang Y, Zhao L, Wang L, Wu Z, Mei
Q, Nie J, Li X, Li Y, Fu X, et al: Whole-exome sequencing of
endometriosis identifies frequent alterations in genes involved in
cell adhesion and chromatin-remodeling complexes. Hum Mol Genet.
23:6008–6021. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zondervan KT, Rahmioglu N, Morris AP,
Nyholt DR, Montgomery GW, Becker CM and Missmer SA: Beyond
endometriosis Genome-wide association study: From genomics to
phenomics to the patient. Semin Reprod Med. 34:242–254. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Montgomery GW and Giudice LC: New lessons
about Endometriosis-somatic mutations and disease heterogeneity. N
Engl J Med. 376:1881–1882. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu J, Zou Y, Luo Y, Guo JB, Liu FY, Zhou
JY, Zhang ZY, Wan L and Huang OP: Prevalence and clinical
significance of mediator complex subunit 12 mutations in 362 Han
Chinese samples with uterine leiomyoma. Oncol Lett. 14:47–54. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tamura K, Dudley J, Nei M and Kumar S:
MEGA4: Molecular evolutionary genetics analysis (MEGA) software
version 4.0. Mol Biol Evol. 24:1596–1599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schwarz JM, Cooper DN, Schuelke M and
Seelow D: MutationTaster2: Mutation prediction for the
deep-sequencing age. Nat Methods. 11:361–362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zighelboim I, Mutch DG, Knapp A, Ding L,
Xie M, Cohn DE and Goodfellow PJ: High frequency strand slippage
mutations in CTCF in MSI-positive endometrial cancers. Hum Mutat.
35:63–65. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ohlsson R, Lobanenkov V and Klenova E:
Does CTCF mediate between nuclear organization and gene expression?
Bioessays. 32:37–50. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bushey AM, Dorman ER and Corces VG:
Chromatin insulators: Regulatory mechanisms and epigenetic
inheritance. Mol Cell. 32:1–9. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fujimoto A, Furuta M, Totoki Y, Tsunoda T,
Kato M, Shiraishi Y, Tanaka H, Taniguchi H, Kawakami Y, Ueno M, et
al: Whole-genome mutational landscape and characterization of
noncoding and structural mutations in liver cancer. Nat Genet.
48:500–509. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aulmann S, Bläker H, Penzel R, Rieker RJ,
Otto HF and Sinn HP: CTCF gene mutations in invasive ductal breast
cancer. Breast Cancer Res Treat. 80:347–352. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kuhn E, Wu RC, Guan B, Wu G, Zhang J, Wang
Y, Song L, Yuan X, Wei L, Roden RB, et al: Identification of
molecular pathway aberrations in uterine serous carcinoma by
genome-wide analyses. J Natl Cancer Inst. 104:1503–1513. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Le Gallo M, O'Hara AJ, Rudd ML, Urick ME,
Hansen NF, O'Neil NJ, Price JC, Zhang S, England BM, Godwin AK, et
al: Exome sequencing of serous endometrial tumors identifies
recurrent somatic mutations in chromatin-remodeling and ubiquitin
ligase complex genes. Nat Genet. 44:1310–1315. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xuan J, Yu Y, Qing T, Guo L and Shi L:
Next-generation sequencing in the clinic: Promises and challenges.
Cancer Lett. 340:284–295. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yao YG, Ogasawara Y, Kajigaya S, Molldrem
JJ, Falcão RP, Pintão MC, McCoy JP Jr, Rizzatti EG and Young NS:
Mitochondrial DNA sequence variation in single cells from leukemia
patients. Blood. 109:756–762. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tsiatis AC, Norris-Kirby A, Rich RG, Hafez
MJ, Gocke CD, Eshleman JR and Murphy KM: Comparison of Sanger
sequencing, pyrosequencing, and melting curve analysis for the
detection of KRAS mutations: Diagnostic and clinical implications.
J Mol Diagn. 12:425–432. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Monzon FA, Ogino S, Hammond ME, Halling
KC, Bloom KJ and Nikiforova MN: The role of KRAS mutation testing
in the management of patients with metastatic colorectal cancer.
Arch Pathol Lab Med. 133:1600–1606. 2009.PubMed/NCBI
|