Introduction
Wilms' tumour (WT), also known as renal embryonal
tumour, is an embryonic cancer of the kidney that comprises
blastemal, stromal and epithelial histological components (1). WT is the fourth most common renal
tumour among paediatric cancers (2), and accounts for about 8% of all solid
neoplasms and 95% of all renal carcinomas in children under 5 years
old (3). Currently, nephrectomy,
followed by chemotherapy and radiotherapy, are the major
therapeutic methods for patients with WT (4). Significant improvements in diagnosis
and treatment have improved the prognosis of patients with WT with
localised disease. However, numerous patients suffer from
metastasis or recurrence of the tumour; therefore, poor therapeutic
outcomes are inevitable (5).
Genetic and epigenetic changes, including the activation of
oncogenes and/or the inhibition of tumour suppressors, are involved
in tumorigenesis and tumour development of WT (6–8).
Therefore, full investigation on the pathogenesis mechanisms of WT,
particularly on the regulatory roles in the rapid growth and
metastasis of WT cells, may be critical for development of novel
therapeutic targets for treating patients with WT.
MicroRNAs (miRNAs) are clusters of noncoding,
evolutionarily conserved and short RNA molecules implicated in gene
expression (9). miRNAs can
regulate gene expression by base pairing with complementary
nucleotides in the 3′-untranslated regions of their target genes,
thereby inducing mRNA degradation or translation suppression
(10). Computational estimations
indicate that over 1000 miRNAs existing in the human genome can
regulate approximately 30% of the human protein-encoding genes
(11). Furthermore, dysregulation
of miRNAs is related with a substantial number of human disorders,
including malignancies (12).
Emerging evidence has highlighted that aberrantly expressed miRNAs
may have significant effects on the occurrence and development of
malignant tumours through acting either as oncogenes or tumour
suppressors (13,14). Upregulation of oncogenic miRNAs
and/or suppression of tumour suppressive miRNAs can regulate
various cancer-related biological features, such as cell
proliferation, cycle, apoptosis, migration, invasion and metastasis
(15). Hence, miRNAs may be
utilised as effective biomarkers for cancer diagnosis, treatments
and prognosis.
miR-199b is dysregulated in various types of human
cancer, such as colorectal cancer (16), acute myeloid leukaemia (17), non-small cell lung cancer (18) and endometrioid endometrial
carcinoma (19). However, miR-199b
expression patterns, possible functions and associated mechanisms
in WT remain predominantly unknown. Hence, miR-199b expression
level in WT was detected in this study, and the regulatory role and
underlying mechanisms of miR-199b on the biological features in WT
were also investigated.
Materials and methods
Human tissue samples
This study was approved by the Ethics Committee of
Yidu Central Hospital of Weifang. Written informed consent was also
obtained from all patients. A total of 24 paired WT and adjacent
normal tissues were collected from the patients diagnosed with WT
and underwent nephrectomy at the Yidu Central Hospital of Weifang
between January 2014 and November 2016. None of these patients with
WT had been treated with chemotherapy or radiotherapy prior to
nephrectomy. All the tissue specimens were immediately frozen and
kept in liquid nitrogen until further RNA isolation.
Cell culture and oligonucleotide
transfection
17.94 and WiT49 cell lines were purchased from
Shanghai ran Tai Biological Technology Co., Ltd. (Shanghai, China),
and was cultured in Dulbecco's modified Eagle's medium (DMEM)
containing 15% heat-inactivated fetal bovine serum (FBS) and 1%
penicillin/streptomycin (all from Gibco, Carlsbad, CA, USA). WiT49
cell line was maintained in DMEM supplemented with 10% FBS and 1%
penicillin/streptomycin. 17.94 and WiT49 All cell lines were grown
in a humidified incubator with 5% CO2 and 95% air at
37°C.
The miR-199b inhibitor and negative control (NC)
miRNA inhibitor were acquired from GenePharma Co., Ltd (Shanghai,
China). The specific small interfering RNA (siRNA) targeting the
expression of Runt-related transcription factor 3 (RUNX3) and
corresponding NC siRNA were provided by RiboBio Co., Ltd.
(Guangzhou, China). Cells were plated in 6-well plates (1.5×105
cells/well) and transfected with miRNA inhibitor or siRNA using
Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), in accordance with the
manufacturer's protocols. Afterwards, the transfected cells were
maintained at 37°C in a cell incubator containing 5%
CO2, and the medium was changed to fresh Maccyo'5
supplemented with 10% FBS at 6 h post-transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA of tissue samples or cells was isolated
with TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions. A
NanoDrop 2000/2000c (Thermo Fisher Scientific, Inc., Wilmington,
DE, USA) was used to detect the concentration of total RNA. For
determination of miR-199b expression level, complementary DNA
(cDNA) was synthesised from total RNA using a TaqMan MicroRNA
Reverse Transcription kit. Subsequently, the qPCR was conducted
with a TaqMan MicroRNA PCR kit (all from Applied Biosystems,
Carlsbad, CA, USA) and performed on Roche Lightcycler 480 Real-time
PCR system (Roche Diagnostics, Basel, Switzerland). The cycling
conditions for qPCR were as follows: 50°C for 2 min, 95°C for 10
min; 40 cycles of denaturation at 95°C for 15 sec; and
annealing/extension at 60°C for 60 sec. To analyse RUNX3 mRNA
level, total RNA underwent reverse transcription into cDNA using a
PrimeScript RT Reagent kit followed by qPCR with a SYBR Premix Ex
Taq™ kit (all from Takara Bio, Dalian, China). The
cycling conditions for qPCR were as follows: 5 min at 95°C,
followed by 40 cycles of 95°C for 30 sec and 65°C for 45 sec. U6
snRNA and GAPDH were employed as internal control for miR-199b and
RUNX3 mRNA, respectively. The primers were designed as follows:
miR-199b forward, 5′-GCCCGCCCAGTGTTTAGACTAT-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′
and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; RUNX3 forward,
5′-GACAGCCCCAACTTCCTCT-3′ and reverse, 5′-CACAGTCACCACCGTACCAT-3′;
and GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. Relative gene expression was
calculated with the 2−ΔΔCt method (20).
Cell counting kit-8 (CCK-8) assay
Transfected cells were collected 24 h
post-transfection and plated onto 96 well plates with a density of
3,000 cells for each well. Following incubation at 37°C for 0, 24,
48 or 72 h, CCK-8 assay was carried out according to the
manufacturer's instructions. After addition of 10 µl CCK-8 reagent
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan) into each
well, the wells were incubated at 37°C with 5% CO2 and
95% air for another 2 h. Afterwards, absorbance was determined at a
wavelength of 450 nm with an automatic multiwell spectrophotometer
(Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell invasion assay
Cell invasion ability was detected using 8 µm pore
Boyden chambers (Corning Inc., Corning, NY, USA) coated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). The transfected
cells were collected, suspended in Maccyo'5 medium without FBS and
counted under a light microscope (IX71; Olympus Co., Tokyo, Japan).
A total of 5×104 cells were seeded into the upper
chambers, whereas the Maccyo'5 medium containing 20% FBS was added
into the lower chambers. Subsequently, the cells were incubated at
37°C with 5% CO2 for 24 h. The noninvaded cells were
removed with a cotton swab, whereas the invaded cells were fixed in
methanol at room temperature for 15 min and stained with 0.5%
crystal violet at room temperature for 15 min. Finally, the stained
cells were photographed and counted under an inverted light
microscope (magnification, ×200) with 5 randomly selected fields
per chamber.
Bioinformatics prediction and
luciferase reporter assay
TargetScan (www.targetscan.org) and microRNA.org
(www.microrna.org/microrna/home.do) were utilised to
predict the putative targets of miR-199b. Luciferase reporter
plasmids, psiCHECK2-RUNX3-3′-UTR wild-type and
psiCHECK2-RUNX3-3′-UTR mutant were synthesised and confirmed by
GenePharma Co., Ltd. The 17.94 and WiT49 cells were transfected
with miR-199b or NC inhibitor in combination with
psiCHECK2-RUNX3-3′-UTR wild-type or psiCHECK2-RUNX3-3′-UTR mutant
using Lipofectamine™ 2000, according to the
manufacturer's instructions. At 48 h post-transfection, luciferase
activity in the cotransfected cells was measured using
Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA),
and renilla luciferase activity was normalised to the firefly
luciferase activity.
Protein extraction and western blot
analysis
Total protein was distilled from tissues or cells
using the RIPA lysis buffer (Beyotime Institute of Biotechnology,
Haimen, China). Afterwards, it was subjected to concentration
determination using a BCA Protein Assay kit (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Equal amounts of the total
protein were resolved using 10% SDS-PAGE and were transferred onto
the polyvinylidene fluoride membranes (EMD Millipore, Billerica,
MA, USA). After blocking with 5% fat-free milk at room temperature
for 1 h, the membranes were incubated with primary antibodies at at
4°C overnight, followed by further incubation with goat anti-rabbit
horseradish peroxidase-conjugated secondary antibody (ab205718;
1:5,000 dilution; Abcam, Cambridge, MA, USA) at room temperature
for 1 h. After washing with Tris-buffered saline with Tween, the
protein signals were visualised using an enhanced chemiluminescence
kit (EMD Millipore). The primary antibodies used in this study
included rabbit anti-human RUNX3 monoclonal antibody (ab49117;
1:1,000 dilution) and rabbit anti-human GAPDH monoclonal antibody
(ab181602; 1:1,000 dilution; both Abcam). GAPDH was used as an
internal control.
Statistical analysis
All data are expressed as the mean ± standard
deviation. SPSS (version 11.0; SPSS, Inc., Chicago, IL, USA) was
used to carry out all the statistical analyses. Differences between
groups were analysed with Student's t-test or one-way ANOVA,
followed by Student-Newman-Keuls test. The association between
miR-199b and RUNX3 mRNA in the WT tissues was evaluated using
Spearman's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-199b is upregulated in the WT
tissues
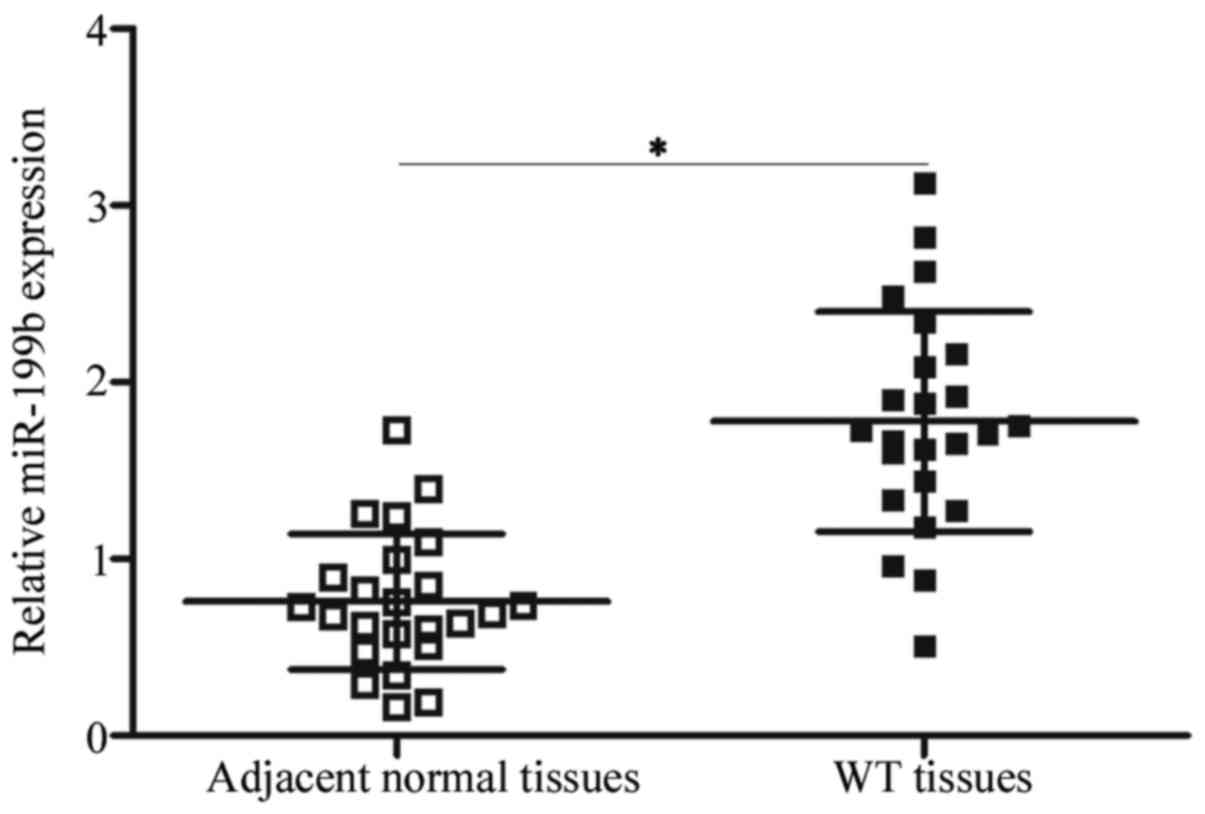
The expression pattern of miR-199b in WT was
determined by detecting its expression in 24 paired WT and adjacent
normal tissues. Using RT-qPCR, miR-199b was significantly
overexpressed in the WT tissues in comparison with that in the
adjacent normal tissues (Fig. 1)
(P<0.05). This result suggests that the dysregulation of
miR-199b may be associated with WT progression.
Downregulation of miR-199b inhibits
cell proliferation and invasion in WT
To characterise the biological functions of miR-199b
expression in WT, the 17.94 and WiT4917.94 and WiT49 cells were
transfected with miR-199b inhibitor to knock down the endogenous
miR-199b expression. Subsequent RT-qPCR analysis revealed that
miR-199b was downregulated in the 17.94 and WiT49 cells that were
transfected with miR-199b inhibitor in comparison with those cells
transfected with NC inhibitor (Fig. 2A
and B) (P<0.05). Moreover, CCK-8 assays were performed to
explore the effect of miR-199b on WT cell proliferation. As shown
in Fig. 2C and D, inhibition of
miR-199b significantly reduced the 17.94 and WiT49 cell
proliferation (P<0.05). Furthermore, the invasion capacity of
the 17.94 and WiT49 cells following transfection with miR-199b or
NC inhibitor was determined using Transwell invasion assay. The
underexpression of miR-199b remarkably weakened the invasion
ability of the 17.94 and WiT49 cells (Fig. 2E) (P<0.05). Therefore, miR-199b
may act as an oncogene in WT progression.
RUNX3 is a direct target of miR-199b
in WT
To understand the molecular mechanism underlying the
oncogenic roles of miR-199b in WT, bioinformatics analysis was
performed to predict the potential targets of miR-199b. RUNX3
(Fig. 3A), which is a well-known
tumour suppressor, was predicted as a major target of miR-199b;
thus, we selected it for further confirmation. To confirm this
hypothesis, we transfected the 17.94 and WiT49 cells with miR-199b
or NC inhibitor in combination with psiCHECK2-RUNX3-3′-UTR
wild-type or psiCHECK2-RUNX3-3′-UTR mutant. The luciferase reporter
assays showed that miR-199b downregulation significantly increased
the luciferase activity of the psiCHECK2-RUNX3-3′-UTR wild-type in
the 17.94 and WiT49 cells (P<0.05), whereas the luciferase
activity was not significantly affected in the
psiCHECK2-RUNX3-3′-UTR mutant (Fig. 3B
and C). In addition, the RUNX3 expression was significantly
increased at both mRNA (Fig. 3D)
(P<0.05) and protein (Fig. 3E)
(P<0.05) levels when miR-199b was knocked down in the 17.94 and
WiT49 cells. Thus, RUNX3 is a direct target gene of miR-199b in
WT.
RUNX3 is underexpressed in WT and is
inversely correlated with the miR-199b level
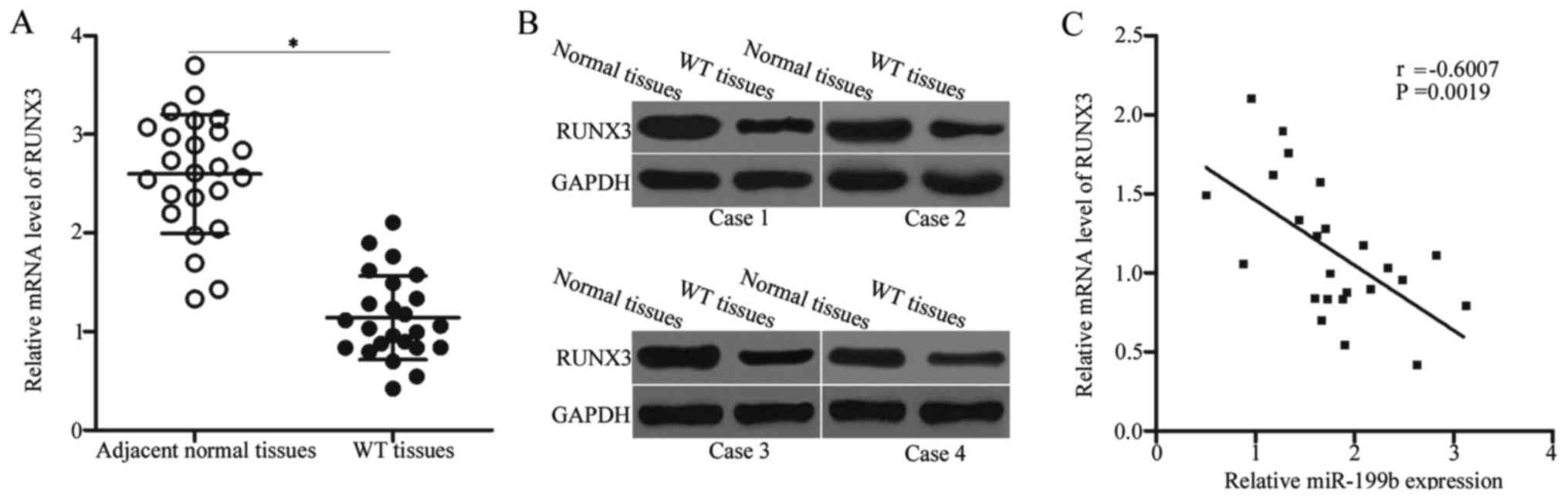
The RUNX3 mRNA level in WT and adjacent normal
tissues were determined to further illustrate the association
between miR-199b and RUNX3 in WT. The expression level of RUNX3
mRNA was lowly expressed in the WT tissues compared with that in
the adjacent normal tissues (Fig.
4A, P<0.05). Western blot analysis also revealed that the
RUNX3 protein was downregulated in the WT tissues (Fig. 4B). Furthermore, Spearman's
correlation analysis identified a negative association between
miR-199b and RUNX3 mRNA levels in WT tissues (Fig. 4C) (r=−0.6007, P=0.0019). The
downregulation of RUNX3 in the WT tissues may at least be partly
due to the upregulation of miR-199b, thereby suggesting that RUNX3
was a direct target of miR-199b.
siRNA interference of RUNX3 reverses
the oncogenic effects of miR-199b in WT cells
To further clarify whether the RUNX3 downregulation
mediated the oncogenic roles of miR-199b in WT, we cotransfected
the miR-199b inhibitor with RUNX3 siRNA or NC siRNA into the 17.94
and WiT49 cells. In comparison with cells cotransfected with the
miR-199b inhibitor and NC siRNA, the RUNX3 protein expression level
was recovered in the 17.94 and WiT49 cells after cotransfection
with RUNX3 siRNA (Fig. 5A,
P<0.05). Next, CCK-8 and Transwell invasion assays showed that
the RUNX3 knockdown counteracted the oncogenic effects of miR-199b
in the proliferation (Fig. 5B and
C) (P<0.05) and invasion (Fig.
5D) (P<0.05) of 17.94 and WiT49 cells. On the basis of the
results mentioned above, miR-199b may serve oncogenic roles in WT,
at least in part, by direct regulation of RUNX3 expression.
Discussion
Emerging evidence has demonstrated that the
deregulation of miRNAs contributes to WT malignant progression
(2,21,22).
Therefore, identifying the essential miRNAs for WT onset and
progression may provide promising therapeutic methods for patients
with this disease. In the current study, the expression of miR-199b
in the WT tissues was significantly higher than that in the
adjacent normal tissue. In addition, miR-199b underexpression
reduced the WT cell proliferation and invasion. More importantly,
RUNX3 was validated as a direct target gene of miR-199b in WT.
Further studies showed that RUNX3 expression is lowly expressed in
the WT tissues; and this expression is inversely associated with
miR-199b level. Moreover, recovered RUNX3 expression counteracted
the oncogenic effects of miR-199b underexpression in the WT cells.
Thus, targeting miR-199b may be a novel therapeutic method for
patients with WT.
In our study, we demonstrated that miR-199b
expression is upregulated in WT. Similar phenomenon was also
observed in osteosarcoma. Highly expressed miR-199b in osteosarcoma
is evidently correlated with tumour grade, metastasis and
recurrence. Osteosarcoma patients with increased miR-199b
expression exhibit poorer overall survival and shorter disease-free
survival relative to those patients with decreased levels (23). However, miR-199b is underexpressed
in several human cancer types. For instance, the miR-199b
expression level is decreased in hepatocellular carcinoma.
Hepatocellular carcinoma patients with low miR-199b levels show
shorter overall survival and progression-free survival rates than
those patients with high miR-199b levels (24,25).
In bladder cancer, miR-199b expression is downregulated, which is
associated with poor prognosis (26). In breast cancer, miR-199b
expression is underexpressed in tumour tissues in comparison with
normal breast tissues. Low miR-199b expression is strongly
associated with TNM stage and lymph node metastasis of patients
with breast cancer. Patients with breast cancer with relatively low
miR-199b levels have poorer overall survival than those with high
miR-199b level. Moreover, miR-199b is an independent prognostic
factor for patients with breast cancer (27). miR-199b is also found to be lowly
expressed in colorectal cancer (16), acute myeloid leukaemia (17), non-small cell lung cancer (18) and endometrioid endometrial
carcinoma (19). These conflicting
findings support the tissue specificity in the expression pattern
of miR-199b and suggest that miR-199b may emerge as a potential
novel biomarker for the diagnosis and prognosis in human
cancers.
miR-199b serves oncogenic roles in WT by promoting
cell proliferation and invasion. Similarly, miR-199b upregulation
increases growth and metastasis of osteosarcoma cells in
vitro (23). Nevertheless,
miR-199b is found to play tumour suppressive roles in multiple
human cancer types. For example, miR-199b overexpression suppresses
cell proliferation and epithelial-mesenchymal transition, and
increases radiosensitivity in hepatocellular carcinoma (24,25).
Fang et al found that miR-199b overexpression restrains cell
growth, colony formation and metastasis (27,28).
Shen et al revealed that restored miR-199b expression
represses the colorectal cancer cell metastasis in vitro and
in vivo; and elevates the cell chemosensitivity to 5-FU and
oxaliplatin (16). Wang et
al found that the miR-199b resumption expression reduces the
cell proliferative, migratory and invasive abilities of non-small
cell lung cancer (18). Shang
et al demonstrated that miR-199b re-expression attenuates
cell proliferation and induces apoptosis in prostate cancer
(29). Koshizuka et al
indicated that enforced expression of miR-199b inhibits cell
migration and invasion of the head and neck cancer (30). Thus, biological functions of
miR-199b in human malignancy have tissue specificity, thereby
suggesting that miR-199b may be an effective therapeutic target for
patients with these types of human malignancy.
Several targets of miR-199b have previously been
identified, including Hif1α (24)
and N-cadherin (25) in
hepatocellular carcinoma, HER2 (28) in breast cancer, SIRT1 (16) in colorectal cancer, ZEB1 (18) in non-small cell lung cancer and
ITGA3 (30) in head and neck
cancer. In this study, RUNX3 was proven to be a direct target of
miR-199b in WT. RUNX3 is downregulated in a variety of human
cancers, including lung cancer (31), bladder cancer (32), breast cancer (33), gastric cancer (34) and colorectal cancer (35). The downregulation of RUNX3 accounts
for the tumour initiation and progression by acting as a tumour
suppressor and regulating numerous cancer-related biological
processes, such as cell proliferation, cell cycle, apoptosis,
metastasis and angiogenesis (36–39).
Furthermore, the deregulation of RUNX3 is associated with malignant
clinical features, thereby influencing the prognosis of patients
with malignant diseases (40). For
example, decreased RUNX3 in breast cancer is significantly
correlated with tumour infiltration, clinical stage, lymph node
metastasis and expressions of oestrogen and progesterone receptors.
In addition, patients with breast cancer with higher RUNX3
expression have higher survival rate than patients with lower RUNX3
(33). These findings suggest that
the ectopic expression of RUNX3 in human cancers may be a promising
therapeutic method.
In conclusion, miR-199b expression is upregulated in
WT. miR-199b inhibition restrains cell proliferation and invasion
in WT by directly targeting RUNX3. These results suggest its
potential as a therapeutic target to treat patients with WT.
However, in our current study, we did not analyse the effects of
miR-199b overexpression in WT cells. This is a limitation of our
study, and we will resolve this in our future experiments.
Moreover, we will explore whether miR-199b could directly targeted
other genes in WT.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZho and HuZ designed the study. HuZ and HaZ
performed the RT-qPCR and western blot analysis, as well as the
CCK-8 and Transwell invasion assays. YZha performed the luciferase
reporter assay. YZho analyzed the data.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Yidu Central Hospital of Weifang. Written informed
consent was obtained from all patients prior to their inclusion
within the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yan-Fang T, Zhi-Heng L, Li-Xiao X, Fang F,
Jun L, Gang L, Lan C, Na-Na W, Xiao-Juan D, Li-Chao S, et al:
Molecular mechanism of the cell death induced by the histone
deacetylase pan inhibitor LBH589 (Panobinostat) in Wilms tumor
cells. PLoS One. 10:e01265662015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cui M, Liu W, Zhang L, Guo F, Liu Y, Chen
F, Liu T, Ma R and Wu R: Over-expression of miR-21 and lower PTEN
levels in Wilms' tumor with aggressive behavior. Tohoku J Exp Med.
242:43–52. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Beckwith JB, Kiviat NB and Bonadio JF:
Nephrogenic rests, nephroblastomatosis, and the pathogenesis of
Wilms' tumor. Pediatr Pathol. 10:1–36. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schenk JP, Günther P, Schrader C, Ley S,
Furtwängler R, Leuschner I, Edelhäuser M, Graf N and Tröger J:
Childhood kidney tumors - the relevance of imaging. Radiologe.
45:1112–1123. 2005.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Routh JC, Grundy PE, Anderson JR, Retik AB
and Kurek KC: B7-h1 as a biomarker for therapy failure in patients
with favorable histology Wilms tumor. J Urol. 189:1487–1492. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Li Y, Ma F, Zhou H, Ding R, Lu B,
Zou L, Li J and Lu R: Inhibitory effect of Par-4 combined with
cisplatin on human Wilms' tumor cells. Tumour Biol.
39:10104283177166892017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wari MN, Vallonthaiel AG, Ahmed A, Saxena
D, Iyer VK, Mathur SR, Agarwala S, Bakhshi S, Srinivas V,
Chattopadhyaya P, et al: Glypican-3 mRNA expression level in Wilms
tumor: Correlation with histological type, stage, and outcome.
Pediatr Surg Int. 33:695–703. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhu S, Liu G, Fu W, Hu J, Fu K and Jia W:
Axl promotes the proliferation, invasion and migration of Wilms'
tumor and can be used as a prognostic factor. Onco Targets Ther.
10:955–963. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baranwal S and Alahari SK: miRNA control
of tumor cell invasion and metastasis. Int J Cancer. 126:1283–1290.
2010.PubMed/NCBI
|
|
11
|
Liu J: Control of protein synthesis and
mRNA degradation by microRNAs. Curr Opin Cell Biol. 20:214–221.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Harada K, Baba Y, Ishimoto T, Shigaki H,
Kosumi K, Yoshida N, Watanabe M and Baba H: The role of microRNA in
esophageal squamous cell carcinoma. J Gastroenterol. 51:520–530.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shen ZL, Wang B, Jiang KW, Ye CX, Cheng C,
Yan YC, Zhang JZ, Yang Y, Gao ZD, Ye YJ and Wang S: Downregulation
of miR-199b is associated with distant metastasis in colorectal
cancer via activation of SIRT1 and inhibition of CREB/KISS1
signaling. Oncotarget. 7:35092–35105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Favreau AJ, McGlauflin RE, Duarte CW and
Sathyanarayana P: miR-199b, a novel tumor suppressor miRNA in acute
myeloid leukemia with prognostic implications. Exp Hematol Oncol.
5:42016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang J, Zhou F, Yin L, Zhao L, Zhang Y and
Wang J: MicroRNA-199b targets the regulation of ZEB1 expression to
inhibit cell proliferation, migration and invasion in non-small
cell lung cancer. Mol Med Rep. 16:5007–5014. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Torres A, Torres K, Pesci A, Ceccaroni M,
Paszkowski T, Cassandrini P, Zamboni G and Maciejewski R:
Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma
coexists with increased expression of mTOR kinase in endometrioid
endometrial carcinoma. BMC Cancer. 12:3692012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang HF, Zhang YY, Zhuang HW and Xu M:
MicroRNA-613 attenuates the proliferation, migration and invasion
of Wilms' tumor via targeting FRS2. Eur Rev Med Pharmacol Sci.
21:3360–3369. 2017.PubMed/NCBI
|
|
22
|
Liu GL, Yang HJ, Liu B and Liu T: Effects
of MicroRNA-19b on the proliferation, apoptosis, and migration of
Wilms' tumor cells via the PTEN/PI3K/AKT signaling pathway. J Cell
Biochem. 118:3424–3434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zeng H, Zhang Z, Dai X, Chen Y, Ye J and
Jin Z: Increased expression of microRNA-199b-5p associates with
poor prognosis through promoting cell proliferation, invasion and
migration abilities of human osteosarcoma. Pathol Oncol Res.
22:253–260. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang C, Song B, Song W, Liu J, Sun A, Wu
D, Yu H, Lian J, Chen L and Han J: Underexpressed microRNA-199b-5p
targets hypoxia-inducible factor-1α in hepatocellular carcinoma and
predicts prognosis of hepatocellular carcinoma patients. J
Gastroenterol Hepatol. 26:1630–1637. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou SJ, Liu FY, Zhang AH, Liang HF, Wang
Y, Ma R, Jiang YH and Sun NF: MicroRNA-199b-5p attenuates
TGF-β1-induced epithelial-mesenchymal transition in hepatocellular
carcinoma. Br J Cancer. 117:233–244. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sakaguchi T, Yoshino H, Yonemori M,
Miyamoto K, Sugita S, Matsushita R, Itesako T, Tatarano S, Nakagawa
M and Enokida H: Regulation of ITGA3 by the dual-stranded
microRNA-199 family as a potential prognostic marker in bladder
cancer. Br J Cancer. 116:1077–1087. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fang C, Wang FB, Li Y and Zeng XT:
Down-regulation of miR-199b-5p is correlated with poor prognosis
for breast cancer patients. Biomed Pharmacother. 84:1189–1193.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fang C, Zhao Y and Guo B: MiR-199b-5p
targets HER2 in breast cancer cells. J Cell Biochem. 114:1457–1463.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shang W, Chen X, Nie L, Xu M, Chen N, Zeng
H and Zhou Q: MiR199b suppresses expression of hypoxia-inducible
factor 1α (HIF-1α) in prostate cancer cells. Int J Mol Sci.
14:8422–8436. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koshizuka K, Hanazawa T, Kikkawa N, Arai
T, Okato A, Kurozumi A, Kato M, Katada K, Okamoto Y and Seki N:
Regulation of ITGA3 by the anti-tumor miR-199 family inhibits
cancer cell migration and invasion in head and neck cancer. Cancer
Sci. 108:1681–1692. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Araki K, Osaki M, Nagahama Y, Hiramatsu T,
Nakamura H, Ohgi S and Ito H: Expression of RUNX3 protein in human
lung adenocarcinoma: Implications for tumor progression and
prognosis. Cancer Sci. 96:227–231. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dodurga Y, Avci CB, Satiroglu-Tufan NL,
Tataroglu C, Kesen Z, Doğan ZO, Yılmaz S and Gündüz C: Detection of
deleted in malignant brain tumors 1 and runt-related transcription
factor 3 gene expressions in bladder carcinoma. Mol Biol Rep.
39:4691–4695. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang Y, Tong D, Lou G, Zhang Y and Geng
J: Expression of RUNX3 gene, methylation status and
clinicopathological significance in breast cancer and breast cancer
cell lines. Pathobiology. 75:244–251. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hsu PI, Hsieh HL, Lee J, Lin LF, Chen HC,
Lu PJ and Hsiao M: Loss of RUNX3 expression correlates with
differentiation, nodal metastasis, and poor prognosis of gastric
cancer. Ann Surg Oncol. 16:1686–1694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng W, Zheng K, Zhong L, Li Q and Huang
Z: Expression of Runx3 and C-myc in human colorectal cancer. Nan
Fang Yi Ke Da Xue Xue Bao. 34:1042–1047. 2014.(In Chinese).
PubMed/NCBI
|
|
36
|
Jili S, Eryong L, Lijuan L and Chao Z:
RUNX3 inhibits laryngeal squamous cell carcinoma malignancy under
the regulation of miR-148a-3p/DNMT1 axis. Cell Biochem Funct.
34:597–605. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen F, Liu X, Cheng Q, Zhu S, Bai J and
Zheng J: RUNX3 regulates renal cell carcinoma metastasis via
targeting miR-6780a-5p/E-cadherin/EMT signaling axis. Oncotarget.
8:101042–101056. 2017.PubMed/NCBI
|
|
38
|
Kim BR, Kang MH, Kim JL, Na YJ, Park SH,
Lee SI, Kang S, Joung SY, Lee SY, Lee DH, et al: RUNX3 inhibits the
metastasis and angiogenesis of colorectal cancer. Oncol Rep.
36:2601–2608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li H, Li D and Meng N: Effects of RUNX3
mediated Notch signaling pathway on biological characteristics of
colorectal cancer cells. Int J Oncol. 50:2059–2068. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wei D, Gong W, Oh SC, Li Q, Kim WD, Wang
L, Le X, Yao J, Wu TT, Huang S and Xie K: Loss of RUNX3 expression
significantly affects the clinical outcome of gastric cancer
patients and its restoration causes drastic suppression of tumor
growth and metastasis. Cancer Res. 65:4809–4816. 2005. View Article : Google Scholar : PubMed/NCBI
|